Abstract
The AU-rich element (AUUUA)n, found in the 3′ noncoding region of many short-lived cytokine and proto-oncogene mRNAs, is sufficient to specifically target these mRNAs for rapid degradation in mammalian cells. The mechanism by which the AU-rich element promotes rapid mRNA decay is not known. Previous studies have shown that release of intracellular stored calcium by ionophore treatment of thymocytes and mast cells inhibits the rapid turnover of AU-rich interleukin mRNAs. Increased cytoplasmic half-life of interleukin mRNAs was linked to calcium-induced activation of the N-terminal c-Jun kinase. In this report we have characterized the calcium-induced stabilization of AU-rich mRNAs. We show that calcium induces stabilization of mRNAs with canonical AU-rich elements in all cell types tested. These results indicate that short-lived mRNA stabilization by calcium is not unique to immune cells nor interleukin mRNAs, but is a widespread default response that includes generic AU-rich mRNAs. Stabilization is shown to be rapid but transient, and to act without altering nuclear transcription or cytoplasmic translation rates. These data support the view that calcium release likely stabilizes short-lived mRNAs by altering trans-acting decay factors that promote AU-rich mRNA turnover.
Keywords: Calcium-induced stabilization, AU-rich element, Short-lived mRNAs
SHORT-LIVED proto-oncogene and cytokine mRNAs typically possess an AU-rich element (ARE) in the 3′ noncoding region (3′NCR) of the mRNA (7), consisting of several closely spaced copies of the pentanucleotide sequence AUUUA. Five contiguous AUUUA copies in the 3′NCR of granulocyte macrophage-colony stimulating factor (GM-CSF) mRNA are sufficient for rapid cytoplasmic degradation (38). Chimeric, experimental mRNAs containing the GM-CSF ARE are also rapidly degraded compared to mRNAs containing a control element in which G and C residues are interspersed in the ARE. The minimal ARE core element of cytokine mRNAs has been established as UUAUUUA(U/A)(U/A) (24,46), although proto-oncogene mRNAs utilize other elements in addition to the ARE [(8), reviewed in (9,10)]. Proto-oncogene and cytokine mRNAs are also differentially regulated. In several murine monocyte-macrophage tumors obtained by random retroviral insertion, c-fos mRNAs were rapidly degraded whereas GM-CSF mRNAs were stabilized, possibly due to inactivation of trans-acting factors (36). Cells that have lost the ability to degrade cytokine mRNAs are generally more transformed, implicating the degradation machinery as antioncogenic [e.g., (3,36)].
The mechanisms by which proto-oncogene and cytokine AREs target an mRNA for selective degradation are not well described, but include sequence-specific trans-acting binding proteins. There are more than 12 different proteins that have been identified as AU-rich sequence binding proteins (AUBPs) by in vitro UV cross-linking to labeled AUUUA sequences [reviewed in (21)]. Some of the AUBPs are classic components of mRNP complexes that participate in splicing, transport, or translation of mRNAs, but several might stabilize or promote ARE mRNA turnover. Of the various AUBPs, two appear to be involved in regulating ARE mRNA turnover. Two members of the elav family of proteins, shown to bind to ARE and U-rich sequences in vitro, are the HuR protein (27,30,31) and Hel-N1 (2). HuR is identical to the nuclear AUBP of Vakalopoulou et al. (42). Binding of HuR and Hel-N1 to AU-rich sequences is now known to correlate with stabilization of an ARE mRNA (14,22). Stabilization of ARE mRNAs was also reported for a complex of three proteins (MW 30–45 kDa) that cross-link in vitro to AUUUA sequences (16,28). One family of related AUBPs, known as AUF1 or hnRNP D proteins, has been strongly implicated as decay-promoting factors in the control of ARE mRNA stability. AUF1 was identified biochemically in cytoplasmic extracts by Brewer as an (AUUUA)n-specific destabilizing complex of proteins that are polysome enriched (5), based on photo-UV cross-linking and cell-free decay extracts. A number of reports support the conclusion that AUF1 is a pivotal destabilizing complex of proteins that promotes ARE mRNA decay. The AUF1 gene is encoded by two loci located to human chromosome 4q21–23 and Xq12 (44). AUF1 consists of four isoform proteins derived by alternative splicing of a single mRNA to produce a p37, p40, p42, and p45 protein (23,44). All four proteins share a common 200-amino acid core region that contains two nonidentical RNA recognition motifs (RRMs). The alternate forms of AUF1 contain a 19-amino acid N-terminal insertion and/or a 49 amino acid C-terminal insertion (23, 44). The insertions are adjacent to the RRMs and might alter the specificity and binding strength of the different AUF1 isoforms for, ARE sequences (23). Several lines of evidence support the conclusion that AUF1 proteins promote ARE mRNA decay, (i) Adherence of monocytes correlates with p37 AUF1 phosphorylation, decreased binding in vitro to (AUUUA)n elements, and stabilization of ARE mRNAs (39). (ii) Cell lines and tissues with reduced expression of p37 and p40 AUF1 proteins only slowly degrade ARE mRNAs compared to cell lines that express higher levels (6,33). (iii) The instability of ARE mRNAs correlates with the strength of p37/p40 AUF1 binding to AREs in vitro (13). Thus, correlative data implicate members of the AUF1 family of proteins in promoting mRNA decay, particularly ARE-containing mRNAs.
