Abstract
The Th2 cytokine IL-4 triggers a signaling cascade which activates transcription by STAT6. The goals of the present study are to define the transcriptomic response of mouse spleen B cells (mSBC) to IL-4 used as single stimulus, its specificity compared to human peripheral blood B cells (hPBBC) and to mouse spleen T cells (mSTC), and the pathways affected. Oligonucleotide-based microarrays were performed using two references, the untreated sample and the cells cultured without IL-4, an experimental design which reduces the potential confounding effect of cellular stress during culture. Specificity was addressed by comparing the response of mSBC and our previously published study on hPBBC, of similar design, and a study by other authors on mSTC. We detected an mSBC-specific response (including novel genes, e.g., Sertad4, Lifr, Pmepa1, Epcam, Tbxas1; and common genes, e.g., Usp2, Cst7, Grtp1, and Casp6), an hPBBC-specific response (e.g., CCL17, MTCL1, GCSAM, HOMER2, IL2RA), and a common mSBC/hPBBC response (e.g., CISH, NFIL3, SOCS1, VDR, CDH1). In contrast, the mSBC and mSTC responses were largely divergent. Gene set enrichment analysis (GSEA) was applied for the first time to identify the pathways affected. Both in mSBC and hPBBC, IL-4 activated Myc, the transcriptional machinery itself, cell cycle, mitochondria and respiratory chain, ribosome, proteasome and antigen presentation, and Wnt signaling, and inhibited GPCR signaling. However, significant differences were found in histone demethylation, Nod signaling, and Rho signaling, which were downregulated in mSBC, and in chromatin condensation, which was downregulated in hPBBC. These findings may have therapeutic implications for the treatment of allergic diseases and parasitic infections.
Abbreviations: Acc. No., accession number; ANOVA, analysis of variance; BCR, B cell receptor; CLL, chronic lymphocytic leukemia; GEO, Gene Expression Omnibus; GSEA, gene set enrichment analysis; hPBBC, human peripheral blood B cells; IL-4, interleukin-4; LPS, lipopolysaccharide; MHC, major histocompatibility complex; moAb, monoclonal antibody; mSBC, mouse spleen B cells; mSTC, mouse spleen T cells; MSigDB, Molecular Signatures database; qRTPCR, quantitative real-time polymerase chain reaction; S.D., standard deviation
Keywords: IL-4, B cells, Microarray, GSEA, Mitochondria, Wnt signaling
Highlights
-
•
The gene expression response of mouse spleen B cells to IL-4 used as single stimulus was identified by microarray.
-
•
Two reference samples were used to calculate net changes and avoid the confounding effect of cell culture stress.
-
•
The functional response of B cells to IL-4 involves Myc, mitochondria, ribosome, proteasome and Wnt, according to GSEA.
-
•
The mouse and human responses to IL-4 display common and specific features.
1. Introduction
B cells act in the humoral immunity component of the adaptive immune response. B cells express B cell receptors (BCRs) on their cell membrane. BCRs allow B cells to bind to specific antigens, against which they will initiate antibody responses. B cells receive help from cognate helper T cells, and differentiate into plasma cells that secrete large amounts of antibodies [1], [2], [3]. IL-4 is a cytokine that induces differentiation of naive helper T cells to Th2 cells. Upon activation by IL-4, Th2 cells produce additional IL-4 in a positive feedback loop. IL-4 is also secreted by NK-T cells, basophils, eosinophils and mast cells. IL-4 stimulates activated B-cell and T-cell proliferation, differentiation of B cells into plasma cells, B-cell class switching to IgE, and MHC class II production [4], [5], [6].
The IL-4 pathway is a signaling cascade initiated by binding of IL-4 to its membrane receptor IL-4R. The activated IL-4R phosphorylates JAK1 and JAK3. JAK1 phosphorylates STAT6 which homodimerizes and enter the nucleus to regulate gene expression. JAK1 and JAK3 lead to activation of the Ras/MAPK pathway and NFκB, which cooperate with STAT6 for transcription [7], [8], [9], [10]. In the first systematic study which addressed the gene expression response of mouse spleen B cells (mSBC) to IL-4, costimulation with lipopolysaccharide (LPS) and anti-CD40 was used [11]. Then, a few studies have reported the gene expression response of human B cells of diverse origin to IL-4 used as single stimulus [12], [13] or with anti-CD40 [14]. Recently, a combined transcriptomic and proteomic study performed in mSBC costimulated with LPS and anti-CD40 established that regulation of protein expression by IL-4 occurs mainly at the transcriptional level [15].
