Abstract
Background:
Drug-induced Liver Injury (DILI) is an important cause of acute liver failure cases in the United States, and remains a common cause of withdrawal of drugs in both preclinical and clinical phases.
Methods:
A structured search of bibliographic databases – Web of Science Core Collection, Scopus and Medline for peer-reviewed articles on models of DILI was performed. The reference lists of relevant studies was prepared and a citation search for the included studies was carried out. In addition, the characteristics of screened studies were described.
Results:
One hundred and six articles about the existing knowledge of appropriate models to study DILI in vitro and in vivo with special focus on hepatic cell models, variations of 3D co-cultures, animal models, databases and predictive modeling and translational biomarkers developed to understand the mechanisms and pathophysiology of DILI are described.
Conclusion:
Besides descriptions of current applications of existing modeling systems, associated advantages and limitations of each modeling system and future directions for research development are discussed as well.
Keywords: Drug evaluation studies, preclinical, evidence-based toxicology, liver injury, drug-induced, side effects
1. INTRODUCTION
Drug-induced Liver Injury (DILI), caused by conventional drugs, herbal medications, dietary supplements, as well as by other xenobiotics, includes liver injury that can sometimes be associated with severe outcomes such as acute liver failure. Therefore, it represents an important cause of morbidity and mortality (ADRs) [1, 2]. About 1000 drugs compounds are estimated to be associated with idiosyncratic hepatotoxicity (iDILI) [3], with central nervous system (CNS) agents and antibiotics being the most common medications that cause DILI [4]. In fact, DILI accounts for the majority of acute liver failure cases in the United States, although drug-induced hepatotoxicity can also mimic many other liver diseases as well [4-6]. DILI is also the primary reason for drug withdrawal from the market [7-9]
Drug-induced hepatotoxicity is a result of complex interactions among genetic, non-genetic and environmental factors [10, 11]. The main event in the pathogenesis of DILI is death of hepatocytes, although cholangiocytes or endothelial cells could also be potential targets [12, 13].
There are three basic types of liver injuries: hepatocellular - presenting as acute hepatitis, cholestatic - presenting as cholestasis, and mixed [14]. However, the dominant liver injury type sometimes changes during illness [15].
As already mentioned, symptoms of DILI may vary, ranging from asymptomatic elevations of liver enzymes to severe hepatitis with jaundice, and only a small fraction of patients develop chronic liver disease. More than 90% of patients with jaundice recover after discontinuation of the drug, and about 10% of patients with severe DILI with jaundice will either die or need liver transplantation [16].
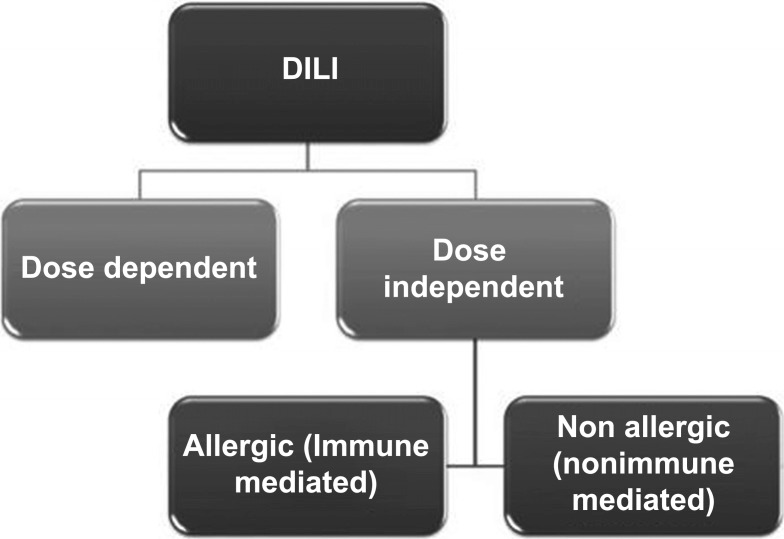
The etiopathogenesis of DILI is complex and involves genetic, metabolic and immune factors. Despite classification difficulties, researchers agree that DILI generally can be divided into two categories. The first category is predictable and dose-dependent. The second is idiosyncratic, mostly dose independent (Fig. 1). Idiosyncratic DILI (iDILI) is further subdivided into two sub-categories: allergic, immune-mediated, and non-allergic, nonimmune-mediated [4, 17-19] as presented in Fig. (1).
Fig. (1).
Division of DILI types based on dose dependency and immune response.
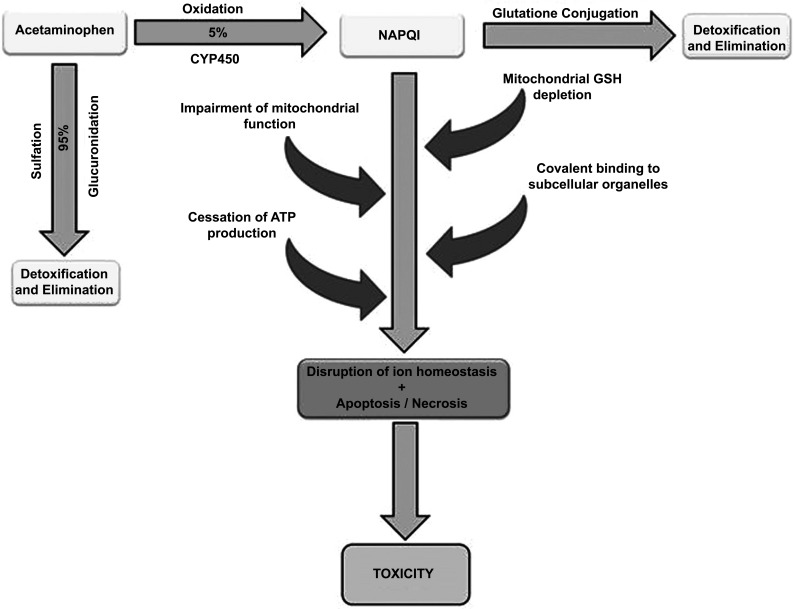
Acetaminophen is one of the most studied drugs that cause a predictable DILI. Its hepatotoxicity is dose-dependent and the hepatic injury occurs due to the activity of the toxic N-acetyl-p-benzoquinone Imine (NAPQI) metabolite, produced by the CYP2E1 pathway of acetaminophen metabolism. Exposure of Reactive Oxygen Species (ROS) to organelles can lead to apoptosis and necrosis of hepatocytes [20] as presented in Fig. (2). Bromfenac, cyclophosphamide and methotrexate are also examples of drugs known for induction of dose-related hepatotoxicity [20].
Fig. (2).
Algorithm of Acetaminophen toxicity in liver. Acetaminophen (APAP) impairs mitochondrial function by the creation of a reactive metabolite N-acetyl-p-benzoquinone Imine (NAPQI), which is induced mostly by the cytochrome P450 enzymes, resulting in depletion of mitochondrial Glutathione (GSH). Once glutathione is depleted, NAPQI binds to subcellular organelles in the cell, causing the binding of APAP to cellular proteins resulting in disruption of calcium homeostasis, mitochondrial dysfunction, oxidative stress, collapses ATP production and may culminate in cell necrosis and death.
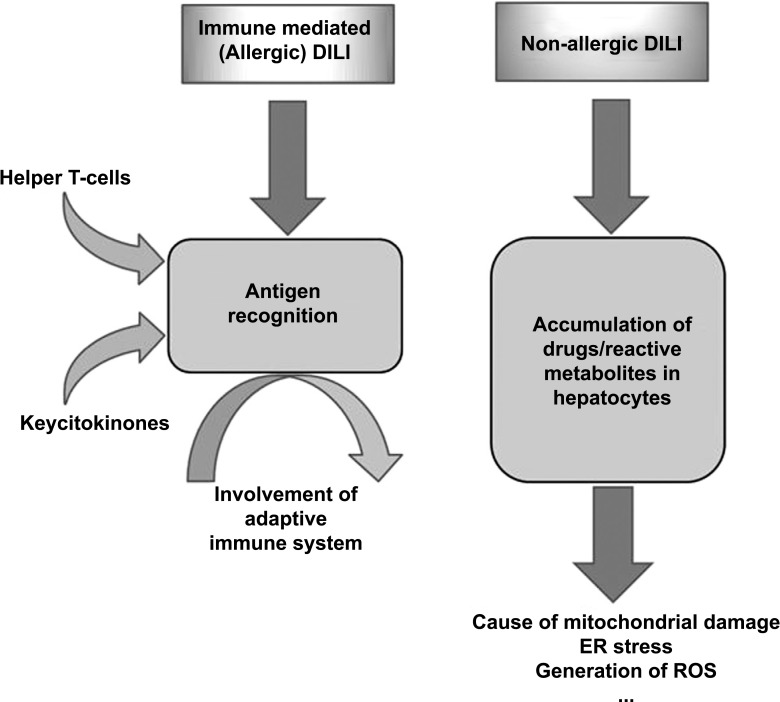
Immune-mediated (allergic) DILI seems to be generated by antigen recognition-mediated by helper T cells, suggesting the influence of the adaptive immune response. It is characterized by a short delay period and fast re-occurrence of liver injury on re-exposure to the medication. For example, hepatotoxicity caused by phenobarbital, often associated with fever, rash and eosinophilia, tends to be more severe with a more rapid onset after re-challenging or re-exposure [21].