Although cytokine mRNAs with ARE sequences are rapidly degraded in most cells, their transient stabilization has been induced in T cells and mast cells by a variety of stimuli that influence calcium release and protein kinase activities. Studies found stabilization of ARE-containing mRNAs upon treatment of these cells with calcium ionophores, lipopolysaccharides, or activators of immunoglobulin receptors [e.g. (4,18–20,26,40,45)]. More recently, two groups have shown that stabilization of IL-2 and IL-3 mRNAs by calcium release in T cells and mast cells involves stimulation of the c-Jun–N-terminal protein kinase (INK) signaling pathway (11,29).
The studies presented here were undertaken to determine whether the calcium-induced stabilization of cytokine ARE mRNAs is ubiquitous, or is restricted to specialized immune cells and interleukin mRNAs. In addition, evidence has been presented that activation of JNK signaling does not correlate tightly with stabilization of the IL-3 mRNA in mast cells (29), in that inhibition of calcium-dependent calcineurin, which is not known to be activated by JNK, also blocks IL-3 mRNA stabilization by calcium. These data indicate that calcium-induced pathways other than those involving JNK also play a role in stabilization of IL-3 mRNA. Here we show that calcium induces stabilization of a reporter mRNA containing the GM-CSF ARE in different tissue culture cell lines. Calcium-induced stabilization of the reporter ARE mRNA is shown to reach maximal levels by 30 min of treatment, and to be of short duration, lasting from 60 to 90 min. ARE mRNA stabilization does not involve new transcription, in that there is no detectable change in the transcription rate of the ARE reporter mRNAs, and it occurs without significantly altering mRNA translation. These results demonstrate that the transient stabilization of ARE mRNAs against decay by calcium release is a ubiquitous default response in most cells that includes canonical AU-rich elements and likely involves the direct action of signal transduction pathways on the decay machinery.
MATERIALS AND METHODS
Reagents
Calcium ionophore A23187 and actinomycin D were obtained from Sigma Chemical Company. Actinomycin D was used at 5 ng/ml to block transcription. A23187 was used at 10 pM for the times indicated.
Constructs
All constructs were derived from plasmids pLSAT and pLSGC as shown in Fig. 1 and described previously (1,12). Plasmids contain the adenovirus (Ad) major late promoter (MLP), tripartite leader 5′ non-coding region, and hepatitis B virus surface antigen (S-antigen) open reading frame (ORF). pLSAT contains the GM-CSF ARE, β-globin 3′NCR, and the SV40 splice-polyadenylation signal (38). pLSGC contains a mutated ARE sequence in which G and C residues have been interspersed (38), constructed into the same genetic background as pLSAT.
FIG. 1.
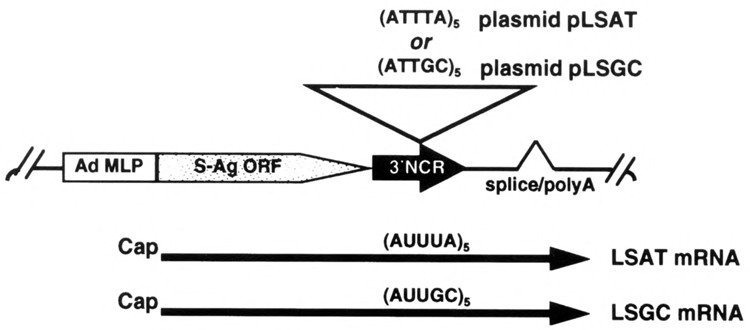
Schematic diagram of plasmids and encoded chimeric mRNAs. mRNAs are transcribed from the adenovirus major late promoter (Ad MLP), express the hepatitis B virus S-antigen open reading frame (S-Ag ORF), and contain a 3′ noncoding region (3′NCR) with either the GM-CSF AU-rich sequence (AUUUA)5 or a control sequence (AUUGC)5.
Cell Culture and DNA Transfections
All cell lines were maintained in Dulbecco modified Eagle’s medium containing 10% supplemented calf serum, 2 mM l-glutamine, and 100 μg/ml gentamicin. Transfection of 10-cm plates of cells was performed in triplicate by the calcium phosphate precipitation technique using 3 μg reporter plasmid and 1 μg of a plasmid expressing β-galactosidase as a control for transfection efficiencies. Cells were harvested after 48 h and transfection efficiencies determined by β-galactosidase assay.
Analysis of Protein and mRNA Levels
Levels of S-antigen protein synthesis were determined in cells labeled with [35S]methionine. Labeling of cells, preparation of extracts, immunoprecipitation of S-antigen, SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and fluorography were carried out as described previously (1). S10 cytoplasmic extracts were prepared by lysis of cells with 0.5% NP40 in 50 mM KCl, 0 mM Tris-HCl, pH 7.5, 1 mM EDTA, clarified by centrifugation at 10,000 × g for 5 min at 4°C. RNAs were isolated from cytoplasmic extracts by guanidine-HCl extraction followed by centrifugation through a CsCl cushion or by phenol-chloroform extraction (12). Northern blot analysis was performed using equal amounts of whole cytoplasmic RNA in formaldehyde-agarose gels, transferred to Nytran membrane, and hybridized to 32P-labeled probes prepared from the S-antigen or GAPDH coding region by the random primer method (15). Levels of mRNAs were normalized to the amount of GADPH mRNA present. Quantitation was obtained by laser densitometry of autoradiographs.
