Abstract
UDP-glycosyltransferases (UGT), catalysing conjugation of UDP-activated sugar donors to small lipophilic chemicals, are widespread in living organisms from bacteria to fungi, plant, or animals. The progress of genome sequencing has enabled an assessment of the UGT multigene family in Haemonchus contortus (family Trichostrongylidae, Nematoda), a hematophagous gastrointestinal parasite of small ruminants. Here we report 32 putative UGT genes divided into 15 UGT families. Phylogenetic analysis in comparison with UGTs from Caenorhabditis elegans, a free-living model nematode, revealed several single member homologues, a lack of the dramatic gene expansion seen in C. elegans, but also several families (UGT365, UGT366, UGT368) expanded in H. contortus only. The assessment of constitutive UGT mRNA expression in H. contortus adults identified significant differences between females and males. In addition, we compared the expression of selected UGTs in the drug-sensitive ISE strain to two benzimidazole-resistant strains, IRE and WR, with different genetic backgrounds. Constitutive expression of UGT368B2 was significantly higher in both resistant strains than in the sensitive strain. As resistant strains were able to deactivate benzimidazole anthelmintics via glycosylation more effectively then the sensitive strain, UGT368B2 enhanced constitutive expression might contribute to drug resistance in H. contortus.
Keywords: UDP-glycosyltransferase, Resistance, Haemonchus contortus, Detoxification
Graphical abstract
Highlights
-
•
UDP-glycosyltransferase family contains 32 members in Haemonchus contortus.
-
•
Constitutive expression of UDP-glycosyltransferases differs between sexes.
-
•
UGT368B2 expression level is significantly higher in resistant strains.
1. Introduction
Haemonchus contortus, a gastrointestinal parasite of small ruminants, has very high propensity to develop resistance to anthelmintic drugs, which causes serious economic losses worldwide (Gilleard and Redman, 2016). The frequent and often indiscriminate administration of anthelmintics in routine farming practice has led to widespread resistance to all available anthelmintics, including the most recently launched drug, monepantel, less than five years after its introduction to the market (Mederos et al., 2014).
The mechanisms of resistance to various anthelmintics have been extensively studied. The principal mechanism of resistance to benzimidazole anthelmintics is a mutation in the target site of isotype 1 β-tubulin in three codons (F167Y, E198A, F200Y) which disables binding of the benzimidazoles to their target molecule (Kotze and Prichard, 2016). However, mutations in the isotype 1 β-tubulin gene locus alone do not fully explain differences in the phenotypic resistance level (Yilmaz et al., 2017) suggesting additional mechanisms are involved. Therefore, non-target mechanisms of resistance have also been studied. Several studies have shown that nematode ATP-binding cassette (ABC) transport proteins may play a role in drug resistance (reviewed in Matouskova et al., 2016). For example, different allele frequencies of P-glycoprotein (Pgp) genes in a cambendazole-selected H. contortus strain (Blackhall et al., 2008) and increased expression levels of several Pgps in a triple-resistant isolate compared to a susceptible isolate were reported (Williamson et al., 2011).
Similarly, an association between drug resistance and expression/activities of drug biotransformation enzymes has been confirmed in various species including helminths (Matouskova et al., 2016). Increased activity and/or expression of cytochrome P450s (CYPs), the main enzymes of phase I drug biotransformation, have been reported to contribute to insecticide resistance (Denecke et al., 2017), herbicide resistance in weeds (Ghanizadeh and Harrington, 2017) and drug resistance in cancer (Azzariti et al., 2011; Rochat, 2005). In the nematode Caenorhabditis elegans, upregulation of CYP35D has been associated with thiabendazole resistance (Jones et al., 2015). Recently, Yilmaz et al. reported increased constitutive expression of one member of the CYP34/35 family in a highly resistant H. contortus isolate in comparison to susceptible or moderately resistant isolates (Yilmaz et al., 2017).
Enzymes involved in phase II drug biotransformation have also been associated with drug resistance in helminths, particularly the UDP-glycosyltransferases (UGTs) (Laing et al., 2013; Vokral et al., 2012, 2013). UGTs, which catalyse conjugation of UDP-activated sugar donors to lipophilic chemicals, are ubiquitous. In plants, secondary metabolites are modified by the binding of various sugars via the action of soluble cytosolic UGTs (Tiwari et al., 2016), while in vertebrates, a substantial number of xenobiotics are conjugated with UDP-glucuronic acid by membrane bound UGTs (Bock, 2003). In insects, UGT enzymes typically use UDP-glucose and have multiple roles including insecticide detoxification (Ahn et al., 2012). In Plutella xylostella, the diamondback moth, UGT2B17 is involved in chlorantraniliprole resistance (Li et al., 2017).
In helminths, detoxification of benzimidazole anthelmintics via glycosylation has been observed (Laing et al., 2010; Vokral et al., 2012, 2013). In addition, anthelmintic glycosidation was more potent in resistant H. contortus strains than in a susceptible one, suggesting involvement of UGTs in resistance to benzimidazole anthelmintics in this nematode (Stuchlíková et al., 2018; Vokral et al., 2012, 2013). Recently, involvement of C. elegans ugt-22 (UGT16C1) in albendazole efficacy was reported (Fontaine and Choe, 2018). Furthermore, UGT enzyme inhibitors increased the activity of naphtalophos, suggesting their direct involvement in naphtalophos biotransformation (Kotze et al., 2014).
Since H. contortus UGTs have not been characterised to date, the present study was designed in order to investigate this family of enzymes. We identified all putative UGT genes in the H. contortus genome and undertook phylogenetic analysis and comparison to the C. elegans UGT family. We have quantitatively compared constitutive transcription levels of UGTs in adults between sexes. Furthermore, the constitutive level of expression of selected UGTs was compared between three isolates with different levels of resistance; the susceptible (ISE), the benzimidazole resistant (IRE) and the multi-drug resistant isolate (WR).
2. Materials and methods
2.1. UGT gene sequences
Putative H. contortus UGT gene sequences (Hc_UGTs) were downloaded from a public database (NCBI GenBank), from the H. contortus draft genome assembly (PRJEB506). Later these sequences were mapped onto a chromosomal assembly of the H. contortus genome made publicly available by the Wellcome Trust Sanger Institute (ftp://ngs.sanger.ac.uk/production/pathogens/Haemonchus_contortus). The Hc_UGTs were sent for naming according to current nomenclature guidelines to the UGT Nomenclature Committee (Mackenzie et al., 2005) and are recorded on their website (http://prime.vetmed.wsu.edu/resources/udp-glucuronsyltransferase-homepage).
The overall protein homology of the Hc_UGT sequences was analysed alongside human UGT2B7 (NP_001065). The deduced amino acid sequences were aligned by Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (Sievers et al., 2011). The output was transferred to BoxShade version 3.21 (https://embnet.vital-it.ch/software/BOX_form.html). Signal peptides and transmembrane domains were predicted by SignalP 4.1 (Petersen et al., 2011) and TMHMM 2.0 on the CBS Prediction Servers (http://www.cbs.dtu.dk/services).