In the present paper, we intend to define the transcriptome response of mSBC to IL-4 as single stimulus, by the use of an experimental design which compares the sample cultured with IL-4 with two references —the untreated sample and the sample cultured without IL-4. Such a design had already been used to define the response to IL-4 of human peripheral blood B cells (hPBBC) from healthy individuals and from chronic lymphocytic leukemia (CLL) B cells [13]. Here, analysis has been optimized by calculating net changes induced by IL-4, which allowed a more stringent identification of the genes genuinely regulated by IL-4. Moreover, in order to shape the core response of B cells to IL-4 and the specific human and mouse responses, we have reanalyzed previous studies and compared their results. Finally, in order to identify the main functional axes activated by the transcriptional response of B cells to IL-4, we have performed for the first time gene set enrichment analysis (GSEA) using the Molecular Signatures database (MSigDB).
2. Material and methods
2.1. Isolation of mSBC and culture with IL-4
C57BL/6 N mice were bred at the animal facility of the University of Murcia under specific-pathogen-free conditions. All the procedures were performed according to the Directive 2010/63/EU on the protection of animals used for scientific purposes and approved by the Ethics Committee for Animal Experimentation of Hospital Clinico Universitario Virgen de la Arrixaca. Mice were killed by carbon dioxide asphyxiation and exanguination at 12 weeks of age. Spleens were excised and placed into a 100 µm cell strainer within an empty Petri dish. Following addition of 1 ml of PBS containing 2% fetal calf serum, 0.06% citrate, and 0.2 I.U. heparin per ml into the strainer, spleens were crushed using the plunger of a syringe. Cell suspensions were treated with ACK Lysing Buffer (Lonza, Basel, Switzerland), and negatively selected using the Pan B Cell Isolation Kit, mouse (Miltenyi Biotec, Bergisch Gladbach, Germany), following the manufacturer's instructions. B cells from 5 mice were cultured for 18 h in RPMI-1640 medium supplemented with 10% FCS (Cambrex, East Rutherford, NJ), 50 U/ml penicillin, 50 U/ml streptomycin, 2.5 μg/ml amphotericin B, and 2 mM L-glutamine with nothing and with adding 10 ng/ml of Pharmingen mouse recombinant IL-4 (BD Biosciences, San Jose, CA). These samples were designated Ctrl and IL-4, respectively, and that obtained before cell culture, sample Pre. Purity of the Pre samples was determined by labeling with monoclonal antibodies (moAb) CD3-FITC, CD19-PE-Cy5, and CD11b-PE and flow cytometry analysis in a BD FACScalibur flow cytometer, using the CellQuest software. Apoptosis of the Ctrl and IL-4 samples was determined using the same procedure after labeling with annexin V-FITC and propidium iodide-PE.
2.2. RNA isolation
Total RNA was isolated using the miRNeasy Mini Kit (Qiagen, Hilden, Germany). RNA samples were quantitated on a NanoDrop 2000 (Thermo Fisher Scientific, Whaltham, MA). RNA quality was examined on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) using the RNA 6000 Nano Kit. Only samples with RIN (RNA Integrity Number) > 7 were further studied.