Transcription Run-On Analysis in Isolated Nuclei
Nuclei were isolated from cells 48 h after transfection as described (43). Nuclei were incubated for 30 min at 33°C in a transcription cocktail containing the following final concentrations: 50 mM Tris-HCl, pH 8.3, 5 mM MgCl2, 20% glycerol, 5 mM DTT, 1 mM each of ATP, CTP, GTP, and 1 μM UTP, 2 μg/ml α-amanitin to inhibit pol II transcription, and 100 μCi of [α-32P]UTP at 800 Ci/mmol. Labeled nuclear RNAs were extracted (17). Slot-blot filters for hybridization were prepared by binding 10 μg of denatured plasmid DNA containing the S-antigen gene to membrane. Prehybridization and hybridization using labeled RNAs were carried out using standard conditions. Slots were excised and quantitated by liquid scintillation counting.
RESULTS
Stabilization of ARE mRNA Calcium Ionophore A23187 Is a Common Cellular Response
Studies were carried out to determine whether the stabilization of IL-2 and IL-3 mRNAs by calcium release is unique to interleukin mRNAs in immune cells, or is a general response of most cells for most ARE mRNAs. Plasmids were previously constructed (1) that express reporter mRNAs containing the hepatitis B virus S-antigen open reading frame (ORF) and a 3′NCR with the minimal GM-CSF ARE (plasmid pLSAT; LSAT mRNA), or a GC-control sequence (plasmid pLSGC; LSGC mRNA) as shown in Fig. 1. HeLa cells (a human cervical carcinoma cell line), CHO cells (Chinese hamster ovary cells), 293 cells (human embryonic kidney cells), and Cos cells (monkey kidney cells) were used for analysis. Cells were transfected with plasmids for 48 h, then duplicate plates were treated with 10 uM A23187 for 3 h, an optimal concentration for release of calcium (20,40). Northern analysis of total cytoplasmic RNA (Fig. 2) demonstrated that the normally unstable and non-abundant LSAT reporter mRNA became as abundant as the stable LSGC mRNA in all cell lines tested, after 3-h treatment with A23187.
FIG. 2.
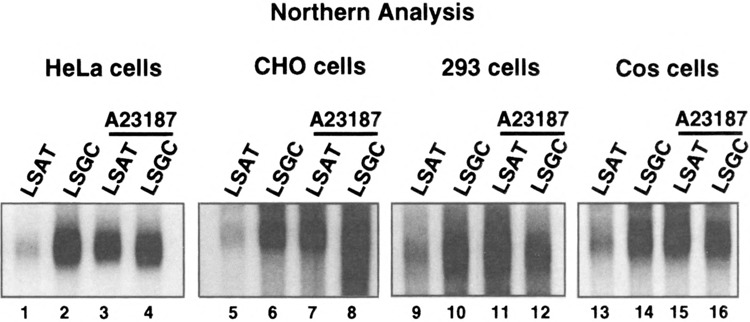
Northern mRNA steady-state analysis of LSAT and LSGC mRNAs in different cell lines. Cells were transfected with pLSGC or pLSAT plasmids, total cytoplasmic RNA was isolated, and equal amounts resolved by Northern analysis. S-antigen mRNA was hybridized to 32P-labeled probes prepared from an S-antigen DNA fragment. Duplicate plates of cells were treated with 10 μM A23187 for 3 h prior to cell lysis.
It was next determined whether the increased abundance of the normally short-lived LSAT mRNA was a result of increased mRNA half-life. HeLa cells were transfected with plasmids pLSGC or pLSAT for 48 h, treated for 3 h with calcium ionophore A23187, followed by 5 μM actinomycin D for the times shown to block new transcription. Half-lives of mRNAs were determined by Northern analysis of total cytoplasmic RNA and densitometry (Fig. 3). The LSAT mRNA, which contains the GM-CSF ARE, normally degrades with a half-life of ∼25 min in HeLa cells, compared to ∼10 h for the stable LSGC control mRNA. Treatment of cells with A23187 for 3 h prior to transcription inhibition resulted in complete stabilization of the LSAT mRNA. Identical results were obtained in the other cell lines tested (data not shown). These results indicate that induced release of calcium stabilizes mRNAs containing the GM-CSF ARE against rapid decay, implicating it as a general default mechanism common to most cells.
FIG. 3.