C. elegans UGTs were obtained from the list of “All UGT Gene Names Currently Approved by the Committee” also recorded on the UGT Nomenclature Committee´s website and sequences were collected from the public database (NCBI GenBank, https://www.ncbi.nlm.nih.gov/).
2.2. Phylogenetic analysis
Predicted amino acid sequences of 32 H. contortus UGTs and 60 C. elegans UGTs were aligned by Muscle and the evolutionary analyses were conducted in MEGA X (Kumar et al., 2018). The evolutionary history was inferred by using the Maximum Likelihood method based on the Le Gascuel 2008 model (Le and Gascuel, 2008). The bootstrap consensus tree inferred from 500 replicates was taken to represent the evolutionary history of the taxa analysed (Felsenstein, 1985). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbour-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. Based on model selection tool a recommended “LG + G + I + F” model was used. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 1.3289)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 0.35% sites). The analysis involved 92 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 432 positions in the final dataset.
2.3. Parasites
Three isolates of H. contortus were used in the present study:
-
(i)
ISE: inbred susceptible-Edinburgh strain (MHco3)
-
(ii)
IRE: inbred resistant-Edinburgh strain (MHco5)
-
(iii)
WR: White River multidrug resistant strain (MHco4)
2.4. Collection of biological material
Nine parasite-free lambs (3–4 months old) were orally infected with 5000 third stage larvae (L3) of H. contortus. Seven weeks after infection the animals were stunned and immediately exsanguinated. The lambs were bred and used in agreement with Czech slaughtering rules for farm animals and according to the protocols, which were evaluated and approved by the Ethics Committee of the Ministry of Education, Youth and Sports (Protocol MSMT-25908/2014-9). Adult nematodes were removed post mortem from sheep abomasum using the agar method (Vanwyk et al., 1980). Freshly isolated living H. contortus adults were washed three times with phosphate-buffered saline (pH 7.4). Females and males were separated manually under a microscope. Three biological replicates of adult males and females separately (ten nematodes per sample) were placed directly to TriReagent® (Molecular Research Centre, OH, USA) for immediate RNA isolation or stored in −80 °C for later use.
2.5. RNA-seq
Our preliminary transcriptome sequencing of the ISE strain was carried out using both female and male samples (one sample per sex only). Standard Illumina TruSeq RNA libraries were prepared and transcriptome sequencing was carried out by GATC Biotech on an Illumina HiSeq2000 Genome Analyzer platform, using single read (1 × 100 bp) technology, yielding approximately 25 million reads for each of the female and male samples. Quality control measures and filtering of all RNA-Seq samples were carried out using CLC Genomics Workbench v8.1 (http://www.clcbio.com) as previously described (Vogel et al., 2014). Digital gene expression analysis was carried out with CLC Genomics Workbench v8.1 to generate BAM mapping files, using the predicted coding sequence dataset of the H. contortus BioProject PRJNA205202 from the WormBase ParaSite website as a reference, and by counting the sequences to estimate expression levels, using previously described parameters for read mapping and normalization (Jacobs et al., 2016). An annotation file was generated using BLAST, Gene Ontology and InterProScan searches implemented in BLAST2GO PRO v2.6.1 (www.blast2go.de). The data are presented as normalized log2-transformed RPKM values (normalizing across the samples, using as calculation the total number of mapped reads and the length of the reference sequence).
2.6. Sample preparation and qPCR screen
RNA was extracted using TriReagent® according to the manufacturer's instructions. Homogenization of the samples (ten adult worms in each sample) was performed using the FastPrep-24 5G Homogenizer (MP Biomedicals, France). RNA yields and purity were determined by measuring the absorbance at 260 and 280 nm using the NanoDrop ND-1000 UV–Vis Spectrophotometer (Thermo Fisher Scientific, MA, USA). Samples were analysed by the Agilent 2100 Bioanalyzer on RNA Nano chips (Agilent Technologies, CA, USA). Only samples with a 260/280 ratio greater than 1.8 and a RIN greater than 6.5 were used for subsequent analysis. Five μg of RNA was treated with DNase I (NEB, UK) to remove genomic DNA contamination at 37 °C for 20 min, inactivated by heat (at 75 °C for 10 min) and diluted to a concentration of 0.2 μg/μl. RNA was stored at −80 °C until further processing. First-strand cDNA was synthesized from 1 μg of total RNA in 20 μl reactions using random hexamer primers and Protoscript® II Reverse Transcriptase (NEB, UK) following the manufacturer's protocol. cDNA was diluted 5× and stored at −20 °C until use in the qPCR assay. Three biological replicates of each sample were analysed.
Primers were designed using Primer3 software (Untergasser et al., 2012) for each gene avoiding the region of hairpin structure as identified by Mfold at 60 °C (Zuker, 2003). All primers were synthesized by Generi Biotech, Czech Republic. The specificity and efficiency of the primers were checked for all qPCR conditions. The primer sequences, amplicon sizes, and efficiencies are listed in Table S1.
The qPCR analyses were performed in QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, CA, USA) using SYBR Green I detection in a final volume of 20 μl. The reaction mixture consisted of qPCR Xceed SG 1-step 2× Mix Lo-ROX (IAB, Czech Republic), both forward and reverse primers (final concentration 100 nM), and 5 μl of diluted cDNA. The PCR reactions were initiated with a denaturation step at 94 °C for 2 min, followed by 40 cycles of two step amplification as follows: denaturation at 95 °C for 10 s, annealing at 60 °C for 40 s. Fluorescence data were acquired during the last step. A dissociation protocol with a gradient (0.5 °C every 30 s) from 65 °C to 95 °C was used to investigate the specificity of the qPCR reaction and presence of primer dimers. Gene-specific amplification was confirmed by a single peak in the melting curve analysis. All samples were measured in duplicates with less than 0.5 Cq difference. Relative expression was calculated using ΔΔCt method (Livak and Schmittgen, 2001), using Hc-ama and Hc-gpd as reference genes as recommended by Lecová et al. (Lecova et al., 2015).
2.7. Statistical analysis
The reported data are expressed as the mean ± S.D. (at least 3 biological replicates of each sample). Statistical comparisons were carried out using the one-way ANOVA with Tukey's post hoc test (GraphPad Prism 7.0). The differences were considered significant at P < 0.05.
3. Results and discussion
Our present project focused on the potential role of UGT enzymes in the glycosylation of anthelmintics, enabling their easier excretion and possibly increasing the resistance of H. contortus. Initially thirty seven putative UGT sequences were identified on 25 scaffolds in the H. contortus genome assembly (Haemonchus_contortus_MHco3-2.0; Bioproject ID: PRJEB506) accessed through WormBase ParaSite website (http://parasite.wormbase.org/index.html). This analysis was repeated with a newer genome version assembled into six chromosomes and a total of 32 UGT sequences were obtained. Five of the initial sequences appeared to be either different alleles or artefacts of the draft assembly. The UGTs were systematically named based on a tree with amino acid sequences. The naming system includes the root symbol UGT (UDP-glycosyltranferase), an Arabic number designating a family and a letter representing family and an Arabic numeral representing individual gene (Table 1). The nomenclature committee recognized 15 UGT families in H. contortus based on the conceptual translations of UGT transcripts.