2.3. RNA labeling, microarray hybridization, and feature extraction
RNA samples were labeled using Agilent Two Color Quick Amp Labeling and RNA Spike-In kits, according to the manufacturer's protocol. The RNA samples from mSBC (Pre, Ctrl, IL-4) were labeled with cyanine 5-CTP and used as tests. A pooled sample, previously described, was labeled with cyanine 3-CTP and used as reference [16]. The labeled cRNAs were mixed together and hybridized onto Agilent Mouse GE 4 × 44 K v2 Microarray Kit targeting 39,430 Entrez Gene RNAs, using the Agilent Gene Expression Hybridization kit. After hybridization, the microarray slides were washed and scanned in an Agilent G2565CA DNA Microarray Scanner. Images were analyzed with the Agilent Feature Extraction software, which computes log ratios (test vs reference) following normalization correction by linear and Lowess methods. Datasets were deposited at the Gene Expression Omnibus (GEO) database under accession number (Acc. No.) GSE73760.
2.4. Microarray analysis
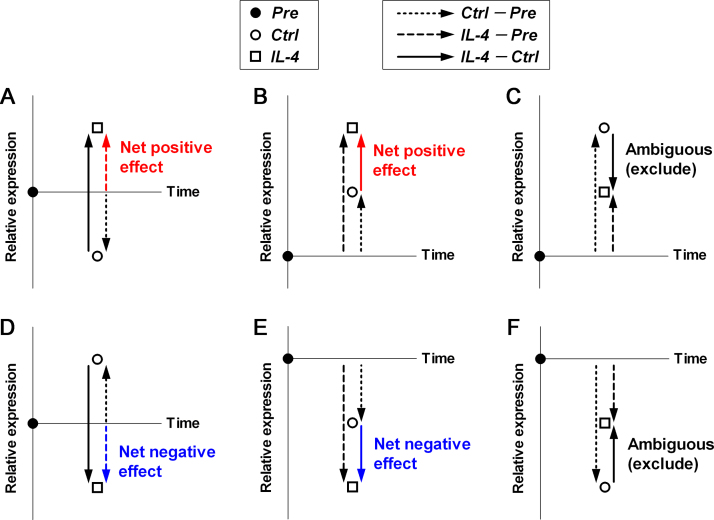
Datasets were transformed into log2 ratios and reference conditions Pre and Ctrl were subtracted to condition IL-4 to generate the IL-4 – Pre and IL-4 – Ctrl lists. The net change induced by IL-4 for each entity was the lowest, if positive (Fig. 1A and B), or the highest, if negative (Fig. 1D and E), of both subtractions, and the corresponding reference value, either Pre or Ctrl, will be called hereafter Ref. Entities taking values with opposite sign on both lists were excluded from further analysis (Fig. 1C and F). Comparison between Ref and IL-4 values was performed using the Student t-test, and p values computed using the Benjamini-Hochberg FDR correction. Fold changes were calculated using the formula 2(IL−4 – Ref) for net positive changes or −2−(IL−4 – Ref) for net negative changes. Entities above 2-fold change (FC) and adjusted (adj.) p < 0.05 were considered significantly regulated by IL-4. The same procedure has been applied for reanalysis of our previously published experiment performed on hPBBC and CLL treated with IL-4 for 18 h [13] (GEO database Acc. No. GSE55288).
Fig. 1.
Schematic of study design. Net changes in gene expression induced by IL-4 in mSBC and hPBBC were calculated following creation of two lists of genes by subtraction of both references, Pre and Ctrl, to IL-4, i.e., the IL-4 – Pre and IL-4 – Ctrl lists, expressed as log2ratios. For entities that took equal sign in both lists, net change was the smaller of the two values in absolute terms, positive (A and B) or negative (D and E). For entities that took opposite sign in both lists (C and F), net change could not be calculated and these entities were excluded. Pre, sample prior to culture; Ctrl and IL-4, samples cultured in parallel for 18 h with nothing and IL-4, respectively.
2.5. GSEA
GSEA was performed to determine whether both conditions (Ref and IL-4) present significant differences for a priori defined set of genes comprised in the MSigDB (http://software.broadinstitute.org/gsea/).