In hepatotoxicity induced by non-allergic DILI drugs, reactive metabolites gradually accumulate in hepatocytes, which could cause mitochondrial impairment (Fig. 3). DILI caused by medications such as ximelagatran, lapatinib, ticlopidine or amoxicillin with clavulanate acid are found to be associated with variants of human leukocyte antigens (HLAs), indicating that adaptive immune response might be significant for hepatotoxicity induced by those drugs. It is, therefore, often a challenge to determine whether DILI is immune-mediated or non-immune-mediated [22, 23].
Fig. (3).
Algorithm of liver damage in allergic and non-allergic DILI models. In the allergic DILI model, drugs or reactive metabolites bind to host proteins (haptenization), and hapten-peptides are processed and represented on HLA binding gutter of antigen-presenting cells (APC) by CD4 T-cells. In non-allergic DILI model drugs or reactive metabolites gradually accumulate in hepatocytes and directly stress mitochondria, enhancing mitochondrial Reactive Oxygen Species (ROS) production. Toxic and metabolic stress activates signal transduction pathways resulting in alterations in mitochondrial. Injured mitochondria relief different contents, as mtDNA and activate cytokines inducing apoptosis or necrosis.
Even though certain HLA haplotypes seem to be associated with the emergence of DILI, other factors, such as the direct hepatotoxicity of medications and their metabolites, or extrinsic factors, such as age, sex, infections and environmental factors, which could also be important in the development of DILI [18].
Hence, the mechanism of drug hepatotoxicity is very challenging to understand. Accordingly, many models to explore the mechanisms of DILI have been developed [24].
However, currently available hepatic model systems are still insufficient to resolve all issues involved in the genesis of DILI. Therefore, application of new technologies in the early drug development is needed.
2. Hepatic cell lines models for evaluation of drug-induced liver injury
There are several models that can be useful for studying the mechanisms and pathophysiology of DILI: primary human hepatocytes cultures, cell lines derived from hepatic carcinoma, liver tissue engineering (engineered hepatic cell lines), human pluripotent stem cells derived hepatic cells and animal models.
2.1. Primary Hepatocyte Culture
Primary human hepatocyte cultures are considered to be the gold standard for the creation of human-relevant liver cell culture models. Moreover, they present significant and predictive results in pharmacological and toxicological in vitro research due to their unstable differentiation [25].
Hepatocytes are differentiated cells expressing many hepatic functions, and retain the expression of both Phases I and II enzymes for a limited time in culture. Thus, tests on primary hepatocyte cultures are often capable of elucidating mechanisms of DILI. For example, many drugs inducing severe DILI have been shown to cause an elevated ROS/ATP ratio in primary human hepatocyte cultures, indicating that oxidative stress. That is followed by hepatic cellular damage and is one of the most important mechanisms of DILI [26].
2.2. Cell Lines Derived from Hepatocellular Carcinomas
Advantages of cell lines derived from hepatocellular carcinoma (hepatoma) include availability, easy handling, stable phenotype and unlimited propagation potential. Nowadays, the applicability of HepG2, or Hep3B, Huh7, hiHeps, SK-Hep-1 or L-02 is limited due to the lack of substantial hepatocyte function, in particular with regard to the biotransformation capacity. On the other hand, HepaRG cells which were generated from the tumor of a female patient suffering from chronic hepatitis C infection and hepatocarcinoma arevery widely used as an in vitro model to assess drug-induced hepatotoxicity.
Numerous studies have shown that HepaRG cells have almost the same expression levels of Phase I and Phase II drug metabolizing enzymes, and nuclear receptors compared to Primary Human Hepatocytes (PHH), and human liver tissue samples. These can be improved with differentiation of the cells induced by 2% Dimethyl Sulfoxide (DMSO). However, the effects decrease on withdrawal of DMSO. Compared to other cell lines, HepaRG cells have been shown to be an adequate tool to study some chronic effects of drugs and other agents in vitro. However, only a few studies exist so far regarding their suitability to detect the effects of drugs requiring a previous activation across the cytochrome P450 (CYP) system. The anti-tuberculosis agent (INH) is a prototypic substance that has a high biotransformation capacity whereby it is metabolized into N-acetylhydrazine which then triggers hepatotoxicity [27].
It is important to emphasize that HepaRG cells displayed a similar response to the effects of acetaminophen as PHH, and a higher activation of genes related to liver damage as compared to HepG2 cells. By contrast, other studies showed a reduced sensitivity of HepaRG cells to the detection of hepatotoxic drugs [25].
However, most hepatoma cell lines contain very low levels of drug metabolizing enzymes, particularly CYP450 enzymes, compared to primary cultured hepatocytes [28]. Several factors are involved in the repression of CYP genes in hepatic cells [29, 30]. Oncogenes (Ha-rasEJ, met, c-Ha-ras) have been reported to be effective in transforming hepatocytes [31-33]. The resulting transformed hepatocytes expressed low levels of drug metabolizing activities [34]. Transfection of expression vectors encoding key hepatic transcription factors into hepatoma cells resulted in significant re-activation of relevant CYPs [35].
2.2.1. Recombinant Cell Models Expressing Drug Biotransformation Enzymes
To date, recombinant models, heterologously expressing cytochrome P450 enzymes, are widely used to study hepatic drug metabolism. These cell culture models have helped acquire information in early phases of drug development process such as drug candidate discovery, and elucidation of metabolism of a given drug [34]. On the other hand, cell culture models constructed by transfecting hepatic and non-hepatic cells with expression vectors encoding human CYPs and drug metabolism genes (GSH-transferases, UDP-glucuronosyltransferases) [36] are useful tools for detection of potential drug-drug interactions, pharmacological characterization, and generation of metabolites [37].
2.2.2. Human Pluripotent Stem Cells
Recent studies have found that hepatic cells derived from Human Pluripotent Stem Cells (hPSCs), like Human Induced Pluripotent Stem Cells (hiPSC), and Human Embryonic Stem Cells (hESC) are more predictive for potential modeling in vitro of drug toxicity compared to existing culture models [38]. Since hPSCs have a primary hepatocyte-like phenotype, tests on hPSCs used for hepatotoxicity testing of drugs could be more accurate than tests on cell lines derived from hepatocarcinomas or animal models [39, 40]. Also, the availability of those cells is unlimited and hPSC from various individuals can yield genotype-specific cell lines [41].
However, hESC differentiation to a fully metabolically competent hepatocyte has still not been successful [42]. This means that there is still a need for progress in the use of hPSC in hepatotoxicity testing, as well as the evolvement of standardized protocols [43, 44].
3. Liver tissue engineering and its usefulness in evaluating DILI
Since data collected from existing in vitro model systems for assessing DILI often fail to predict hepatotoxicity of the tested drugs, the urgent need for novel and better in vitro models exists [45]. As mentioned earlier, primary hepatocyte cultures lose their phenotypic characteristics and viability. Furthermore, hepatocytes in monocultures were found to be more sensitive to hepatotoxins due to accumulation of waste products [46]. Co-culturing primary hepatocytes with Non-parenchymal Cells (NPC), such as Kupffer, stellate, or sinusoid endothelial cells resulted in the long term maintenance of hepatocyte phenotype and viability [45].
Hepatocytes co-cultured with Non-parenchymal Cells (NPCs) organized into a 3D tissue structure provided a better model for the evaluation of DILI due to a higher level of similarity with in vivo tissue architecture. For example, in acetaminophen hepatotoxicity models, Schyschka et al. showed differences in specific protein expression between 2D and 3D models. Thus, 3D models preserved certain metabolic functions absent in 2D cultures [47].