Table 1.
Summary of Haemonchus contortus UGT sequences.
| Name | gene model numbera | gene model numberb | chromozome location | STRAND | note |
|---|---|---|---|---|---|
| UGT10B1 | Hcoi01037200 | g1131 | ch4 | F | |
| UGT21C1 | Hcoi01817600 | g3162 | ch4 | F | no TM |
| UGT22A2 | Hcoi00058000 | g5477 | ch5 | F | |
| UGT24B1 | Hcoi00244800 | g4156 | ch4 | F | f |
| UGT24C1 | Hcoi00868700 | g3647 | ch4 | F | |
| UGT24D1 | Hcoi01592800 | g4156 | ch4 | F | f, no TM |
| UGT24D2 | Hcoi00244900 | g4328 | ch4 | R | no TM, no Sp |
| UGT26A2 | Hcoi01238200 | g22728 | ch3 | R | |
| UGT27B1 | Hcoi01933800 | g5051 | ch5 | R | |
| UGT365A1 | Hcoi01462600 | g4004 | ch4 | F | |
| UGT365B1 | Hcoi01462400 | g3995 | ch4 | F | |
| UGT365B2 | Hcoi01461700 | n.d. | ch4 | R | |
| UGT365B3 | Hcoi01461900 | g3996 | ch4 | R | |
| UGT365B4 | Hcoi01462000 | g3997 | ch4 | R | |
| UGT365B5 | Hcoi01462200 | g3999 | ch4 | R | |
| UGT365B6 | Hcoi01462100 | g3998 | ch4 | R | no Sp |
| UGT366A1 | Hcoi02015300 | g3385 | ch4 | F | no TM |
| UGT366B1/B2 | Hcoi00320100 | g3390 | ch4 | F | f, (B2 no Sp) |
| UGT366C1 | Hcoi01255100 | g3398 | ch4 | F | |
| UGT366D1 | Hcoi01255000 | g3397 | ch4 | F | |
| UGT367A1 | Hcoi00538800 | g23518 | chX (6) | R | |
| UGT368A1 | Hcoi01078900 | g10480 | ch2 | F | |
| UGT368A2 | Hcoi01452100 | g10479 | ch2 | F | no TM |
| UGT368B1 | Hcoi00785300 | g10478 | ch2 | F | |
| UGT368B2 | Hcoi00785400 | g10477 | ch2 | F | |
| UGT369A1 | Hcoi00240800 | g4727 | ch4 | R | |
| UGT370A1 | Hcoi01632300 | n.d. | ch5 | F | no TM |
| UGT370B1 | Hcoi01632400 | g5172 | ch5 | F | no TM |
| UGT371A1 | Hcoi01917400 | g1755 | ch4 | F | no TM |
| UGT372A1 | Hcoi01985800 | g9057 | ch5 | F | |
| UGT373A1 | Hcoi01651100 | g10412 | ch2 | R |
no TM -translated sequence lack predicted transmembrane helix on the C-terminus.
no Sp -translated sequence lack predicted signal peptide.
f -potential fusion transcript.
n.d.-not detected in assosiated transcriptom, but present in the genome V3.
-Gene model numbers corresponding to Haemonchus contortus genome assembly PRJEB506.
-Gene model numbers corresponding to Haemonchus contortus genome V3.
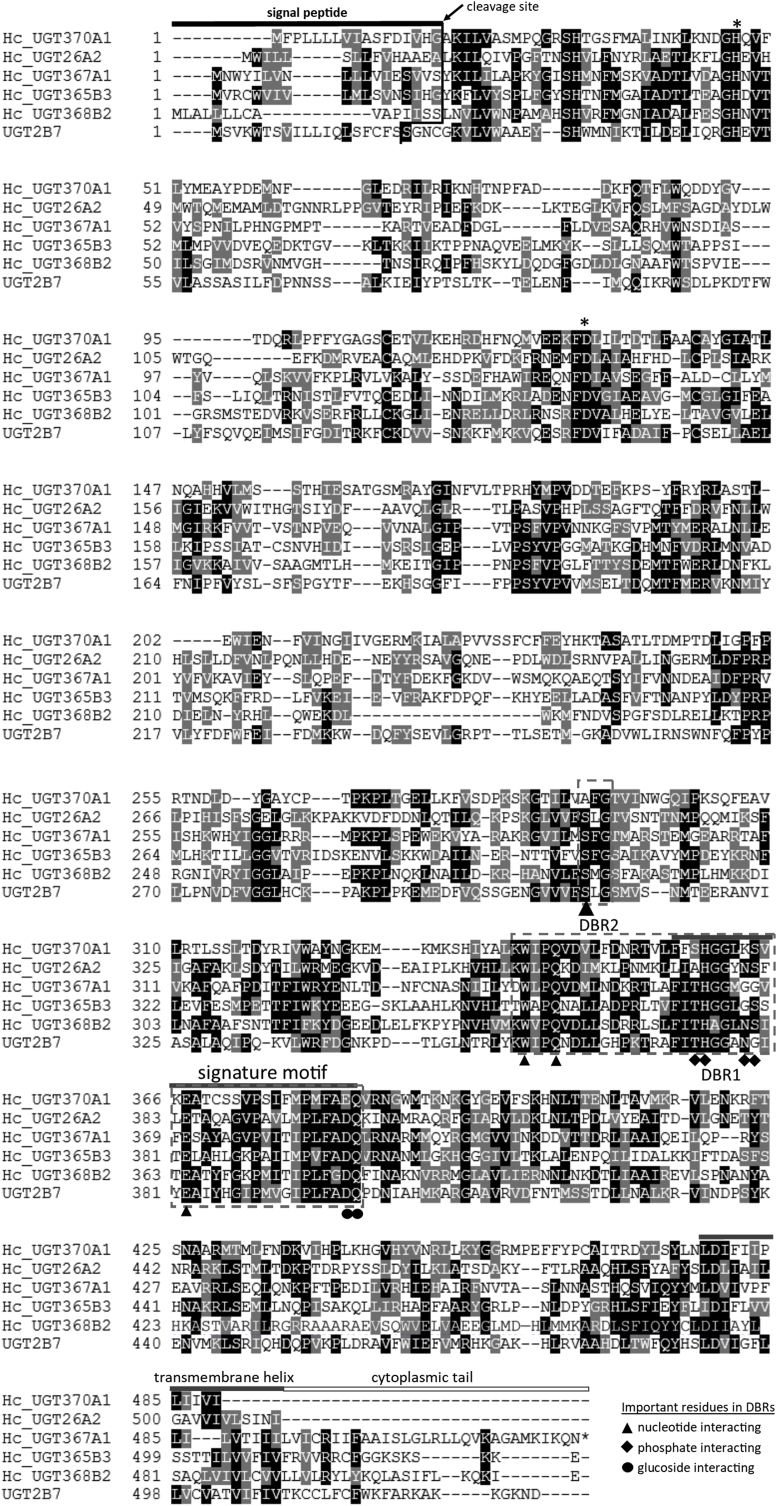
The UGT superfamily is defined by a common protein structure and signature sequence of 44 amino acids responsible for binding the UDP moiety of the sugar donor (Bock, 2016; Mackenzie et al., 2005), (Meech et al., 2012). All the identified Hc_UGTs comply with this condition (see Fig. 1 and Fig 1S). Since the mammalian and insect UGTs contain a transmembrane helix which binds the UGT to the endoplasmic reticulum (ER) we have analysed the Hc_UGT sequences for the presence of ER signal peptides and transmembrane helixes. However, a few sequences lack both predicted characteristics (Table 1). In humans, such shorter UGT1 proteins, lacking the transmembrane segment, are functionally inactive, but can heterodimerize with full-length UGT forms and inhibit their activity (Bellemare et al., 2010). At present, it is unknown if in H. contortus these short UGT sequences are correct, whether this is an artefact of transcript identification, or an indication for the presence of UGT pseudogenes. It is also unclear if such truncated UGTs could have any enzyme activity. However, all of these sequences contain the amino acids that are important for catalytic activity, as derived from the only crystal structure