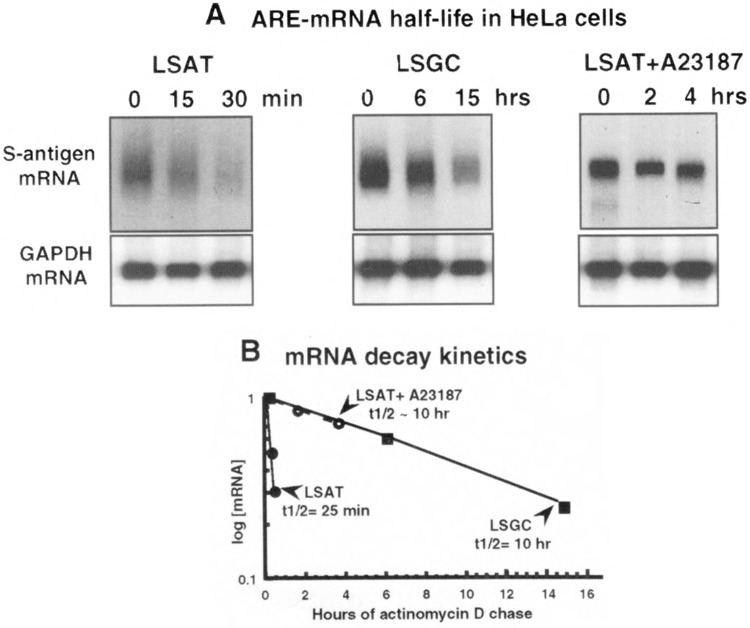
Half-life of LSAT and LSGC mRNAs in HeLa cells treated with A23187. Cells were transfected with pLSGC or pLSAT plasmids, treated with 5 μM actinomycin D for the times indicated to block transcription. A23187 (10 μM) was added with actinomycin D to deplete plates of LSAT mRNA for 3 h. (A) Total cytoplasmic RNA was isolated, equal amounts resolved by Northern analysis and hybridized to 32P-labeled probes specific to the S-antigen or GAPDH coding region. Data were derived by averaging the densitometric plots of two independent trials. (B) Data are plotted relative to the untreated control mRNA (time 0), which was set to 1.0 for normalization.
Calcium Stabilization of ARE mRNAs Is Posttranscriptional, Cytoplasmic, and Unrelated to mRNA Translation
A number of studies have shown that translation of ARE-containing mRNAs can activate or facilitate their rapid decay (1,12,34,35). Calcium release can also impair translation elongation through calmodulin-kinase III phosphorylation of elongation factor eEF2, particularly in neuronal cells [reviewed in (32)], which could block mRNA decay. Alternatively, calcium release can also stimulate translation initiation rates by promoting initiation factor eIF2 activation through dephosphorylation [reviewed in (32)]. Thus, calcium can have either no effect on translation or opposing effects, or it can stimulate or inhibit protein synthesis in different systems, which might account for the effects on ARE mRNA turnover. The effect, if any, of calcium on reporter ARE mRNA translation was assessed by comparing the level of reporter S-antigen protein levels for the normally long-lived LSGC mRNAs. As shown in Fig. 2, treatment of HeLa cells with calcium ionophore A23187 did not alter the abundance of the control LSGC mRNA. Any effect of calcium release on mRNA translation can therefore be readily detected. As shown in Fig. 4, S-antigen reporter translation rates were measured by labeling cells with [35S]methionine during 3-h calcium release in transfected HeLa and Cos cells. S-antigen was immunoprecipitated and resolved by SDS-PAGE and fluorography. A23187-induced calcium release stimulated S-antigen protein synthesis only very slightly, consistent with previous reports showing some increase in translation initiation rates. These results therefore exclude inhibition of translation by calcium release as a mechanism for ARE mRNA stabilization.
FIG. 4.
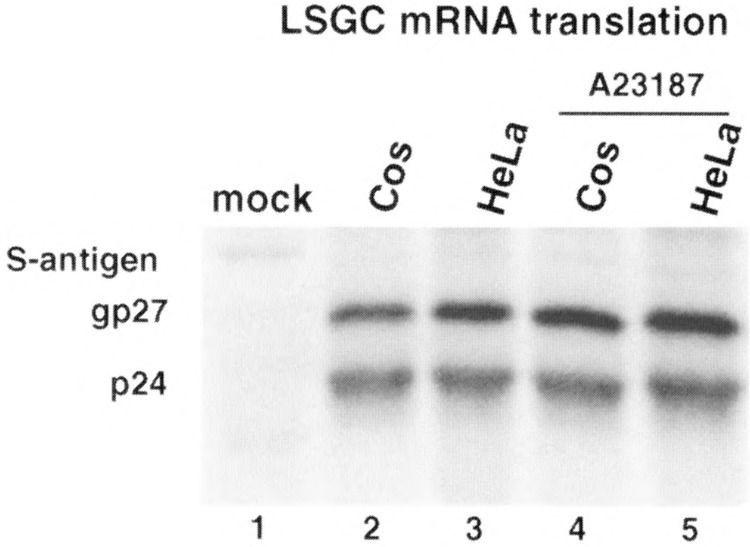
Effect of calcium ionophore A23187 on translation rates of reporter LSGC S-antigen mRNAs. Transfected HeLa and Cos cells, with and without 3-h treatment with 10 μM A23187, were labeled during treatment with 50 μCi of [35S]methionine per milliliter. Lysates were prepared and S-antigen immunoprecipitated from equal protein amounts and resolved by SDS-PAGE as described previously (1). S-antigen polypeptides consist of a 24-kDa species and a 27-kDa glycosylated form, as indicated.