A recent study showed that the proteome of PHH spheroids remained similar to that of donor hepatic tissue. Therefore, it could potentially reflect inter-individual differences and be used to study hepatotoxicity on hepatocytes with a particular genotype of interest [48]. They also seem to be suitable for studying chronic DILI since extended exposure to known hepatotoxic drugs resulted in increased hepatotoxicity in vitro [48].
Although 3D cultures appear to have numerous advantages compared to previously discovered in vitro models, there is a need for further studies to extensively evaluate the sensitivity and specificity of hepatotoxicity in such cultures. These 3D cultures still do not entirely resemble in vivo conditions. For instance, different cell types of vascular structures and the biliary tract still cannot be generated in 3D culture models. However, these missing cell types do contribute to the complex liver morphology and physiology of the human liver, and their absence could also alter the results of toxicity studies using such models. Furthermore, liver tissue engineering using 3D models have not solved the problem of lack of an adaptive immune response which plays a role in DILI in humans [49].
On the other hand, efforts to develop portions of human organs on chips to regenerate attributes of organ-level functioning have made impressive advances in the past few years. Hence, in 2014, the National Institutes of Health (NIH) funded numerous institutions for the second phase of the Tissue Chip for Drug Screening program. Taylor et al. developed a tissue chip that replicated the liver acinus and sinusoids. They constructed two liver acinus chips that included all the essential cell types found in the liver, including hepatocytes, endothelial cells, stellate cells and Kupffer cells. Over a 28-day period, the chips mimicked key liver functions including metabolism, clearance, protein synthesis, and urea detoxification. Furthermore, they have focused on induced and embryonic pluripotent stem cells (iPSC) and their proliferation and differentiation time. Consequently, this approach permitted an easily sharable cell population with unlimited growth potential that can mimic preliminary clinical trials in vitro, hopefully permitting application to other human investigations [50].
4. Animal models for evaluation of DILI
Experimental animals are useful models for studying DILI and its pathogenesis [51]. Despite the generation of new predictive cell culture model systems, experiments in animal models are an unavoidable part of the pre-clinical drug development [52, 53]. This fact relies on the assumption that basic processes are comparable among different species [54].
Animal testing is required because liver tissue culture models are not completely representative of liver function in the human body. This is especially true with regard to the lack of an immune system which plays a major role in the development of DILI.
The animal models used for DILI include not only rodents (rats, mice, rabbits and guinea pigs), but also larger non-rodent animals as pigs, sheep and monkeys [55].
There is also a need for critical analysis of the correlation between findings of DILI in animal models and humans. At present, because of the shortage of these types of studies, there are no satisfactory data to reliably estimate the relevance of pre-clinical animal investigations to anticipate DILI in humans [55, 56].
In fact, the two drug-induced idiosyncratic toxicities in humans which most often led to termination in the clinical phase of the drug-development process, DILI and hypersensitivity reactions, also showed the poorest correlation with animal studies [55].
There was no evidence for a stronger correlation of DILI in humans between non-rodent and rodent animal models [57]. However, a comprehensive retrospective review of published information suggested that a finding of adverse drug reactions in dogs was a better predictor of DILI in humans, compared to the results of tests using rodent and non-human primates (monkeys) [55, 58]. However, in some therapeutic classes of drugs such as anticancer drugs, rodents provided valid information for assessment of DILI risk for humans [58]. Also, novel advances in technology seemed to make animal models for drug toxicity testing more similar to humans. In fact, primary human hepatocytes, as well as iPSCs, have been used in immunodeficient TK-NOG mice, to produce a humanized mouse model. The aim was to increase the physiological resemblance to human conditions and, therefore, make animal testing more relevant [59-61]. These findings suggest that there is a need to examine DILI in both rodent and non-rodent animals.
In addition, it has been shown that tests in animal experimental models such as mice, are of greater value for studying Type A, predictable hepatotoxicity, than they are for studying Type B, idiosyncratic hepatotoxicity (IDILI) in humans [62].
One of the best studied DILI in animal models, an example of predictable and direct hepatotoxicity, is the toxicity of acetaminophen [63, 64].
In contrast, as a result of the infrequent occurrence and unpredictable nature of IDILI in animal models, it is quite impractical to implement such studies, since the number of animals used in pre-clinical tests makes it difficult to detect such rare hepatotoxic events [55, 56]. Despite this struggle, animal models are crucial to examine the hypotheses of IDILI, leading to a better understanding of specific mechanisms involved. In the end, a better understanding of specific IDILI mechanisms may result in the development of more predictable animal models for IDILI studying. Due to the lack of sufficient non-clinical predictive models for testing IDILI, there is still a need for the development of valid animal models. In the ideal scenario, an animal model should produce IDILI at a high frequency, at a reasonable price, and have the ability to differentiate drugs that cause idiosyncratic hepatic injury from the drugs that do not [65]. IDILI mechanistic theories assume that inflammatory stress, drug disposition polymorphisms, inadequate adaptation to moderate injury, mitochondrial dysfunction and adaptive immune system response, might play a significant role in the genesis of hepatotoxicity. Therefore, emerging animal models to test these hypotheses are being developed [65].
The mechanism of IDILI is poorly understood because of the lack of a valid animal model. Therefore, Uetrecht et al. tried to develop the first animal model of IDILI analogous to human IDILI by inhibiting immune tolerance. They developed an animal model in which treatment of female C57BL/6 mice with Amodiaquine (AQ) resulted in a slight liver injury with a delayed beginning and resolution regardless of continued treatment. Furthermore, this investigation demonstrated that AQ treatment of Cbl-b2/2 and PD-12/2 mice have damaged immune tolerance and consequently resulted in a modestly greater injury. Also, cotreatment of C57BL/6 with AQ and anti-CTLA4 showed an increase in ALT compared to the treatment with AQ alone. An increase in T regulatory cells and T helper cells expressing PD-1 and CTLA4 suggested that there was an induction of immune tolerance in these mice, and the ALT activity was normalized despite continued treatment [66].
Also, the development of other animal models in the future might lead to a discovery of yet unknown biomarkers and methods for prevention and treatment strategies of IDILI [67].
5. Databases and Predictive Modeling
Since DILI can be associated with acute liver failure [5], it is important to develop methods and procedures that will reveal possible hepatotoxicity among drug candidates as soon as possible [68].
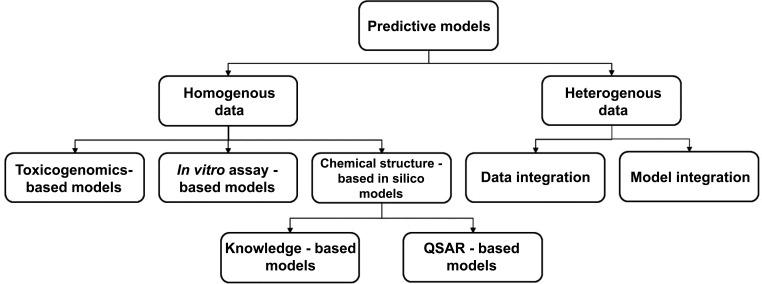
Currently, the poor in-depth understanding of DILI pathogenesis and mechanisms is the limiting factor in developing predictive models for DILI. However, research has led to the development of a wide range of predictive models for DILI. Predictive models can commonly be divided into two basic: predictive models: from homogenous data and from heterogeneous data (reviewed in depth in [69]. The first group is subdivided into three categories based on the type of data utilized: chemical structure-based in silico (or computational) models, in vitro assay-based models and toxicogenomics-based models. A schematic presentation of possible data sources used for development of predictive models for drug-induced liver injury is presented in Fig. (4).
Fig. (4).
Schematic presentation of possible data sources used for development of predictive models for DILI models. There are two basic groups; predictive models from homogenous data and from heterogeneous data. The first group is subdivided into three categories: chemical structure-based in silico (or computational) model, in vitro assay-based models and toxicogenomics-based models. The second group is subdivided into two categories: the first category is data integration-based models which use multiple sources of data for developing one predictive model. The second category - model integration uses multiple individually developed models from multiple data sources.
The first category of predictive models allows rapidly screening a large number of chemicals at minimal cost [69]. The main assumption is that similar chemical structures have similar properties and toxicity profiles [70].
Chemical structure-based in silico models encompass the knowledge-based or expert system models and QSAR-based (quantitative structure-activity relationship-based) models, using structure alerts and molecular descriptors for DILI risk assessment. Expert system models are usually developed with commercial software (such as Derek for Windows) and require expert opinion for evaluation of the validity of the structural alerts [69]. In contrast, QSAR-based models do not require the opinion of experts, and consequently can produce results more rapidly [69].