of the C-terminal domain of human UGT2B7 available that revealed detailed information on the sugar donor-binding site, implicated in binding UDP-glucuronic acid (Miley et al., 2007). In the free living nematode C. elegans, conjugates of the anthelmintic drug albendazole (ABZ) with glucose were identified (Laing et al., 2010). Similarly, in H. contortus all glycosylated metabolites of anthelmintics that have been identified are conjugates of glucose or another hexose (Stuchlíková et al., 2018). Therefore, we believe that Hc_UGTs use UDP-glucose (hexose) as the major sugar donor rather than UDP-glucuronic acid, hence the binding topology could be slightly different. However, the important residues identified in humans are also conserved in the majority of H. contortus UGTs (Fig. 1, S1). On the other hand, prediction of a putative UGT function based on sequence homology can be unreliable, as closely related sequences may express different catalytic activities (Malik and Black, 2012) and functional analysis of interesting UGTs are warranted.
Fig. 1.
Comparison of selected Hc_UGTs with human UGT2B7. Multiple alignment was performed by using Clustal Omega. Signal peptide in N-terminus predicted by Signal P4.1 is shown as black bar, signature motif and transmembrane domain in C-terminus half in grey and cytoplasmic tail in white above the alignment. Important catalytic residues H and D are indicated by asterisks (*) above the alignment. DBR refers to donor binding region, and several important residues interacting with sugar donor are indicated by symbols under the alignment.
3.1. Constitutive UGT expression differs between sexes
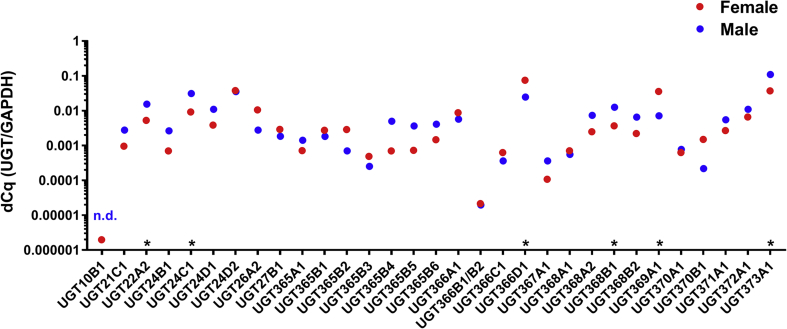
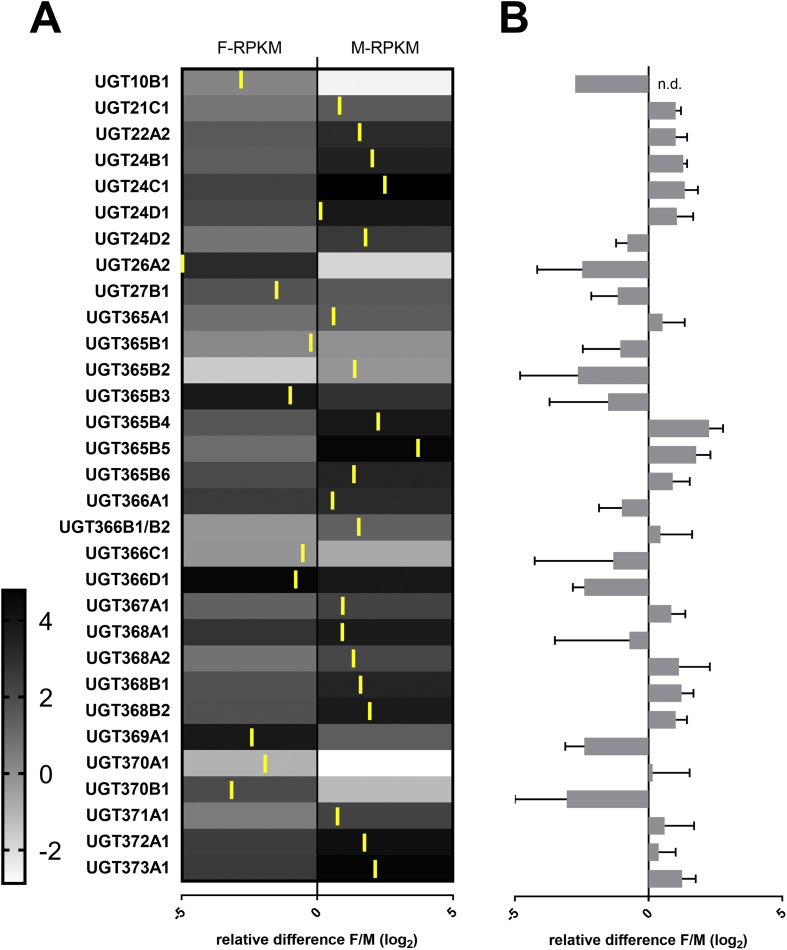
We have designed primers for 32 UGT transcripts, based on the reference gene set and preliminary RNA sequencing of H. contortus adults of the anthelmintic-sensitive isolate. All these gene transcript levels were tested by qPCR and were compared between males and females. Fig. 2 shows the relative quantity (ΔCq, UGTs/Hc_gpd) in males and females. Six genes had significantly different expression between sexes; four genes had higher expression in males (UGT22A2, 24C1, 368B1 amd 373A1) and two in females (UGT366D1, 369A1). Only UGT10B1 was below the detection limit in males. The UGTs with the highest expression in females were UGT369A1 and UGT366D1 and in males, UGT373A1 and UGT22A2. The preliminary RNA-seq data usually corresponded well with the results from the qPCR method (Fig. 3). In five cases (UGT24D2, 365B2, 366A1, 368A1, 370A1) the ratio of expression in males and females differs in RNA-seq and qPCR. This discrepancy can be attributed on one hand to the lack of replicates in the preliminary RNA-seq and big variation in biological replicates in several qPCR on the other hand. In our recent metabolism experiments, females and males were tested separately and showed major sex-differences in the metabolism of all anthelmintics tested (Stuchlíková et al., 2018). Mostly, a higher amount of metabolites were found in females than in males and three detected metabolites of flubendazole (N-glycosides) were female-specific. Only one N-glycoside of albendazole was detected in a higher amount in males than in females. Such discrepancies may be attributed to the different roles of UGTs in females and males apart from the xenobiotic function.