2.6. Quantitative real-time polymerase chain reaction (qRTPCR)
RNA samples were subjected to reverse transcription with the iScript cDNA Synthesis Kit (Bio-Rad), following the manufacturer's instructions. QRTPCR was performed with the SYBR Premix Ex Taq (Takara Bio, Mountain View, CA) in an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). QuantiTect primer assays Mm_Casp6_2 (QT00494921), Mm_Cbr3_2 (QT02239174), Mm_ Cdh1_1 (QT00121163), Mm_Cst7_1 (QT00114548), Mm_Epcam_2 (QT02304456), Mm_Grtp1_2 (QT01196237), Mm_Lifr_1 (QT00140980), Mm_Pmepa1_1 (QT00128779), Mm_Pole2_1 (QT00113386), Mm_Sdc1_2 (QT01751029), Mm_Sertad4_1 (QT00111482), Mm_Slc39a8_1 (QT00113092), Mm_Tbxas1_1 (QT01037106), Mm_Usp2_1 (QT00164906), Mm_Vdr_1 (QT00149345) and, as reference, Mm_Gapdh_3 (QT01658692) (Qiagen), were used. The relative expression values, expressed as fold change relative to the reference, were calculated using the formula 2−ΔΔCt as described elsewhere [13]. Comparisons between conditions Pre, Ctrl and IL-4 were performed using 1-way analysis of variance (ANOVA) with Tukey's multiple comparison post test.
3. Results and discussion
3.1. Gene expression response of mSBC to IL-4 in vitro
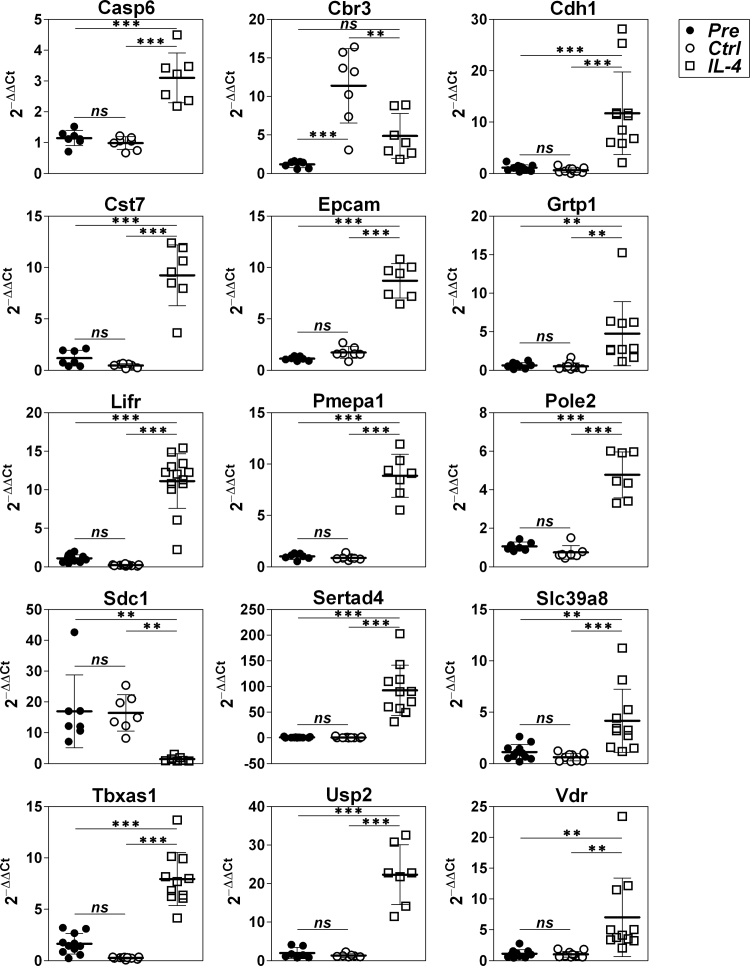
The purified mSBC Pre samples included 94.1 ± 2.9% of CD3–CD19 + cells (mean ± S.D), 2.2 ± 2.3% of CD3 + CD19– cells (T cells) and 1.4 ± 0.5% of CD3–CD19–CD11b+ cells (myeloid cells). Cell culture affected survival of mSBC negatively (40.1 ± 16.4% survival in Ctrl). IL-4 exerted a slight but significant positive effect (44.6 ± 17.2% survival in IL-4, Student's t-test p < 0.05 for Ctrl vs IL-4). These data indicate that cell culture causes cell stress and should be treated as a confounder for analysis of gene expression regulation by IL-4 in mSBC. To minimize bias induced by cell culture, net changes induced by IL-4 were calculated, and entities for which IL-4 – Pre and IL-4 – Ctrl had opposite sign were excluded. A dataset which comprised the log2ratios of the Ref samples (either Pre or Ctrl, the one used to calculate net change for each entity) and the IL-4 samples from 22,338 entities that passed the filter was generated, and then analyzed using the Student t-test and GSEA. Using Benjamini-Hochberg with adj. p < 0.05, 284 probes, corresponding to 212 known genes, were positively regulated by IL-4 > 2-fold (Supplemental table 1), and 335 probes, corresponding to 227 known genes, were negatively regulated by IL-4 > 2-fold (Supplemental table 2). As will be mentioned later when referring to specific genes, qRTPCR validations confirmed significant differences between IL-4 compared to Pre and Ctrl in 14 out of 15 genes (Fig. 2), with the only exception of Cbr3, which behaved by qRTPCR rather as a gene of the type exemplified in Fig. 1C.