In vitro assay-based models (Fig. 4) have been widely used in preclinical testing as conventional cytotoxicity assays with single endpoints. However, they did not predict DILI reliably, and had a sensitivity of less than 25% [69, 71].
Nowadays, they are used as High Content Screening (HCS) assays that simultaneously measure multiple end points in live cells. They enable high-throughput screening with higher sensitivity [72]. However, they require drug exposure data which are generally underestimated [73] and consequently, the predictions are not reliable.
The third category of predictive models uses microarray-based technology to measure alterations in gene expression induced by hepatotoxins. The main disadvantage of this predictive model is that assessed end points (altered gene expression) are not necessarily associated with DILI risk. In the last few years, two toxicogenomic data sets have been published: the Japanese toxicogenomics database [74] and Drug Matrix [75]. Toxicogenomics data sets could enable progress in developing DILI prediction models [76].
Predictive models from heterogeneous data are subdivided into two categories- both using an integrative approach to improve the predictive power of the models. The first category is a data integration-based model based on multiple sources of data for developing one predictive model. The second category - model integration uses multiple individually developed models from multiple data sources (Fig. 4).
In conclusion, it should be noted that desired predictive model should integrate several factors: chemical structure and toxicogenomics data, cellular endpoints and multiple data sources. It is considered to be highly specific (between 90-95%) with moderate sensitivity (about 50%). However, predictive models that are currently in use are still not superior to animal toxicity studies [69].
6. Translational biomarkers for DILI
The ideal hepatic biomarker should detect as well as predict, idiosyncratic DILI in experimental animals with translational application to human toxicity. Because of the poorly understood pathogenesis of idiosyncratic DILI, it is extremely challenging to precisely detect hepatic biomarkers in this case [77].
By monitoring the levels of a serum biomarker, we should be able to track the severity of hepatic injury and the recovery of normal liver function. For instance, enzymes that leak rapidly from damaged hepatocytes allow us to detect and/or monitor DILI. So far, the most commonly used enzymes are alanine aminotransferase (ALT) and Aspartate Aminotransferase (AST) [78].
Furthermore, ALT, which itself is a highly sensitive hepatic injury biomarker; in combination with serum total bilirubin can be a reference marker for idiosyncratic DILI. These markers are used in clinical practice as predictors of severe DILI in humans [11].
It has been proven that even in the absence of liver injury, in cases when inhibitory factors such as vitamin B12 cofactor deficiency are present, serum ALT values can be lower than expected. Furthermore, in some metabolic disorders such as type 1 diabetes and nonalcoholic liver disease, increased ALT activity is present without histologic evidence of liver injury [79, 80].
Therefore, further experiments and novel biomarkers are needed to enhance the ALT specificity. So far, some enzyme biomarkers such paraoxonase (PON) 1, Malate Dehydrogenase (MDH), and purine-nucleoside phosphorylase (PNP) have shown potential to address these limitations in ALT measurement. Additional testing is required in order to confirm the full validity of these biomarkers [79, 81, 82].
Recently, investigations have demonstrated that miRNAs can be used as sensitive and specific biomarkers for DILI. Plasma levels of miRNA correlated relatively well with serum ALT [83].
Based on research, two miRNAs (miR-122 and miR-192) were proposed as novel DILI biomarkers because of significant levels in serum. The miRNA levels were stable, and not subjected to post-processing modifications, which allowed them to be accurately extracted from serum or plasma [83]. Also, miRNAs in the blood could be useful biomarkers for human chemical hepatotoxicity, such as acetaminophen liver injury [84]. Moreover, recently studies demonstrated that changes in the serum profile of miRNAs occurred in individuals experiencing acetaminophen-induced DILI [85-88]. In fact, serum miR-122 were detected earlier than ALT elevations in patients admitted to the hospital after acetaminophen overdose [89].
Additionally, there is a possibility of using noninvasive urinary miRNAs as DILI biomarkers. This method is less invasive than the usual blood extraction. However, due to its lack of organ-specificity, further studies are required in order to verify their true value [87, 90-92].
Cytokines may be potential biomarkers for DILI because of their role in the inflammatory process; the most highly expressed were interleukin-1β, tumor necrosis factor α and interleukin-6. However, their short half-life and almost undetectable serum levels still represent difficulties in using them as biomarkers [93, 94]. Along with cytokines, changes in the plasma metabolome can be tracked, e.g. for amino acids, complex lipids and fatty acids, which can indicate liver toxicity [95].
In the future, detecting biomarkers that are capable of identifying subjects prior to liver injury will be crucial in order to reduce the incidence of DILI. Still, biomarkers cannot yet provide unambiguous evidence for DILI. Therefore, new experimental approaches and biomarkers assessments are needed to overcome these limitations [96].
7. THE Use of genomic studies TO provide individualized prediction of potential drug toxicity
Significant advances in understanding of the pharmacogenomics and immunopathogenesis of severe immunologically-mediated Adverse Drug Reactions (ADRs) have been made in the last few years.
Currently, there are many reasons to investigate the genetic basis of ADRs, ranging from personal interest to the fact that the ADR may present a public health problem. The most important is to examine whether it will be possible to identify the specific genetic markers as predictors of ADR and apply individualized pharmacotherapy accordingly. Many investigators have worked on specific drugs and their capacity to cause ADRs and using Genome-wide Association Studies (GWAS) to identify the ADR-specific genetic predisposing factors [97].
An association with HLADRB1*15:01 and amoxicillin/clavulanate DILI has been reported by a numerous research groups. Additionally, the HLA-B*35:02 allele was found to have a significant association with minocycline DILI, and the presence of the HLA-B*57:01 allele has been linked with an 81-fold increased risk of flucloxacillin DILI [98, 99].
Moreover, a modest number of HLA alleles have been found to have overlapping associations with many adverse reactions involving DILI, drug-induced pancreatitis and cutaneous hypersensitivity [100].
Also, many HLA alleles linked with DILI have a very high negative predictive value, and their usage can rule out hepatotoxicity due to distinct drugs. Some investigations used N-acetyltransferase 2 genotyping to define adequate doses of isoniazid in an anti-tuberculosis therapeutic regimen showed that pharmacogenetic-based clinical algorithms have the possibility to improve the efficiency of a drug and reduce DILI [101, 102].
To date, it is possible that with the development of personalized medicine, various genes for disease detection, adverse reactions and drug efficacy could be tested. Consequently, this would personalize healthcare and reduce DILI risk by omitting medications in patients with specific HLA alleles. This would have an influence on cost-effectiveness paradigms too [98].
Although, the connection results with non-HLA genes have been less replicated than the HLA associations, it is confirmed that drug investigations have the ability to identify the correct agent underlying DILI, in particular when the patient had been exposed to more than one medication possibly causing DILI. Recent metabolism genes, for instance UGT2B7 and NAT2 have been shown to contribute to different forms of DILI [103].
Moreover, CYP2C9 and CYP2C19 have major role in the metabolism of various drugs associated with DILI, and the possibility that CYP2C9 and CYP2C19 genotypes might be general risk factors for DILI has been investigated. However, no associations were found.
In addition, the role for CYP2C9 in oxidative metabolism of diclofenac has been established. However, for patients who had suffered diclofenac-related DILI, there was no significant difference in the frequency of CYP2C9 various alleles compared with controls [104].
Numerous investigations have established that some common genetic variants are strongly associated with DILI. Hence, in the future DNA sequencing may have a major role in the identification of rare variants that contribute to DILI.
T-cell-mediated ADRs such as DILI, Stevens-Johnson Syndrome/Toxic Epidermal Necrolysis (SJS/TEN), and other drug hypersensitivity syndromes have been considered to be mediated through various interactions with many class I and II HLA alleles. The ability to analyze the whole genome in an objective manner has led to some outstanding discoveries of the role of the HLA genes as genomic biomarkers of ADRs -predisposition [105].
Hypersensitivity was associated with abacavir and the predisposition was linked to HLA- B*57:01 which was confirmed in a clinical investigation. Moreover, implementation has shown to decrease the tendency of hypersensitivity in a cost-effective manner. Associations have also been demonstrated for carbamazepine- (HLA- B*1502 and HLA- A*3101) and allopurinol- (HLA- B*58:01) induced severe cutaneous ADR.