Fig. 2.
Relative abundancies of UGT genes from females and males of sensitive ISE strain. For each UGT the mean of relative expression is displayed as ΔCq, calculated as efficiency (UGT)−Cq(UGT)/efficiency (Hc_gpd)−Cq(Hc_gpd), * indicates significant difference between female and male at P < 0.05, N = 3.
Fig. 3.
Comparison of constitutive expression of UGTs in females and males of adult Haemonchus contortus. Preliminary RNA-seq expression, heat map (A) indicating transcript abundances of UGTs expressed as log2 transformed normalized values (RPKM) across H. contortus female (F-RPKM) or male (M-RPKM) samples (N = 1). Relative expression (Female/Male, log2) of Hc_UGTs mRNA from quantitative real-time PCR assay (B), (N = 3). n.d. not detected. Yellow border lines and the X-axis below the heat map correspond to the ratio of Female/Male log2 relative transcript abundancies for direct comparison with the qPCR results. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Phylogenetic analysis of the Hc_UGT family
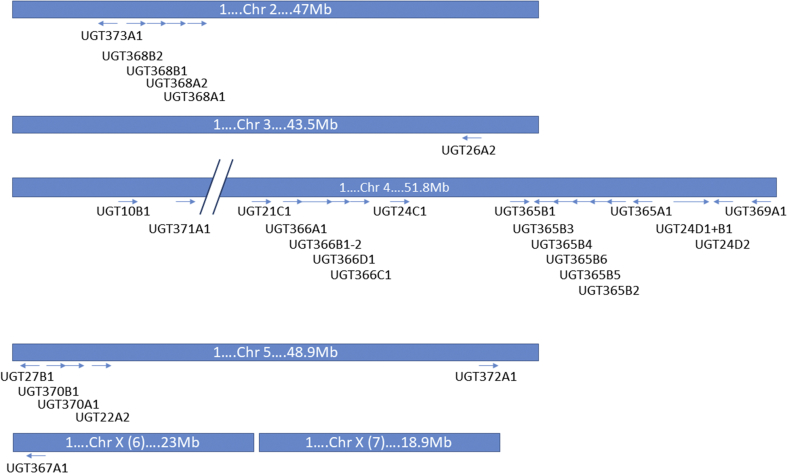
As deduced from the nomenclature, the Hc_UGTs are divided into 15 families with different phylogenetic patterns. The distribution of UGTs across H. contortus chromosomes is highly uneven (Fig. 4). Chromosome 3 and X contain only one UGT sequence each, chromosome 5 and 2 contain five sequences each. Most of the UGT sequences (20) are located on chromosome 4, which was previously shown to have a higher recombination rate compared to other autosomes (Doyle et al., 2018). Some of the identified UGTs belong to single membered families and some represent “blooms” such as UGT365 family. All the genes from this family are located on chromosome 4. Interestingly, only UGT365B1 is encoded on the forward strand with 14 identified exons and the sequence separating the next gene is approximately 20 kb long. All other UGT365B genes are located on the reverse strand next to each other (within a 40 kb region) and UGT365A1 is located one gene downstream, with all genes containing 13 exons. It appears that this gene cluster might have arisen through initial gene duplication (A and B subfamily) followed by expansion of the UGT365Bs into six genes. From the preliminary RNA-seq analysis, only four genes out of this seven-member family are transcribed at moderate levels in adults, and none of them was differentially expressed among tested strains. Gene duplications in Drosophila are enriched for genes implicated in drug metabolism (Cardoso-Moreira et al., 2016). Some duplication hotspots present in the Drosophila genome contain clusters of genes that have been independently duplicated more than once. Similarly in H. contortus, such gene duplications may represent an ecological adaptation. Apart from UGT365 family two other clusters of multi-copy genes were found in the H. contortus genome. The UGT368 family contains four genes (A1, A2 and B1 and B2) that are clustered in close proximity close toward the end of chromosome 2, with increased recombination rate (Doyle et al., 2018), and the UGT366 family with four genes including one potential fusion transcript (A1, B1/2, C1, D1).
Fig. 4.
Chromozome localization of each UGT gene. The arrow direction indicates location on forward (→) or reverse (←) strand.
Two identified transcripts appear to arise from fusion of two UGTs (UGT366B1 and B2 and UGT24A1 and 24B2) in the novel version of the genome. Two UGT domains were identified in the conceptual translation containing all the catalytically important residues. Surprisingly, in the old version another UGT fusion transcript was present. Although RNA-seq data should enable the identification of fusion transcripts (Kumar et al., 2016), our preliminary RNA-seq data did not fully support their presence and it should be verified by another method. At this stage, we can only speculate whether these fusion transcripts represent incorrectly spliced transcripts, pseudogenes, new genes encoding heterodimeric UGTs or just a sequencing error. Nevertheless, this finding is quite intriguing as a possibility for an evolved UGT as accumulating evidence suggest that UGTs operate as oligomeric complexes (Bock and Kohle, 2009; Finel and Kurkela, 2008).
3.3. Comparative analysis of UGT family in H. contortus and C. elegans
Fig. 5 shows a neighbour-joining tree of conceptual translations of the H. contortus UGT genes aligned with all UGTs from C. elegans. The C. elegans genome encodes 60 UGTs divided into 17 families of which seven are single-member genes (Wormbase gene names are added to the official UGT names in Supplemntary Table S2 to link up with other reports involving C. elegans UGTs). Homologues of four single-membered UGTs in C. elegans are present in H. contortus also as single copy genes (UGT10B1, 22A2, 26A2, 27B1). H. contortus lacks genes that are homologous to any of the dramatically expanded families in C. elegans (UGT14, 15, 18). Similarly, in the family of CYPs, the comparison of H. contortus and C. elegans genes revealed a lack of expansion in the parasite genome, possibly explained by lower exposure to environmental toxins compared to the free living nematodes (Laing et al., 2013). However, the exposure of larval stages might contribute to local duplication of some of the UGT families. Three families are expanded in H. contortus; the UGT365 family with seven members, UGT366 with four members and UGT366 with five members. Among the transcription factor SKN-1 responsive genes the C. elegans ugt-22 was recently identified as detoxication gene involved in albendazole efficacy (Fontaine and Choe, 2018). This gene ugt-22 (Ce_UGT16C1), belongs to Ce_UGT16 family, which is closest to the seven member family Hc_UGT365, suggesting a role in benzimidazole detoxification for one of the member in this family. The ugt-22 (Ce_UGT16C1) overexpression increased spontaneous motility on 11 μM albendazole and EC50 was increased by 2.4 fold. A rescue experiment using some of the Hc_UGT in C. elegans ugt-22- mutant would be intriguing.