Fig. 2.
QRTPCR validations of transcriptome changes in mSBC in response to IL-4. QRTPCR analysis of 15 genes, 14 tested for upregulation (Casp6, Cbr3, Cdh1, Cst7, Epcam, Grtp1, Lifr, Pmepa1, Pole2, Sertad4, Slc39a8, Tbxas1, Usp2, and Vdr) and 1 for downregulation (Sdc1). All the genes validated microarray results except Cbr3. QRTPCR data are expressed as 2–ΔΔCt. Pre, sample prior to culture; Ctrl and IL-4, samples cultured in parallel for 18 h with nothing and IL-4, respectively. Bars indicate mean ± S.D., ANOVA with Tukey's multiple comparison post test, ** p < 0.01, ***p < 0.001.
Our previous studies using hPBBC and CLL [13], of similar design to the present study, were reanalyzed to calculate net changes. This approach resulted slightly more stringent with regards to the previously published lists of genes significantly regulated by IL-4, since they decreased from 123 genes to 95 genes (87 up and 8 down) in hPBBC and from 189 to 170 genes (130 up and 40 down) in CLL. In addition, recent studies by Xue et al. [14] and Mokada-Gopal et al. [15], performed on human tonsil B cells treated for 24 h with anti-CD40 ± IL-4 and mSBC treated for 96 h with LPS + anti-CD40 ± IL-4, respectively, were retrieved from the GEO database (acc. nos. GSE71810 and GSE84075, respectively). The GEO2R tool, linked to the GEO website (https://www.ncbi.nlm.nih.gov/geo/geo2r/), was used to analyse the tonsil response, which consisted in 591 and 811 genes significantly upregulated and downregulated > 2-fold with adj. p < 0.05, respectively, according to the Student's t-test. Analysis of the mSBC study resulted in 469 and 344 genes. These figures are higher than those of our studies, likely partly due to false positives that could have been excluded if the baseline values had been measured and the net changes calculated. Nonetheless, these signatures, together with Lu IL-4 Signaling Up of the MSigDB, composed by 94 genes upregulated by IL-4 in hPBBC after 4 h of stimulation [12], were compared with signatures from our mSBC and hPBBC studies in order to define mouse-specific, human-specific, or common gene expression responses of B cells to IL-4 (Supplemental table 3). Around 55% and ~66% of the genes upregulated and downregulated by IL-4, respectively, identified in mSBC in the present study were novel. The upregulated genes Sertad4, Lifr, Pmepa1, Epcam, Tbxas1, and the downregulated gene Sdc1, were validated by qPCR (Fig. 2).