Moreover, the association with HLA- B*1502 is present in certain South- Eastern Asian populations, and the predisposition has specific phenotypes (only for SJS/TEN). On the other hand, the association with HLA- A*3101 was seen in different ethnic groups, and predisposed to bland as well more serious cutaneous reactions associated with carbamazepine [97].
Significantly, strong HLA allele-drug specific associations with DILI such as HLA-A*33:03 for ticlopidine, HLA-B*57:01 for flucloxacillin and HLA-DQA1*02:01 for lapatinib have been found. These facts indicate that the immune system has one of the major roles in the pathogenesis of other forms of drug-induced organ toxicity.
Furthermore, even more efforts should be made to discover more genetic markers and to achieve high predictability for ADRs, with the goal of successful implementation of individualized pharmacotherapy into clinical practice [106].
Conclusion
In this article, we have considered the existing understanding about DILI, various methods and approaches to design appropriate models to study DILI in vitro and in vivo, and provided a discussion of current applications and future directions for research of each modeling system. Despite much effort and extensive research in the field, the prediction of DILI using in vitro and in vivo models remains very unreliable. One of the important reasons for that lies in the genesis of DILI itself because of the complex interactions among genetic, non-genetic and environmental factors. Given the multiplicity of factors involved in the onset of DILI, it is reasonable to assume that addressing safety in drug-development programs will include multiple modalities of modeling in the future as well. Development of models that allow the monitoring of onset, progression and reversibility of drug-induced toxicity remains an ultimate goal as such models represent desirable tools for the design of better and safer drugs.
Consent for Publication
Not applicable.
Acknowledgements
The study was funded by grants from Croatian Ministry of Science, Education and Sports number VIF-2016-MEFOS-17 (to MS).
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Hamilton L.A., Collins-Yoder A., Collins R.E. Drug-induced liver injury. AACN Adv. Crit. Care. 2016;27(4):430–440. doi: 10.4037/aacnacc2016953. [DOI] [PubMed] [Google Scholar]
- 2.Vuppalanchi R., Liangpunsakul S., Chalasani N. Etiology of new-onset jaundice: how often is it caused by idiosyncratic drug-induced liver injury in the United States? Am. J. Gastroenterol. 2007;102(3):558–562. doi: 10.1111/j.1572-0241.2006.01019.x. [DOI] [PubMed] [Google Scholar]
- 3.Reuben A., Koch D.G., Lee W.M., Group A.L.F.S. Drug-induced acute liver failure: Results of a U.S. multicenter, prospective study. Hepatology. 2010;52(6):2065–2076. doi: 10.1002/hep.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat. Rev. Drug Discov. 2005;4(6):489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 5.Ostapowicz G., Fontana R.J., Schiødt F.V., Larson A., Davern T.J., Han S.H., McCashland T.M., Shakil A.O., Hay J.E., Hynan L., Crippin J.S., Blei A.T., Samuel G., Reisch J., Lee W.M., U.S. Acute Liver Failure Study Group Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann. Intern. Med. 2002;137(12):947–954. doi: 10.7326/0003-4819-137-12-200212170-00007. [DOI] [PubMed] [Google Scholar]
- 6.Halegoua-De Marzio D., Navarro V. Drug-induced hepatotoxicity in humans. Curr. Opin. Drug Discov. Devel. 2008;11(1):53–59. [PubMed] [Google Scholar]
- 7.Fung M., Thornton A., Mybeck K., Wu J.H-H., Hornbuckle K., Muniz E. Evaluation of the characteristics of safety withdrawal of prescription drugs from worldwide pharmaceutical markets-1960 to 1999. Drug Inf. J. 2001;1(35):24. [Google Scholar]
- 8.Chen M., Vijay V., Shi Q., Liu Z., Fang H., Tong W. FDA-approved drug labeling for the study of drug-induced liver injury. Drug Discov. Today. 2011;16(15-16):697–703. doi: 10.1016/j.drudis.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Almario E.E., Borlak J., Suzuki A., Chen M. Drug-induced liver injury. BioMed Res. Int. 2017;2017:2461694. doi: 10.1155/2017/2461694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLeve L.D., Wang X. Role of oxidative stress and glutathione in busulfan toxicity in cultured murine hepatocytes. Pharmacology. 2000;60(3):143–154. doi: 10.1159/000028359. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerman H.J. Drug-induced liver disease. Clin. Liver Dis. 2000;4(1):73–96. doi: 10.1016/s1089-3261(05)70097-0. [DOI] [PubMed] [Google Scholar]
- 12.DeLeve L.D., Wang X., Kuhlenkamp J.F., Kaplowitz N. Toxicity of azathioprine and monocrotaline in murine sinusoidal endothelial cells and hepatocytes: The role of glutathione and relevance to hepatic venoocclusive disease. Hepatology. 1996;23(3):589–599. doi: 10.1002/hep.510230326. [DOI] [PubMed] [Google Scholar]
- 13.Odin J.A., Huebert R.C., Casciola-Rosen L., LaRusso N.F., Rosen A. Bcl-2-dependent oxidation of pyruvate dehydrogenase-E2, a primary biliary cirrhosis autoantigen, during apoptosis. J. Clin. Invest. 2001;108(2):223–232. doi: 10.1172/JCI10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussaini S.H., Farrington E.A. Idiosyncratic drug-induced liver injury: An overview. Expert Opin. Drug Saf. 2007;6(6):673–684. doi: 10.1517/14740338.6.6.673. [DOI] [PubMed] [Google Scholar]
- 15.Andrade R.J., Lucena M.I., Kaplowitz N., García-Muņoz B., Borraz Y., Pachkoria K., García-Cortés M., Fernández M.C., Pelaez G., Rodrigo L., Durán J.A., Costa J., Planas R., Barriocanal A., Guarner C., Romero-Gomez M., Muņoz-Yagüe T., Salmerón J., Hidalgo R. Outcome of acute idiosyncratic drug-induced liver injury: Long-term follow-up in a hepatotoxicity registry. Hepatology. 2006;44(6):1581–1588. doi: 10.1002/hep.21424. [DOI] [PubMed] [Google Scholar]
- 16.Björnsson E., Olsson R. Outcome and prognostic markers in severe drug-induced liver disease. Hepatology. 2005;42(2):481–489. doi: 10.1002/hep.20800. [DOI] [PubMed] [Google Scholar]
- 17.Holt M., Ju C. Drug-induced liver injury. Handb. Exp. Pharmacol. 2010;196:3–27. doi: 10.1007/978-3-642-00663-0_1. [DOI] [PubMed] [Google Scholar]
- 18.Ulrich R. Idiosyncratic toxicity: A convergence of risk factors. Annu. Rev. Med. 2007;58:17–34. doi: 10.1146/annurev.med.58.072905.160823. [DOI] [PubMed] [Google Scholar]
- 19.Han D., Shinohara M., Ybanez M.D., Saberi B., Kaplowitz N. Signal transduction pathways involved in drug-induced liver injury. Handb. Exp. Pharmacol. 2010;196:267–310. doi: 10.1007/978-3-642-00663-0_10. [DOI] [PubMed] [Google Scholar]
- 20.Lee W.M. Drug-induced hepatotoxicity. N. Engl. J. Med. 2003;349(5):474–485. doi: 10.1056/NEJMra021844. [DOI] [PubMed] [Google Scholar]
- 21.Bryant A.E., Dreifuss F.E. Hepatotoxicity associated with antiepileptic drug therapy. CNS Drugs. 1995;4(2):99–113. [Google Scholar]
- 22.Daly A.K., Day C.P. Genetic association studies in drug-induced liver injury. Semin. Liver Dis. 2009;29(4):400–411. doi: 10.1055/s-0029-1240009. [DOI] [PubMed] [Google Scholar]
- 23.Hirata K., Takagi H., Yamamoto M., Matsumoto T., Nishiya T., Mori K., Shimizu S., Masumoto H., Okutani Y. Ticlopidine-induced hepatotoxicity is associated with specific human leukocyte antigen genomic subtypes in Japanese patients: A preliminary case-control study. Pharmacogenomics J. 2008;8(1):29–33. doi: 10.1038/sj.tpj.6500442. [DOI] [PubMed] [Google Scholar]