Fig. 5.
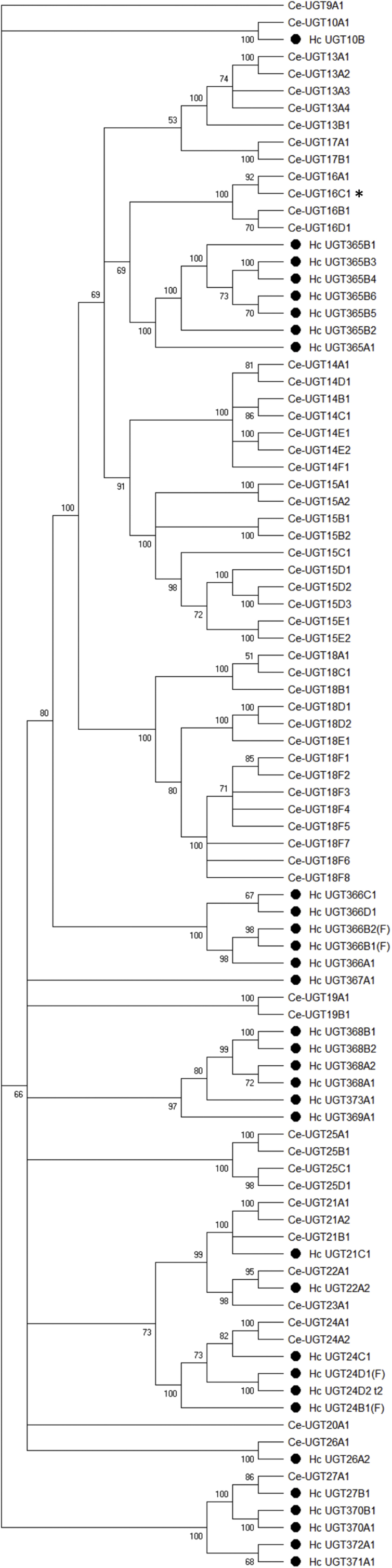
Phylogenetic tree of Haemonchus contortus (marked by black dots) and Caenorhabditis elegans UDP-glycosyltransferases (UGTs). A consensus phylogenetic tree was constructed using the Maximum Likelihood method based on the Le Gascuel 2008 model. The bootstrap consensus tree was calculated (500 replicates). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. UGT from C. elegans with distinct role in resistance is marked by * (Fontaine and Choe, 2018).
3.4. Constitutive UGT expression in different H. contortus strains
One of the major aims of our study was to analyse the possible differences in constitutive expression of UGTs between the drug-susceptible and two drug-resistant strains of H. contortus. Constitutive expression of selected UGTs separately in females and males of H. contortus adults from the ISE, IRE and WR strains was tested. The ISE (MHco3) strain is fully susceptible to anthelmintics and has been adopted as the standard genome strain for the H. contortus sequencing project at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/Projects/H_contortus/). The anthelmintic resistant IRE (MHco5) strain was derived from the ISE strain by selection with thiabendazole (Roos et al., 2004) and the multi-resistant strain WR (MHco4) which has a different origin and is genetically divergent (Gilleard and Redman, 2016). We selected genes for comparison in the three isolates with different levels of resistance based on normalized expression values from the RNA-seq (log2 transformed RPKM >2, reads per kilobase of transcript per million mapped reads).
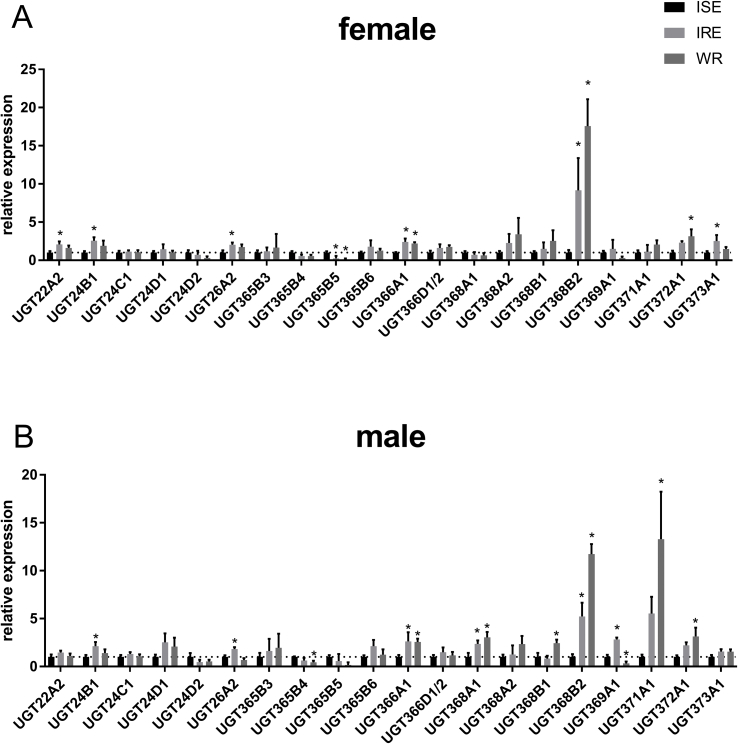
Out of 32 transcripts in total, 19 transcripts in males had RPKM >2, but only ten transcripts in females met the same condition. Three and four genes had significantly lower expression in WR males and females, respectively. Since this isolate has a divergent genetic background those genes may not be present in its genome as representatives of recent duplication (UGT365B4 and B5) or have become pseudogenes (UGT369A1) and are not present or transcribed in this isolate. Several UGT isoforms had higher expression levels in resistant strains, either in females or in males. UGT371A1 transcript levels were significantly higher in both strains in males only. UGT368B2 transcript levels were higher in males and females of both resistant isolates, e.g. they displayed 9.16 and 17.58 times higher expression levels in IRE and WR females in comparison to ISE females and similarly 5.23 and 11.72 times higher expression in IRE and WR males, respectively (Fig. 6). This finding is in agreement with previously mentioned studies of anthelmintic metabolism and of UGT activity toward the model substrate p-nitrophenol, when the amount of glucosides formed ex vivo and in vitro was higher in H. contortus resistant strains than in the sensitive strain (Vokral et al., 2012), (Stuchlíková et al., 2018). An important aspect is that the ISE and IRE isolates included in our study have a similar genetic background as described above (Kwa et al., 1993) (Roos et al., 2004). Comparison of basal transcript levels in these two isolates showed subtle differences in many UGT genes that could be accounted for by within population genetic diversity. However, the significant difference in the constitutive transcription level of UGT368B2 in both resistant strains supports the possibility of an important role for this UGT in resistance in general. However, since the reasons why this transcript is constitutively upregulated are not known, the upregulation can have different origin. Additionally, it remains to be elucidated whether this specific UGT is involved in the biotransformation of benzimidazole anthelmintics and plays a significant role in resistance.