The effect of calcium release on S-antigen reporter mRNA transcription rate was determined by measuring precommitted transcription elongation rates in nuclei isolated from transfected HeLa cells. Table 1 presents the results of nuclear run-on analysis. There was no significant difference in transcription rates of LSAT or LSGC mRNAs from untreated or A23187-treated cells. These results therefore demonstrate that the increased cytoplasmic abundance of the GM-CSF ARE containing mRNA is a result of increased cytoplasmic stability, not from changes in nuclear transcription or cytoplasmic translation rates.
TABLE 1.
NUCLEAR TRANSCRIPTION RATES
| Construct | Treatment | Nuclear Transcription Rate |
|---|---|---|
| pLSGC | none | 1.0 |
| pLSAT | none | 1.1 |
| pLSGC | A23187 | 1.2 |
| pLSAT | A23187 | 1.2 |
Transcription rates were measured by run-on analysis using isolated nuclei obtained from transfected cells, with or without 3-h treatment of 10 μM A23187. Rates were calculated by comparing values from liquid scintillation counting of slot-blot filter bands. Untreated pLSGC values were normalized to 1.0 and rates expressed as the ratio of the indicated sample to normalized LSGC.
Calcium Induces Rapid But Short-Lived Stabilization of ARE mRNAs
The kinetics of mRNA stabilization during calcium release or the duration of stabilization have not been previously addressed. This is an important consideration in that it has already been noted that the profile of calcium-dependent JNK activation may not coincide with that of IL-2 or IL-3 mRNA stabilization in T cells or mast cells. Thus, although inhibition of the MEKK1–JNK pathway blocks the stabilization of IL-2 or IL-3 mRNAs during calcium release, it is likely that factors other than JNK are at play.
HeLa cells were transfected with pLSGC or pLSAT plasmids, treated with 10 μM A23187 for the times indicated, and steady-state S-antigen mRNA levels examined by Northern analysis (Fig. 5A). Maximum inhibition of ARE mRNA decay was achieved within 30 min of treatment with A23187, with no change during 3 more hours of treatment. Finer kinetic analysis revealed that treatment of cells with A23187 for 15 min was not sufficient, and only slightly stabilized the reporter ARE mRNA (unpublished results). To determine the duration of stabilization of the ARE mRNA, transfected HeLa cells were treated with A23187 for 30 min, medium was changed, and A23187 was chased for the times indicated (Fig. 5B). By 90 min following initial treatment, LSAT mRNA levels were reduced to that of untreated controls. These data therefore indicate that the calcium-induced stabilization of a reporter mRNA containing the GM-CSF ARE is short-lived, with a half-life of about 45 min.
FIG. 5.
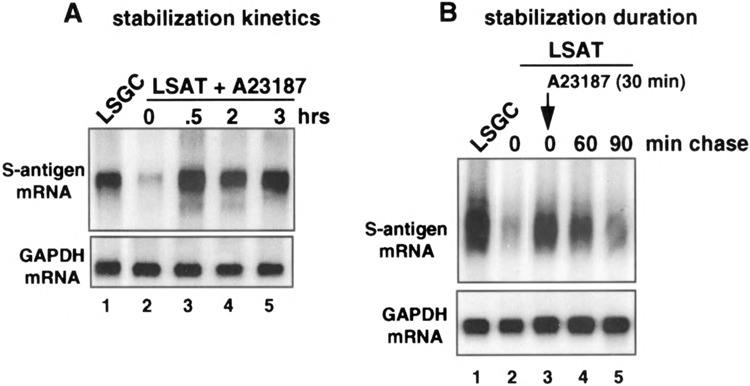
Kinetics and duration of LSAT mRNA stabilization by ionophore treatment of cells. HeLa cells were transfected with pLSGC or pLSAT plasmids. (A) Cells were treated with 10 μM A23187 for the times indicated. (B) Cells were treated with 10 μM A23187 for 30 min followed by a chase of new medium without A23187 for the times indicated. Total cytoplasmic RNA was purified, equal amounts resolved by Northern analysis and hybridized to probes for S-antigen or GAPDH mRNAs.
Calcium-Induced Stabilization of ARE mRNAs Does Not Require New Transcription
Calcium activation of JNK can either act indirectly to stabilize ARE mRNAs by stimulating new transcription, by generating the Jun component of the Fos/Jun transcription factor AP-1, or by directly phosphorylating and altering the activity of proteins involved in ARE mRNA decay. Experiments were therefore carried out to determine whether calcium-induced stabilization of ARE mRNAs involves new transcription. Previous studies demonstrating calcium-induced stabilization of IL-2 and IL-3 mRNAs did not directly address this issue (11,29). HeLa cells were transfected with pLSGC or pLSAT plasmids, then transcription was blocked by addition of actino-mycin D for 15 min prior to addition of A23187 to release calcium. If new transcription is required to stabilize the ARE reporter mRNA, then addition of actinomycin D should prevent stabilization. However, if no new transcription is required for stabilization, then the progressive depletion of the ARE mRNA should be halted upon addition of the calcium ionophore, resulting in maintenance of the lower level of ARE mRNA. Northern mRNA analysis of total cytoplasmic mRNA (Fig. 6) showed that there was an expected 50% decrease in LSAT mRNA level following 15-min actinomycin D treatment (compare lanes 2 and 3), which was stabilized at this level following treatment with A23187. These results indicate that calcium stabilization of the reporter ARE mRNA does not require new transcription.