- 24.Yuan L., Bambha K. Bile acid receptors and nonalcoholic fatty liver disease. World J. Hepatol. 2015;7(28):2811–2818. doi: 10.4254/wjh.v7.i28.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeilinger K., Freyer N., Damm G., Seehofer D., Knöspel F. Cell sources for in vitro human liver cell culture models. Exp. Biol. Med. (Maywood) 2016;241(15):1684–1698. doi: 10.1177/1535370216657448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., Doshi U., Suzuki A., Chang C.W., Borlak J., Li A.P., Tong W. Evaluation of multiple mechanism-based toxicity endpoints in primary cultured human hepatocytes for the identification of drugs with clinical hepatotoxicity: Results from 152 marketed drugs with known liver injury profiles. Chem. Biol. Interact. 2016;255:3–11. doi: 10.1016/j.cbi.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Mann A., Pelz T., Rennert K., Mosig A., Decker M., Lupp A. Evaluation of HepaRG cells for the assessment of indirect drug-induced hepatotoxicity using INH as a model substance. Hum. Cell. 2017;30(4):267–278. doi: 10.1007/s13577-017-0175-9. [DOI] [PubMed] [Google Scholar]
- 28.Donato M.T., Lahoz A., Castell J.V., Gómez-Lechón M.J. Cell lines: A tool for in vitro drug metabolism studies. Curr. Drug Metab. 2008;9(1):1–11. doi: 10.2174/138920008783331086. [DOI] [PubMed] [Google Scholar]
- 29.Castell J.V., Jover R., Martínez-Jiménez C.P., Gómez-Lechón M.J. Hepatocyte cell lines: Their use, scope and limitations in drug metabolism studies. Expert Opin. Drug Metab. Toxicol. 2006;2(2):183–212. doi: 10.1517/17425255.2.2.183. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez-Antona C., Donato M.T., Boobis A., Edwards R.J., Watts P.S., Castell J.V., Gómez-Lechón M.J. Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: Molecular mechanisms that determine lower expression in cultured cells. Xenobiotica. 2002;32(6):505–520. doi: 10.1080/00498250210128675. [DOI] [PubMed] [Google Scholar]
- 31.Fischbach M., Cao H.W., Diez Ibanez M., Tsaconas C., Alouani S., Montandon F., El Baraka M., Padieu P., Dreano M., Chessebeuf-Padieu M. Maintenance of liver function in long term culture of hepatocytes following in vitro or in vivo Ha-rasEJ transfection. Cell Biol. Toxicol. 1991;7(4):327–345. doi: 10.1007/BF00124069. [DOI] [PubMed] [Google Scholar]
- 32.Höhne M.W., Zieroth S., Veser U., Kahl G.F., Schwarz L.R. Carcinogen-induced diploid hepatocytes: sensitive target cells for transformation by mutated c-Ha-ras oncogene. Mol. Carcinog. 1993;7(3):180–189. doi: 10.1002/mc.2940070308. [DOI] [PubMed] [Google Scholar]
- 33.Sirica A.E. Immortalizing hepatocytes with truncated MET: a little bit of gene goes a long way. Hepatology. 1997;26(2):510–512. doi: 10.1002/hep.510260241. [DOI] [PubMed] [Google Scholar]
- 34.Donato M.T., Jover R., Gómez-Lechón M.J. Hepatic cell lines for drug hepatotoxicity testing: limitations and strategies to upgrade their metabolic competence by gene engineering. Curr. Drug Metab. 2013;14(9):946–968. doi: 10.2174/1389200211314090002. [DOI] [PubMed] [Google Scholar]
- 35.Benet M., Lahoz A., Guzmán C., Castell J.V., Jover R. CCAAT/enhancer-binding protein alpha(C/EBPalpha) and hepatocyte nuclear factor 4alpha(HNF4alpha) synergistically cooperate with constitutive androstane receptor to transactivate the human cytochrome P450 2B6(CYP2B6) gene: application to the development of a metabolically competent human hepatic cell model. J. Biol. Chem. 2010;285(37):28457–28471. doi: 10.1074/jbc.M110.118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crespi C.L., Miller V.P. The use of heterologously expressed drug metabolizing enzymes-state of the art and prospects for the future. Pharmacol. Ther. 1999;84(2):121–131. doi: 10.1016/s0163-7258(99)00028-5. [DOI] [PubMed] [Google Scholar]
- 37.Prakash C., Sharma R., Gleave M., Nedderman A. In vitro screening techniques for reactive metabolites for minimizing bioactivation potential in drug discovery. Curr. Drug Metab. 2008;9(9):952–964. doi: 10.2174/138920008786485209. [DOI] [PubMed] [Google Scholar]
- 38.Sison-Young R.L., Kia R., Heslop J., Kelly L., Rowe C., Cross M.J., Kitteringham N.R., Hanley N., Park B.K., Goldring C.E. Human pluripotent stem cells for modeling toxicity. Adv. Pharmacol. 2012;63:207–256. doi: 10.1016/B978-0-12-398339-8.00006-9. [DOI] [PubMed] [Google Scholar]
- 39.Sirenko O., Hesley J., Rusyn I., Cromwell E.F. High-content assays for hepatotoxicity using induced pluripotent stem cell-derived cells. Assay Drug Dev. Technol. 2014;12(1):43–54. doi: 10.1089/adt.2013.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J.H., Wang M., Lee J., Park H.J., Han C., Hong H.S., Kim J.S., An G.H., Park K., Park H.K., Zhu S.F., Sun X.B., Kim J.H., Woo D.H. Prediction of hepatotoxicity for drugs using human pluripotent stem cell-derived hepatocytes. Cell Biol. Toxicol. 2018;34(1):51–64. doi: 10.1007/s10565-017-9392-y. [DOI] [PubMed] [Google Scholar]
- 41.Anson B.D., Kolaja K.L., Kamp T.J. Opportunities for use of human iPS cells in predictive toxicology. Clin. Pharmacol. Ther. 2011;89(5):754–758. doi: 10.1038/clpt.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bale S.S., Vernetti L., Senutovitch N., Jindal R., Hegde M., Gough A., McCarty W.J., Bakan A., Bhushan A., Shun T.Y., Golberg I., DeBiasio R., Usta O.B., Taylor D.L., Yarmush M.L. In vitro platforms for evaluating liver toxicity. Exp. Biol. Med. (Maywood) 2014;239(9):1180–1191. doi: 10.1177/1535370214531872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmgren G., Sjögren A.K., Barragan I., Sabirsh A., Sartipy P., Synnergren J., Björquist P., Ingelman-Sundberg M., Andersson T.B., Edsbagge J. Long-term chronic toxicity testing using human pluripotent stem cell-derived hepatocytes. Drug Metab. Dispos. 2014;42(9):1401–1406. doi: 10.1124/dmd.114.059154. [DOI] [PubMed] [Google Scholar]
- 44.Takayama K., Morisaki Y., Kuno S., Nagamoto Y., Harada K., Furukawa N., Ohtaka M., Nishimura K., Imagawa K., Sakurai F., Tachibana M., Sumazaki R., Noguchi E., Nakanishi M., Hirata K., Kawabata K., Mizuguchi H. Prediction of interindividual differences in hepatic functions and drug sensitivity by using human iPS-derived hepatocytes. Proc. Natl. Acad. Sci. USA. 2014;111(47):16772–16777. doi: 10.1073/pnas.1413481111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dash A., Inman W., Hoffmaster K., Sevidal S., Kelly J., Obach R.S., Griffith L.G., Tannenbaum S.R. Liver tissue engineering in the evaluation of drug safety. Expert Opin. Drug Metab. Toxicol. 2009;5(10):1159–1174. doi: 10.1517/17425250903160664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richert L., Binda D., Hamilton G., Viollon-Abadie C., Alexandre E., Bigot-Lasserre D., Bars R., Coassolo P., LeCluyse E. Evaluation of the effect of culture configuration on morphology, survival time, antioxidant status and metabolic capacities of cultured rat hepatocytes. Toxicol. In Vitro. 2002;16(1):89–99. doi: 10.1016/s0887-2333(01)00099-6. [DOI] [PubMed] [Google Scholar]
- 47.Schyschka L., Sánchez J.J., Wang Z., Burkhardt B., Müller-Vieira U., Zeilinger K., Bachmann A., Nadalin S., Damm G., Nussler A.K. Hepatic 3D cultures but not 2D cultures preserve specific transporter activity for acetaminophen-induced hepatotoxicity. Arch. Toxicol. 2013;87(8):1581–1593. doi: 10.1007/s00204-013-1080-y. [DOI] [PubMed] [Google Scholar]