Fig. 6.
Comparison of constitutive expression in different Haemonchus contortus strains analysed by qPCR. Relative expression of selected Hc_UGTs mRNA from females (A) and males (B). * indicates significant difference between ISE and WR or ISE and IRE at P < 0.05, N = 3.
4. Conclusions
Adults of H. contortus are able to form many glycosylated anthelmintic metabolites. We have investigated the UGT family in this parasite genome and identified 32 genes that might be responsible for such glycosylation. Interestingly, comparison with the C. elegans UGT family revealed gene diversification of specific UGT subfamilies in H. contortus. We have found significant sex differences in the constitutive expression levels of several UGTs. Furthermore, comparison of the constitutive expression of UGTs in sensitive and resistant strains revealed that UGT368B2 was highly expressed in both females and males of resistant strains only. Further work should be aimed at testing this UGT in worm isolates to confirm or disprove its broader involvement in the resistance of H. contortus to anthelmintics. The importance of our finding in anthelmintic resistance is supported by the fact that higher expression of UGT368B2 was found in both resistant strains, which have a different genetic background, one from inbred selection and one from field isolation. This means that, like the ß-tubulin point mutation, the change affecting the level of expression must have occurred multiple times. Further studies are warranted and testing of other H. contortus strains and isolates is required.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgement
The work was supported by the Czech Science Foundation [17-11954Y] and by Charles University in Prague [PRIMUS/17/SCI/4, UNCE/18/SCI/012 and SVV 260 416]. Lucie Raisová Stuchlíková was supported by the project EFSA-CDN [CZ.02.1.01/0.0/0.0/16_019/0000841] co-funded by ERDF. Roz Laing is funded by a BBSRC strategic LoLa [BB/M003949/1].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2018.09.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ahn S.-J., Vogel H., Heckel D.G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 2012;42:133–147. doi: 10.1016/j.ibmb.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Azzariti A., Porcelli L., Quatrale A.E., Silvestris N., Paradiso A. The coordinated role of CYP450 enzymes and P-gp in determining cancer resistance to chemotherapy. Curr. Drug Metabol. 2011;12:713–721. doi: 10.2174/138920011798357042. [DOI] [PubMed] [Google Scholar]
- Bellemare J., Rouleau M., Girard H., Harvey M., Guillemette C. Alternatively spliced products of the UGT1A gene interact with the enzymatically active proteins to inhibit glucuronosyltransferase activity in vitro. Drug Metab. Dispos. 2010;38:1785–1789. doi: 10.1124/dmd.110.034835. [DOI] [PubMed] [Google Scholar]
- Blackhall W.J., Prichard R.K., Beech R.N. P-glycoprotein selection in strains of Haemonchus contortus resistant to benzimidazoles. Vet. Parasitol. 2008;152:101–107. doi: 10.1016/j.vetpar.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Bock K.W. Vertebrate UDP-glucuronosyltransferases: functional and evolutionary aspects. Biochem. Pharmacol. 2003;66:691–696. doi: 10.1016/s0006-2952(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Bock K.W., Kohle C. Topological aspects of oligomeric UDP-glucuronosyltransferases in endoplasmic reticulum membranes: advances and open questions. Biochem. Pharmacol. 2009;77:1458–1465. doi: 10.1016/j.bcp.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Bock K.W. The UDP-glycosyltransferase (UGT) superfamily expressed in humans, insects and plants: animal-plant arms-race and co-evolution. Biochem. Pharmacol. 2016;99:11–17. doi: 10.1016/j.bcp.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Cardoso-Moreira M., Arguello J.R., Gottipati S., Harshman L.G., Grenier J.K., Clark A.G. Evidence for the fixation of gene duplications by positive selection in Drosophila. Genome Res. 2016;26:787–798. doi: 10.1101/gr.199323.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke S., Fusetto R., Martelli F., Giang A., Battlay P., Fournier-Level A., O'Hair R.A., Batterham P. Multiple P450s and variation in neuronal genes underpins the response to the insecticide imidacloprid in a population of Drosophila melanogaster. Sci. Rep. 2017;7:11338. doi: 10.1038/s41598-017-11092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.R., Laing R., Bartley D.J., Britton C., Chaudhry U., Gilleard J.S., Holroyd N., Mable B.K., Maitland K., Morrison A.A., Tait A., Tracey A., Berriman M., Devaney E., Cotton J.A., Sargison N.D. A genome resequencing-based genetic map reveals the recombination landscape of an outbred parasitic nematode in the presence of polyploidy and polyandry. Genome Biol Evol. 2018;10:396–409. doi: 10.1093/gbe/evx269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Finel M., Kurkela M. The UDP-glucuronosyltransferases as oligomeric enzymes. Curr. Drug Metabol. 2008;9:70–76. doi: 10.2174/138920008783331158. [DOI] [PubMed] [Google Scholar]
- Fontaine P., Choe K. The transcription factor SKN-1 and detoxification gene ugt-22 alter albendazole efficacy in Caenorhabditis elegans. Int J Parasitol Drugs Drug Resist. 2018;8:312–319. doi: 10.1016/j.ijpddr.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanizadeh H., Harrington K.C. Non-target site mechanisms of resistance to herbicides. Crit. Rev. Plant Sci. 2017;36:24–34. [Google Scholar]
- Gilleard J.S., Redman E. Vol. 93. 2016. pp. 31–68. (Genetic diversity and population structure of Haemonchus contortus. Haemonchus Contortus and Haemonchosis - Past, Present and Future Trends). [DOI] [PubMed] [Google Scholar]
- Jacobs C.G., Steiger S., Heckel D.G., Wielsch N., Vilcinskas A., Vogel H. Sex, offspring and carcass determine antimicrobial peptide expression in the burying beetle. Sci. Rep. 2016;6:25409. doi: 10.1038/srep25409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L.M., Flemming A.J., Urwin P.E. NHR-176 regulates cyp-35d1 to control hydroxylation-dependent metabolism of thiabendazole in Caenorhabditis elegans. Biochem. J. 2015;466:37–44. doi: 10.1042/BJ20141296. [DOI] [PubMed] [Google Scholar]
- Kotze A.C., Ruffell A.P., Ingham A.B. Phenobarbital induction and chemical synergism demonstrate the role of UDP-glucuronosyltransferases in detoxification of naphthalophos by Haemonchus contortus larvae. Antimicrob. Agents Chemother. 2014;58:7475–7483. doi: 10.1128/AAC.03333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotze A.C., Prichard R.K. Vol. 93. 2016. pp. 397–428. (Anthelmintic resistance in Haemonchus contortus: history, mechanisms and diagnosis. Haemonchus Contortus and Haemonchosis - Past, Present and Future Trends). [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Vo A.D., Qin F.J., Li H. Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci. Rep. 2016;6:21597. doi: 10.1038/srep21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa M.S.G., Veenstra J.G., Roos M.H. Molecular characterization of beta-tubulin genes present in benzimidazole resistant populations of Haemonchus contortus. Mol. Biochem. Parasitol. 1993;60:133–144. doi: 10.1016/0166-6851(93)90036-w. [DOI] [PubMed] [Google Scholar]