The mouse-specific response to IL-4 also included ~28% upregulated genes and ~19% downregulated genes that overlapped with the signatures of the Mokada-Gopal's study but not with human signatures. Within this group, Usp2 activates Wnt signaling [17]; Rab35 is a GTPase that regulates phosphoinositides and F-actin on endosomes [18]; Grtp1 is a Rab-GAP with broad specificity [19]; and Casp6 regulates B cell activation and differentiation into plasma cells by modifying cell cycle entry [20]; among the downregulated genes, Gpr34 is a transmembrane G protein-coupled receptor (GPCR). The upregulated genes Usp2, Cst7 (cystatin), Grtp1, and Casp6 were validated by qRTPCR (Fig. 2). Other highly regulated genes within this group included St7, Klhdc2, Gadd45g, or Insm1, upregulated, and Anxa1, downregulated.
The remaining ~17% upregulated and ~15% downregulated genes have also been found in at least a human signature. This group includes 15 genes, 14 upregulated and 1 downregulated, that coincided in the present study in mSBC and our previous study in hPBBC. Among the upregulated genes, Cish and Socs1 are protein kinase inhibitors that take part in negative feedback loops to attenuate cytokine signaling [21]; Nfil3 and Xbp1 are bZIP transcriptional regulators involved in Ig class switching and high-level Ig secretion by plasma cells [22], [23]; Vdr (vitamin D3 receptor) controls serum IgE levels [24]; IL4i1, a L-phenylalanine oxidase, controls BCR-dependent activation [25]; Dock10, a Rac1 and Cdc42 guanine-nucleotide exchange factor (GEF), is involved in B cell development [16]; Auh, an RNA-binding hydratase, is involved in mitochondrial protein synthesis [26]; the Vmp1 gene hosts miR-21 [27]; and Il4r takes part in a positive feedback loop to amplify IL-4 signaling [4]. Vdr, Cdh1 (E-cadherin), and Slc39a8 were validated by qRTPCR in mSBC (Fig. 2).
An hPBBC response to IL-4, not shared by mSBC, was also detected. Among the upregulated genes, CCL17 plays a role in class switch recombination [22]; GCSAM induces Syk phosphorylation and negatively regulates cell motility through Rho activation [28], [29]; and IL2RA drives differentiation of B cells towards plasma cells [30]. Other highly upregulated genes of this group included RASL10A, MTCL1, SLC37A3, HS3ST1, MELTF, ZBTB8A, CLEC4A, HOMER2, GNG8, QSOX1, or PALLD.
Finally, to assess which responses are specific to B cells, the responses of mSBC and hPPBC were compared to those of mouse splenic T cells (mSTC) and human cord blood T cells (UCTC), reported by Chen et al. [31] and Kanduri et al. [32], respectively (Supplemental table 3). In these studies, CD4 cells were treated with anti-CD3 + anti-CD28 ± IL-4, for 48 h and 72 h, respectively. The mSTC study identified 31 upregulated and 90 downregulated genes, of which only 8 (Cish, Nfil3, Il4i1, Casp6, Lilrb4, Hipk2, Gp49a, and Il4ra) and 4 (Ifng, Ltb, Trim30b, Ifi203) overlapped with our present mSBC study, respectively. The UCTC study included two references, allowing calculate net changes (GEO acc. no. GSE71575), and analysis using GEO2R resulted in 67 and 25 genes, of which only 7 upregulated (GFI1, IGSF3, SOCS1, NFIL3, LRRC32, SPINT2, and ITPRIPL2) overlapped with hPBBC. Therefore, most of the IL-4 response of B cells is specific.