- 48.Bell C.C., Hendriks D.F., Moro S.M., Ellis E., Walsh J., Renblom A., Fredriksson Puigvert L., Dankers A.C., Jacobs F., Snoeys J., Sison-Young R.L., Jenkins R.E., Nordling Å., Mkrtchian S., Park B.K., Kitteringham N.R., Goldring C.E., Lauschke V.M., Ingelman-Sundberg M. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 2016;6:25187. doi: 10.1038/srep25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersson T.B. Evolution of novel 3D culture systems for studies of human liver function and assessments of the hepatotoxicity of drugs and drug candidates. Basic Clin. Pharmacol. Toxicol. 2017;121(4):234–238. doi: 10.1111/bcpt.12804. [DOI] [PubMed] [Google Scholar]
- 50.Beckwitt C.H., Clark A.M., Wheeler S., Taylor D.L., Stolz D.B., Griffith L., Wells A. Liver ‘organ on a chip’. Exp. Cell Res. 2018;363(1):15–25. doi: 10.1016/j.yexcr.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uetrecht J. Evaluation of which reactive metabolite, if any, is responsible for a specific idiosyncratic reaction. Drug Metab. Rev. 2006;38(4):745–753. doi: 10.1080/03602530600959615. [DOI] [PubMed] [Google Scholar]
- 52.Anders H.J., Vielhauer V. Identifying and validating novel targets with in vivo disease models: Guidelines for study design. Drug Discov. Today. 2007;12(11-12):446–451. doi: 10.1016/j.drudis.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 53.Morgan S.J., Elangbam C.S. Animal Models of Disease for Future Toxicity Predictions. New York: John Wiley & Sons, Inc.; 2016. [Google Scholar]
- 54.Varga O., Hansen A., Sandoe P., Olsson I. Validating animal models for preclinical research: A scientific and ethical discussion. Altern. Lab. Anim. 2010;38(3):245–248. doi: 10.1177/026119291003800309. [DOI] [PubMed] [Google Scholar]
- 55.Olson H., Betton G., Robinson D., Thomas K., Monro A., Kolaja G., Lilly P., Sanders J., Sipes G., Bracken W., Dorato M., Van Deun K., Smith P., Berger B., Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000;32(1):56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- 56.Ballet F. Hepatotoxicity in drug development: Detection, significance and solutions. J. Hepatol. 1997;26:26–36. doi: 10.1016/s0168-8278(97)80494-1. [DOI] [PubMed] [Google Scholar]
- 57.Committee for Medicinal Products for Human Use (CHMP) Reflection paper on non-clinical evaluation of druginduced liver injury(DILI). EMA; 2010. pp. 1–12. [Google Scholar]
- 58.Greaves P., Williams A., Eve M. First dose of potential new medicines to humans: How animals help. Nat. Rev. Drug Discov. 2004;3(3):226–236. doi: 10.1038/nrd1329. [DOI] [PubMed] [Google Scholar]
- 59.Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.R., Ueno Y., Zheng Y.W., Koike N., Aoyama S., Adachi Y., Taniguchi H. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499(7459):481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 60.Hasegawa M., Kawai K., Mitsui T., Taniguchi K., Monnai M., Wakui M., Ito M., Suematsu M., Peltz G., Nakamura M., Suemizu H. The reconstituted ‘humanized liver’ in TK-NOG mice is mature and functional. Biochem. Biophys. Res. Commun. 2011;405(3):405–410. doi: 10.1016/j.bbrc.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pridgeon S.C., Zhang F., Heslop J.A., Nugues C.M.L., Kitteringham N.R., Park B.K., Goldring C.E.P. Application of pluripotent stem cells in drug ‐ induced liver injury safety assessment. In: Will Y.J., McDuffie E., Jeffy B.D., editors. Drug Discovery Toxicology: From Target Assessment to Translational Biomarkers. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2016. p. 333. [Google Scholar]
- 62.Madariaga M. Drug-related hepatotoxicity. N. Engl. J. Med. 2006;354(20):2192–2192. [PubMed] [Google Scholar]
- 63.Dahlin D.C., Miwa G.T., Lu A.Y.H., Nelson S.D. N-acetyl-para-benzoquinone imine: A cytochrome-p-450-mediated oxidation-product of acetaminophen. Proc. Natl. Acad. Sci. USA. 1984;81(5):1327–1331. doi: 10.1073/pnas.81.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng W., Lobach A.R., Zhu X., Chen X., Liu F., Metushi I.G., Sharma A., Li J., Cai P., Ip J., Novalen M., Popovic M., Zhang X., Tanino T., Nakagawa T., Li Y., Uetrecht J. Animal models of idiosyncratic drug reactions. Adv. Pharmacol. 2012;63:81–135. doi: 10.1016/B978-0-12-398339-8.00003-3. [DOI] [PubMed] [Google Scholar]
- 65.Roth R.A., Ganey P.E. Animal models of idiosyncratic drug-induced liver injury-current status. Crit. Rev. Toxicol. 2011;41(9):723–739. doi: 10.3109/10408444.2011.575765. [DOI] [PubMed] [Google Scholar]
- 66.Metushi I.G., Hayes M.A., Uetrecht J. Treatment of PD-1(-/-) mice with amodiaquine and anti-CTLA4 leads to liver injury similar to idiosyncratic liver injury in patients. Hepatology. 2015;61(4):1332–1342. doi: 10.1002/hep.27549. [DOI] [PubMed] [Google Scholar]
- 67.Deng X., Luyendyk J.P., Ganey P.E., Roth R.A. Inflammatory stress and idiosyncratic hepatotoxicity: Hints from animal models. Pharmacol. Rev. 2009;61(3):262–282. doi: 10.1124/pr.109.001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Przybylak K.R., Cronin M.T. In silico models for drug-induced liver injury-current status. Expert Opin. Drug Metab. Toxicol. 2012;8(2):201–217. doi: 10.1517/17425255.2012.648613. [DOI] [PubMed] [Google Scholar]
- 69.Chen M., Bisgin H., Tong L., Hong H., Fang H., Borlak J., Tong W. Toward predictive models for drug-induced liver injury in humans: Are we there yet? Biomarkers Med. 2014;8(2):201–213. doi: 10.2217/bmm.13.146. [DOI] [PubMed] [Google Scholar]
- 70.Hewitt M., Enoch S.J., Madden J.C., Przybylak K.R., Cronin M.T. Hepatotoxicity: A scheme for generating chemical categories for read-across, structural alerts and insights into mechanism(s) of action. Crit. Rev. Toxicol. 2013;43(7):537–558. doi: 10.3109/10408444.2013.811215. [DOI] [PubMed] [Google Scholar]
- 71.Xu J.J., Diaz D., O’Brien P.J. Applications of cytotoxicity assays and pre-lethal mechanistic assays for assessment of human hepatotoxicity potential. Chem. Biol. Interact. 2004;150(1):115–128. doi: 10.1016/j.cbi.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 72.Tolosa L., Pinto S., Donato M.T., Lahoz A., Castell J.V., O’Connor J.E., Gómez-Lechón M.J. Development of a multiparametric cell-based protocol to screen and classify the hepatotoxicity potential of drugs. Toxicol. Sci. 2012;127(1):187–198. doi: 10.1093/toxsci/kfs083. [DOI] [PubMed] [Google Scholar]
- 73.Poulin P., Jones H.M., Jones R.D., Yates J.W., Gibson C.R., Chien J.Y., Ring B.J., Adkison K.K., He H., Vuppugalla R., Marathe P., Fischer V., Dutta S., Sinha V.K., Björnsson T., Lavé T., Ku M.S.PhR.M.A. CPCDC initiative on predictive models of human pharmacokinetics, part 1: Goals, properties of the PhRMA dataset, and comparison with literature datasets. J. Pharm. Sci. 2011;100(10):4050–4073. doi: 10.1002/jps.22554. [DOI] [PubMed] [Google Scholar]
- 74.Uehara T., Ono A., Maruyama T., Kato I., Yamada H., Ohno Y., Urushidani T. The Japanese toxicogenomics project: Application of toxicogenomics. Mol. Nutr. Food Res. 2010;54(2):218–227. doi: 10.1002/mnfr.200900169. [DOI] [PubMed] [Google Scholar]
- 75.Ganter B., Snyder R.D., Halbert D.N., Lee M.D. Toxicogenomics in drug discovery and development: Mechanistic analysis of compound/class-dependent effects using the DrugMatrix database. Pharmacogenomics. 2006;7(7):1025–1044. doi: 10.2217/14622416.7.7.1025. [DOI] [PubMed] [Google Scholar]
- 76.Chen M., Zhang M., Borlak J., Tong W. A decade of toxicogenomic research and its contribution to toxicological science. Toxicol. Sci. 2012;130(2):217–228. doi: 10.1093/toxsci/kfs223. [DOI] [PubMed] [Google Scholar]