- Laing R., Kikuchi T., Martinelli A., Tsai I.J., Beech R.N., Redman E., Holroyd N., Bartley D.J., Beasley H., Britton C., Curran D., Devaney E., Gilabert A., Hunt M., Jackson F., Johnston S.L., Kryukov I., Li K., Morrison A.A., Reid A.J., Sargison N., Saunders G.I., Wasmuth J.D., Wolstenholme A., Berriman M., Gilleard J.S., Cotton J.A. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14:R88. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing S.T., Ivens A., Laing R., Ravikumar S., Butler V., Woods D.J., Gilleard J.S. Characterization of the xenobiotic response of Caenorhabditis elegans to the anthelmintic drug albendazole and the identification of novel drug glucoside metabolites. Biochem. J. 2010;432:505–514. doi: 10.1042/BJ20101346. [DOI] [PubMed] [Google Scholar]
- Le S.Q., Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lecova L., Ruzickova M., Laing R., Vogel H., Szotakova B., Prchal L., Lamka J., Vokral I., Skalova L., Matouskova P. Reliable reference gene selection for quantitative real time PCR in Haemonchus contortus. Mol. Biochem. Parasitol. 2015;201:123–127. doi: 10.1016/j.molbiopara.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Li X.X., Zhu B., Gao X.W., Liang P. Over-expression of UDP-glycosyltransferase gene UGT2B17 is involved in chlorantraniliprole resistance in Plutella xylostella (L.) Pest Manag. Sci. 2017;73:1402–1409. doi: 10.1002/ps.4469. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mackenzie P.I., Bock K.W., Burchell B., Guillemette C., Ikushiro S., Iyanagi T., Miners J.O., Owens I.S., Nebert D.W. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenetics Genom. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- Malik V., Black G.W. Structural, functional, and mutagenesis studies of UDP-glycosyltransferases. Structural and Mechanistic Enzymology: Bringing Together Experiments and Computing. 2012;87:87–115. doi: 10.1016/B978-0-12-398312-1.00004-4. [DOI] [PubMed] [Google Scholar]
- Matouskova P., Vokral I., Lamka J., Skalova L. The role of xenobiotic-metabolizing enzymes in anthelmintic deactivation and resistance in helminths. Trends Parasitol. 2016;32:481–491. doi: 10.1016/j.pt.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Mederos A.E., Ramos Z., Banchero G.E. First report of monepantel Haemonchus contortus resistance on sheep farms in Uruguay. Parasites Vectors. 2014;7:598. doi: 10.1186/s13071-014-0598-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meech R., Rogers A., Zhuang L., Lewis B.C., Miners J.O., Mackenzie P.I. Identification of residues that confer sugar selectivity to UDP-glycosyltransferase 3A (UGT3A) enzymes. J. Biol. Chem. 2012;287:24122–24130. doi: 10.1074/jbc.M112.343608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miley M.J., Zielinska A.K., Keenan J.E., Bratton S.M., Radominska-Pandya A., Redinbo M.R. Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J. Mol. Biol. 2007;369:498–511. doi: 10.1016/j.jmb.2007.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Rochat B. Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance - focus on tamoxifen, paclitaxel and imatinib metabolism. Clin. Pharmacokinet. 2005;44:349–366. doi: 10.2165/00003088-200544040-00002. [DOI] [PubMed] [Google Scholar]
- Roos M.H., Otsen M., Hoekstra R., Veenstra J.G., Lenstra J.A. Genetic analysis of inbreeding of two strains of the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2004;34:109–115. doi: 10.1016/j.ijpara.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W.Z., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchlíková L.R., Matoušková P., Vokřál I., Lamka J., Szotáková B., Sečkařová A., Dimunová D., Nguyen L.T., Várady M., Skálová L. Metabolism of albendazole, ricobendazole and flubendazole in Haemonchus contortus adults: sex differences, resistance-related differences and the identification of new metabolites. Int J Parasitol Drugs Drug Resist. 2018;8:50–58. doi: 10.1016/j.ijpddr.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari P., Sangwan R.S., Sangwan N.S. Plant secondary metabolism linked glycosyltransferases: an update on expanding knowledge and scopes. Biotechnol. Adv. 2016;34:714–739. doi: 10.1016/j.biotechadv.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwyk J.A., Gerber H.M., Groeneveld H.T. A technique for the recovery of nematodes from ruminants by migration from gastrointestinal ingesta gelled in agar-large scale application Onderstepoort. J Vet Res. 1980;47:147–158. [PubMed] [Google Scholar]
- Vogel H., Badapanda C., Knorr E., Vilcinskas A. RNA-sequencing analysis reveals abundant developmental stage-specific and immunity-related genes in the pollen beetle Meligethes aeneus. Insect Mol. Biol. 2014;23:98–112. doi: 10.1111/imb.12067. [DOI] [PubMed] [Google Scholar]
- Vokral I., Bartikova H., Prchal L., Stuchlikova L., Skalova L., Szotakova B., Lamka J., Varady M., Kubicek V. The metabolism of flubendazole and the activities of selected biotransformation enzymes in Haemonchus contortus strains susceptible and resistant to anthelmintics. Parasitology. 2012;139:1309–1316. doi: 10.1017/S0031182012000595. [DOI] [PubMed] [Google Scholar]
- Vokral I., Jirasko R., Stuchlikova L., Bartikova H., Szotakova B., Lamka J., Varady M., Skalova L. Biotransformation of albendazole and activities of selected detoxification enzymes in Haemonchus contortus strains susceptible and resistant to anthelmintics. Vet. Parasitol. 2013;196:373–381. doi: 10.1016/j.vetpar.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Williamson S.M., Storey B., Howell S., Harper K.M., Kaplan R.M., Wolstenholme A.J. Candidate anthelmintic resistance-associated gene expression and sequence polymorphisms in a triple-resistant field isolate of Haemonchus contortus. Mol. Biochem. Parasitol. 2011;180:99–105. doi: 10.1016/j.molbiopara.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Yilmaz E., Ramunke S., Demeler J., Krucken J. Comparison of constitutive and thiabendazole-induced expression of five cytochrome P450 genes in fourth-stage larvae of Haemonchus contortus isolates with different drug susceptibility identifies one gene with high constitutive expression in a multi-resistant isolate. Int J Parasitol Drugs Drug Resist. 2017;7:362–369. doi: 10.1016/j.ijpddr.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.