FIG. 6.
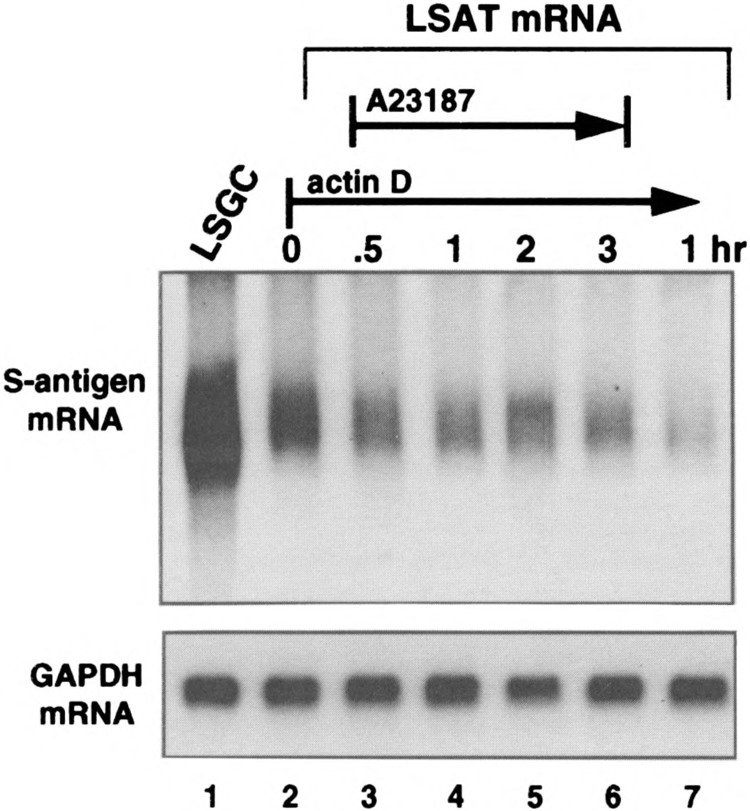
Stabilization of LSAT mRNA by A23187 does not involve new transcription. HeLa cells were transfected with pLSGC or pLSAT plasmids, treated with 5 μM actinomycin D for 30 min to block new transcription, then treated with 10 μM A23187 for the times indicated (lanes 1–6). In lane 7, transcription was blocked for 1 h with actinomycin D without A23187 treatment. Total cytoplasmic RNA was purified, equal amounts resolved by Northern analysis and hybridized to probes for S-antigen or GAPDH.
DISCUSSION
In this study we have used model reporter mRNAs containing the GM-CSF ARE or a control sequence to characterize the effects of calcium release on shortlived ARE mRNA stability. As observed by others for IL-2, IL-3, and GM-CSF mRNAs in immune cells (11,20,29,40), the chimeric ARE reporter mRNA used in our study was fully stabilized against rapid decay by calcium ionophore treatment of different cell lines (Fig. 2). Increased abundance of the reporter ARE mRNA was shown to be a result of increased cytoplasmic stability (Fig. 3), and not due to changes in mRNA translation rates (Fig. 4), which can influence mRNA stability. Nuclear transcription rates were also not detectably altered by calcium release (Table 1). Thus, calcium-induced stabilization of normally short-lived ARE mRNAs is a widespread default response common to most cells and ARE mRNAs, rather than a specialized function of certain cytokine mRNAs in immune cells. Moreover, the minimal cis-acting element required to confer calcium-induced stabilization is the ARE. In two previous reports, one study found that the ARE was sufficient to confer mRNA stabilization in response to calcium release (29), whereas another found that additional 5′NCR sequences were also required (11). It is possible that in the latter study that calcium-induced specific translational inhibition of the IL-2 mRNA, which might explain the requirement fort 5′ sequences in stabilization. The results of our study are in agreement with those of Ming et al. (29), and argue for a single cis-acting mechanism.
The mechanism of calcium stabilization of ARE mRNAs is not known. Although inhibition of the MEKK1–JNK signaling pathway blocks calcium-induced stabilization of ARE mRNAs, the kinetics of JNK activation are not always consistent with those of mRNA stabilization (29). In our study, it was clear that ARE mRNA stabilization required 30 min of continuous calcium ionophore treatment (Fig. 5), whereas JNK activation typically occurs within minutes of treatment (41). In addition, it has been reported that calcium release mediated by ionophore A23187, or other agents, stimulates JNK in thymocytes but not in HeLa cells and many epithelial and fibroblast cell lines (25,37,41). Therefore, whereas JNK activation appears to play a role in ARE mRNA stabilization in certain cells, it is likely that there are distinct but redundant calcium-dependent pathways to mRNA stabilization independent of JNK.