- 77.Shenton J.M., Chen J., Uetrecht J.P. Animal models of idiosyncratic drug reactions. Chem. Biol. Interact. 2004;150(1):53–70. doi: 10.1016/j.cbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 78.Ozer J., Ratner M., Shaw M., Bailey W., Schomaker S. The current state of serum biomarkers of hepatotoxicity. Toxicology. 2008;245(3):194–205. doi: 10.1016/j.tox.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 79.Ozer J.S., Chetty R., Kenna G., Palandra J., Zhang Y., Lanevschi A., Koppiker N., Souberbielle B.E., Ramaiah S.K. Enhancing the utility of alanine aminotransferase as a reference standard biomarker for drug-induced liver injury. Regul. Toxicol. Pharmacol. 2010;56(3):237–246. doi: 10.1016/j.yrtph.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Ramaiah S.K. A toxicologist guide to the diagnostic interpretation of hepatic biochemical parameters. Food Chem. Toxicol. 2007;45(9):1551–1557. doi: 10.1016/j.fct.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 81.Amacher D.E., Adler R., Herath A., Townsend R.R. Use of proteomic methods to identify serum biomarkers associated with rat liver toxicity or hypertrophy. Clin. Chem. 2005;51(10):1796–1803. doi: 10.1373/clinchem.2005.049908. [DOI] [PubMed] [Google Scholar]
- 82.Schomaker S., Warner R., Bock J., Johnson K., Potter D., Van Winkle J., Aubrecht J. Assessment of emerging biomarkers of liver injury in human subjects. Toxicol. Sci. 2013;132(2):276–283. doi: 10.1093/toxsci/kft009. [DOI] [PubMed] [Google Scholar]
- 83.Wang K., Zhang S., Marzolf B., Troisch P., Brightman A., Hu Z., Hood L.E., Galas D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA. 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding X., Ding J., Ning J., Yi F., Chen J., Zhao D., Zheng J., Liang Z., Hu Z., Du Q. Circulating microRNA-122 as a potential biomarker for liver injury. Mol. Med. Rep. 2012;5(6):1428–1432. doi: 10.3892/mmr.2012.838. [DOI] [PubMed] [Google Scholar]
- 85.Ward J., Kanchagar C., Veksler-Lublinsky I., Lee R.C., McGill M.R., Jaeschke H., Curry S.C., Ambros V.R. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc. Natl. Acad. Sci. USA. 2014;111(33):12169–12174. doi: 10.1073/pnas.1412608111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krauskopf J., Caiment F., Claessen S.M., Johnson K.J., Warner R.L., Schomaker S.J., Burt D.A., Aubrecht J., Kleinjans J.C. Application of high-throughput sequencing to circulating microRNAs reveals novel biomarkers for drug-induced liver injury. Toxicol. Sci. 2015;143(2):268–276. doi: 10.1093/toxsci/kfu232. [DOI] [PubMed] [Google Scholar]
- 87.Yang X., Salminen W.F., Shi Q., Greenhaw J., Gill P.S., Bhattacharyya S., Beger R.D., Mendrick D.L., Mattes W.B., James L.P. Potential of extracellular microRNAs as biomarkers of acetaminophen toxicity in children. Toxicol. Appl. Pharmacol. 2015;284(2):180–187. doi: 10.1016/j.taap.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McGill M.R., Jaeschke H. MicroRNAs as signaling mediators and biomarkers of drug- and chemical-induced liver injury. J. Clin. Med. 2015;4(5):1063–1078. doi: 10.3390/jcm4051063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Antoine D.J., Dear J.W., Lewis P.S., Platt V., Coyle J., Masson M., Thanacoody R.H., Gray A.J., Webb D.J., Moggs J.G., Bateman D.N., Goldring C.E., Park B.K. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013;58(2):777–787. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang X., Greenhaw J., Shi Q., Su Z., Qian F., Davis K., Mendrick D.L., Salminen W.F. Identification of urinary microRNA profiles in rats that may diagnose hepatotoxicity. Toxicol. Sci. 2012;125(2):335–344. doi: 10.1093/toxsci/kfr321. [DOI] [PubMed] [Google Scholar]
- 91.Yang X., Li Z., Su Z., Davis K., Chen T., Mendrick D. Urinary microRNAs as noninvasive biomarkers for acetaminophen-induced liver injury. J. Postgenom. Drug Biomark. Develop. 2011;1:101. [Google Scholar]
- 92.Sanjay S., Girish C. Role of miRNA and its potential as a novel diagnostic biomarker in drug-induced liver injury. Eur. J. Clin. Pharmacol. 2017;73(4):399–407. doi: 10.1007/s00228-016-2183-1. [DOI] [PubMed] [Google Scholar]
- 93.Laverty H.G., Antoine D.J., Benson C., Chaponda M., Williams D., Kevin Park B. The potential of cytokines as safety biomarkers for drug-induced liver injury. Eur. J. Clin. Pharmacol. 2010;66(10):961–976. doi: 10.1007/s00228-010-0862-x. [DOI] [PubMed] [Google Scholar]
- 94.Tarrant J.M. Blood cytokines as biomarkers of in vivo toxicity in preclinical safety assessment: Considerations for their use. Toxicol. Sci. 2010;117(1):4–16. doi: 10.1093/toxsci/kfq134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Ravenzwaay B., Cunha G.C., Leibold E., Looser R., Mellert W., Prokoudine A., Walk T., Wiemer J. The use of metabolomics for the discovery of new biomarkers of effect. Toxicol. Lett. 2007;172(1-2):21–28. doi: 10.1016/j.toxlet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 96.Goodsaid F.M., Frueh F.W., Mattes W. Strategic paths for biomarker qualification. Toxicology. 2008;245(3):219–223. doi: 10.1016/j.tox.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 97.Pirmohamed M. Genetics and the potential for predictive tests in adverse drug reactions. Chem. Immunol. Allergy. 2012;97:18–31. doi: 10.1159/000335613. [DOI] [PubMed] [Google Scholar]
- 98.Clare K.E., Miller M.H., Dillon J.F. Genetic factors influencing drug-induced liver injury: Do they have a role in prevention and diagnosis? Curr. Hepatol. Rep. 2017;16(3):258–264. doi: 10.1007/s11901-017-0363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O’Donohue J., Oien K.A., Donaldson P., Underhill J., Clare M., MacSween R.N., Mills P.R. Co-amoxiclav jaundice: Clinical and histological features and HLA class II association. Gut. 2000;47(5):717–720. doi: 10.1136/gut.47.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grove J.I., Aithal G.P. Human leukocyte antigen genetic risk factors of drug-induced liver toxicology. Expert Opin. Drug Metab. Toxicol. 2015;11(3):395–409. doi: 10.1517/17425255.2015.992414. [DOI] [PubMed] [Google Scholar]
- 101.Aithal G.P. Pharmacogenetic testing in idiosyncratic drug-induced liver injury: Current role in clinical practice. Liver Int. 2015;35(7):1801–1808. doi: 10.1111/liv.12836. [DOI] [PubMed] [Google Scholar]
- 102.Cai Y., Yi J., Zhou C., Shen X. Pharmacogenetic study of drug-metabolising enzyme polymorphisms on the risk of anti-tuberculosis drug-induced liver injury: a meta-analysis. PLoS One. 2012;7(10):e47769. doi: 10.1371/journal.pone.0047769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Urban T.J., Daly A.K., Aithal G.P. Genetic basis of drug-induced liver injury: Present and future. Semin. Liver Dis. 2014;34(2):123–133. doi: 10.1055/s-0034-1375954. [DOI] [PubMed] [Google Scholar]
- 104.Aithal G.P., Day C.P., Leathart J.B., Daly A.K. Relationship of polymorphism in CYP2C9 to genetic susceptibility to diclofenac-induced hepatitis. Pharmacogenetics. 2000;10(6):511–518. doi: 10.1097/00008571-200008000-00004. [DOI] [PubMed] [Google Scholar]
- 105.Karlin E., Phillips E. Genotyping for severe drug hypersensitivity. Curr. Allergy Asthma Rep. 2014;14(3):418. doi: 10.1007/s11882-013-0418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaniwa N., Saito Y. Pharmacogenomics of severe cutaneous adverse reactions. Pharmacogenomics. 2013;14(6):595–598. doi: 10.2217/pgs.13.27. [DOI] [PubMed] [Google Scholar]