Of the two families of ARE binding proteins involved in regulating mRNA turnover, there is evidence for phosphorylation-dependent decay involving the AUF1 proteins. Studies have shown that the AUF1 isoforms are phosphorylated, and have linked stronger ARE binding to AUF1 phosphorylation, as well as to accelerated turnover of ARE mRNAs (39). It is not known whether phosphorylation of HuR, the other ARE binding protein associated with ARE mRNA stabilization when overexpressed, plays a role in its activity. It is clear, however, that studies now need to direct attention to identification of trans-acting protein factors involved in mRNA decay, and their regulation by phosphorylation.
ACKNOWLEDGMENTS
This work was supported by NIH grant CA-42357 to R.J.S., and used the Kaplan Cancer Center core facilities. A.M.C. was supported by NIH award 5T32-AI-107180 from NIAID. We thank G. Laroia and B. Sarkar for critical reading of the manuscript.
REFERENCES
- 1. Aharon T.; Schneider R. J. Selective destabilization of short-lived mRNAs with the granulocyte-macro-phage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol. Cell. Biol. 13:1971–1980; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antic D.; Keene J. D. Messenger ribonucleoprotein complexes containing human ELAV proteins: Interactions with cytoskeleton and translational apparatus. J. Cell. Sci. 111:183–197; 1998. [DOI] [PubMed] [Google Scholar]
- 3. Banholzer R.; Nair A. P. K.; Hirsch H. H.; Ming X-F.; Moroni C. Rapamycin destabilizes interleukin-3 mRNA in autocrine tumor cells by a mechanism requiring an intact 3′ untranslated region. Mol. Cell. Biol. 17:3254–3260; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bickel M.; Iwai Y.; Pluznik D. H.; Cohen R. B. Binding of sequence-specific proteins to the adenosine-plus uridine-rich sequences of the murine granulocyte/ macrophage colony stimulating factor mRNA. Proc. Natl. Acad. Sci. USA 89:10001–10005; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brewer G. An A+U rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 11:2460–2466; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buzby J. S.; Lee S. M.; Van Winkle P. V.; DeMaria C. T.; Brewer G.; Cairo M. S. Increased granulocyte-macrophage colony stimulating factor mRNA instability in cord versus adult mononuclear cells is translation-dependent and associated with increased levels of A+U rich element binding factor. Blood 88:2889–2897; 1996. [PubMed] [Google Scholar]
- 7. Caput D.; Beuder B.; Hartog K.; Thayer R.; Brown-Shimer S.; Cerami A. Identification of a common nu-cleotide sequence in the 3′untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Nad. Acad. Sci. USA 83:1670–1674; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C-Y.; Shyu A-B. Selective degradation of early response-gene mRNAs: Functional analyses of sequence features of the AU-rich elements. Mol. Cell. Biol. 14:8471–8482; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen C-Y. A.; Shyu A-B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465–170; 1995. [DOI] [PubMed] [Google Scholar]
- 10. Chen C. Y.; Chen T. M.; Shyu A. B. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabi-lizing function. Mol. Cell. Biol. 14:416-426; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen C. Y.; Del Gatto-Konczak F.; Wu Z.; Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science 280:1945–1949; 1998. [DOI] [PubMed] [Google Scholar]
- 12. Curatola A. M.; Nadal M.; Schneider R. J. Selective mRNA degradation controlled by the GM-CSF ARE is activated by ribosome transit into the 3′ noncoding region. Mol. Cell. Biol. 15:6331–6340; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeMaria C. T.; Brewer G. AUF1 binding affinity to A+U rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 271:12179–12184; 1996. [DOI] [PubMed] [Google Scholar]
- 14. Fan X. C.; Steitz J. A. Overexpression of HuR, a nu-clear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448–3460; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feinberg A. P.; Vogelstein B. A technique for radio-labeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6–13; 1983. [DOI] [PubMed] [Google Scholar]
- 16. Gillis P.; Malter J. S. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine and oncogene mRNAs. J. Biol. Chem. 266:3172–3177; 1991. [PubMed] [Google Scholar]
- 17. Greenberg M. E.; Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 311:433–437; 1984. [DOI] [PubMed] [Google Scholar]
- 18. Hahn S.; Wodnar-Filipowicz A.; Nair A. P. K.; Moroni C. Ras oncogenes amplify lymphokine (interleu-kin 3, granulocyte-macrophage colony-stimulating factor) induction by calcium ionophore. Oncogene 6:2327–2332; 1991. [PubMed] [Google Scholar]
- 19. Hirsch H. H.; Backenstoss V.; Moroni C. Impaired interleukin-3 mRNA decay in autocrine mast cell tumors after transient calcium ionophore stimulation. Growth Factors 13:99–110; 1996. [DOI] [PubMed] [Google Scholar]
- 20. Iwai Y.; Akahane K.; Pluznik D. H.; Cohen R. B. Ca2+ ionophore A23187-dependent stabilization of granulocyte-macrophage colony-stimulating factor messenger RNA in murine thymoma EL-4 cells is mediated tyhrough two distinct regions in the 3′-untrans-lated region. J. Immunol. 150:4386–4394; 1993. [PubMed] [Google Scholar]
- 21. Jacobson A.; Peltz S. W. Interrelationships of the pathways of mRNA decay and translation in eukaryo-tic cells. Annu. Rev. Biochem. 65:693–739; 1996. [DOI] [PubMed] [Google Scholar]
- 22. Jain R. G.; Andrews L. G.; McGowan K. M.; Pekala P. H. Keene J. D. Ectopic expressson of Hel-Nll an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol. Cell. Biol. 17:954–962; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kajita Y.; Nakayama J-I.; Aizawa M.; Ishikawa F. The UUAG-specific RNA binding protein, heterolo-gous nuclear ribonucleoprotein DO. J. Biol. Chem. 270:22167–22175; 1995. [DOI] [PubMed] [Google Scholar]
- 24. Lagnado C. A.; Brown C. Y.; Goodall G. J. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: The functional sequence within AU-rich elements may be UUAUUUA(U/A)U/ A). Mol. Cell. Biol. 14:7984–7995; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X.; Yu H.; Graves L. M.;Earp H. S. Protein ki-nase C and protein kinase A inhibit calcium-dependent but not stress-dependent c-Jun N-terminal kinase activation in rat liver epithelial cells. J. Biol. Chem. 272:14996–5002; 1997. [DOI] [PubMed] [Google Scholar]
- 26. Lindstein T.; June C. H.; Lednetter J. A.; Stella G.; Thompson C. B. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244:339–343; 1989. [DOI] [PubMed] [Google Scholar]
- 27. Ma W-J.; Cheng S.; Campbell C.; Wright A.; Fur-neaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144–8151; 1996. [DOI] [PubMed] [Google Scholar]
- 28. Maker J. Identification of an AUUUA-specific messenger RNA binding protein. Science 246:664–666; 1989. [DOI] [PubMed] [Google Scholar]
- 29. Ming X. F.; Kaiser M.; Moroni C. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 17:6039–6048; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Myer V. E.; Fan X. C.; Steitz J. A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 16:2130–2139; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Myer V. E.; Lee S. I.; Steitz J. A. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc. Natl. Acad. Sci. USA 89:1296–1300; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palfrey H. C.; Na C. (1995) Calcium-dependent regulation of protein synthesis. Adv. Second Messenger Phosphoprotein Res. 30:191–223; 1995. [DOI] [PubMed] [Google Scholar]
- 33. Pende A.; Tremmel K. D.; DeMaria C. T.; Blaxall B. C.; Minobe W. A.; Sherman J. A.; Bisognano J. D.; Bristow M. R.; Brewer G.; Port J. D. Regulation of the mRNA-binding protein AUF1 by activation of the p-adrenergic receptor signal transduction pathway. J. Biol. Chem. 271:8493–8501; 1996. [DOI] [PubMed] [Google Scholar]
- 34. Raymond V.; Atwater J. A.; Verma I. M. Removal of an mRNA destabilizing element correlates with the increased oncogenicity of proto-oncogene fos. Onco-gene Res. 4:861–865; 1989. [PubMed] [Google Scholar]
- 35. Savant-Bhonsale S.; Cleveland D. W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 6:1927–1939; 1992. [DOI] [PubMed] [Google Scholar]
- 36. Schuler G. D.; Cole M. D. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell 55:1115–1122; 1988. [DOI] [PubMed] [Google Scholar]
- 37. Scimeca J. C.; Servant M. J.; Dyer J. O.; Meloche S. Essential role of calcium in the regulation of MAP kinase phosphatase-1 expression. Oncogene 15:717–725; 1997. [DOI] [PubMed] [Google Scholar]
- 38. Shaw G.; Kamen R. A conserved AU sequence from the 3’untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659–667; 1986. [DOI] [PubMed] [Google Scholar]
- 39. Sirenko O. I.; Lofquist A. K.; DeMaria C. T.; Morris J. S.; Brewer G.; Haskill S. Adhesion-dependent regulation of an A+U rich element-binding activity associated with AUF1. Mol. Cell. Biol. 17:3898–3906; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stoeklin G.; Hahn S.; Moroni C. Functional hierarchy of AUUUA motifs in mediating rapid interleukin-3 mRNA decay. J. Biol. Chem. 269:28591–28597; 1994. [PubMed] [Google Scholar]
- 41. Su B.; Jacinto E.; Hibi M.; Kallunki T.; Karin M.; Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell 77:727–736; 1994. [DOI] [PubMed] [Google Scholar]
- 42. Vakalopoulou E.; Schaack J.; Shenk T. A 32 kilodal-ton protein binds to AU rich domains in the 3’untranslated regions of rapidly degraded mRNAs. Mol. Cell. Biol. 11:3355–3364; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Velcich A.; Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 311:433–438; 1984. [DOI] [PubMed] [Google Scholar]
- 44. Wagner B.; DeMaria C. T.; Sun Y.; Wilson G. M.; Brewer G. Structure and genomic organization of the human AUF1 gene: Alternative pre-mRNA splicing generates four isoforms. Genomics 48:195–202; 1998. [DOI] [PubMed] [Google Scholar]
- 45. Wodner-Filipowicz A.; Moroni C. Regulation of in-terleukin 3 mRNA expression in mast cells occurs at the posttranscriptional level and is mediated by calcium ions. Proc. Natl. Acad. Sci. USA 87:777–781; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zubiaga A. M.; Belasco J. G.; Greenberg M. E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 15:2219–2230; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