3.2. GSEA
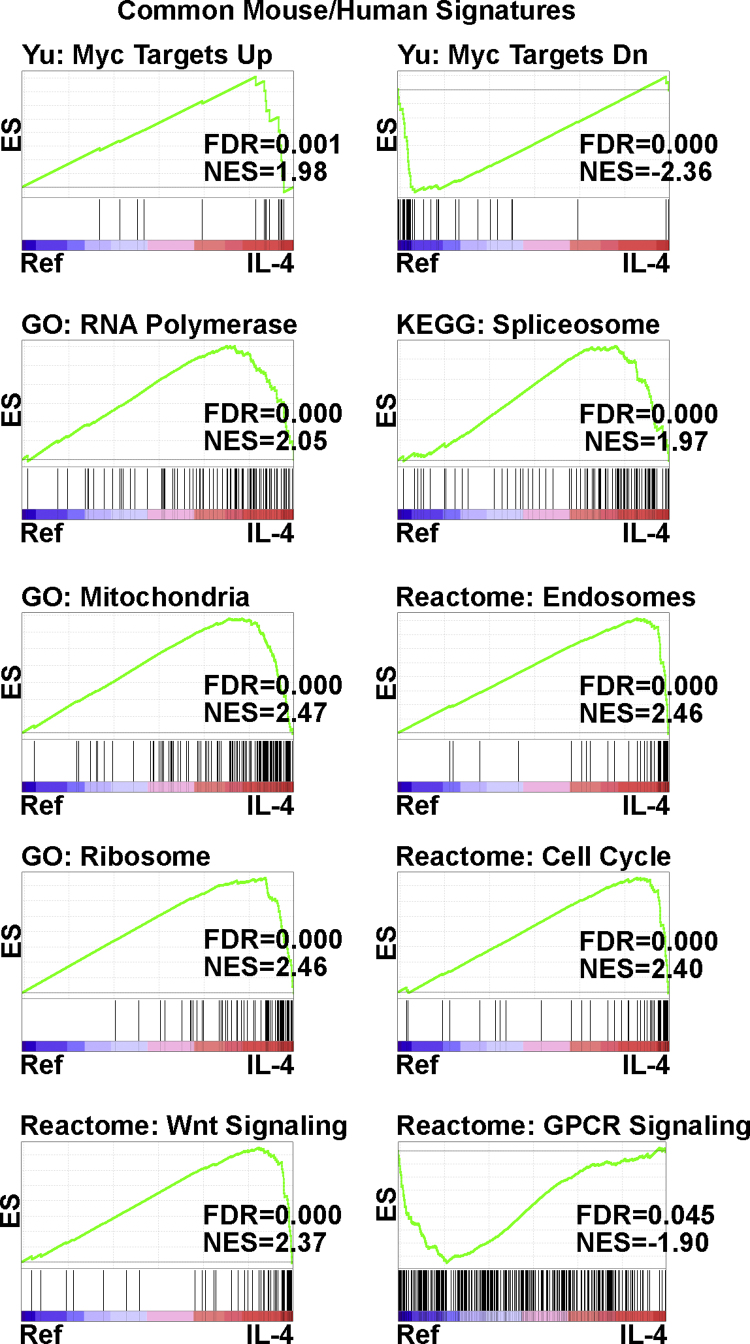
To gain insights into the functional meaning of gene expression changes induced by IL-4, we performed GSEA in mSBC and hPBBC. We first summarize significant pathways found both in mSBC and hPBBC (Fig. 3). The Chemical and Genetic Perturbations collection of the MSigDB comprises numerous pairs of up and down signatures. We judge that significance of a signature is higher when the up and down gene sets are both significant, with opposite sign, with respect to conditions Ref and IL-4, or vice versa. The most significant and recurrent pairs were found for the upregulated and downregulated targets of Myc, associated to IL-4 and Ref, respectively, which strongly suggests that IL-4 potentiates Myc function. In addition, GSEA using the Canonical Pathways and Cellular Components collections revealed changes induced by IL-4 in the transcriptional machinery itself (RNA polymerase, spliceosome), stimulation of the cell cycle machinery (DNA replication, mitotic spindle), mitochondrial activity (oxidative phosphorylation, mitochondrial matrix, mitochondrial envelope), the ribosome, and the proteasome (antigen presentation, endosomes), positive regulation of Wnt signaling, and negative regulation of GPCR signaling.
Fig. 3.
Common pathways and cellular components regulated by IL-4 in mSBC and mPBBC according to GSEA. Histograms depict results of comparisons IL-4 vs Ref in mSBC. The headers of some histograms are shortened designations for the following gene sets: GO: RNA polymerase complex, GO: Mitochondrial protein complex, Reactome: Cross-presentation of soluble exogenous antigens – Endosomes, GO: Organellar ribosome, Reactome: Regulation of mitotic cell cycle, Reactome: Signaling by Wnt, and Reactome: Signaling by GPCR. ES, enrichment score; FDR, false discovery rate; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score.
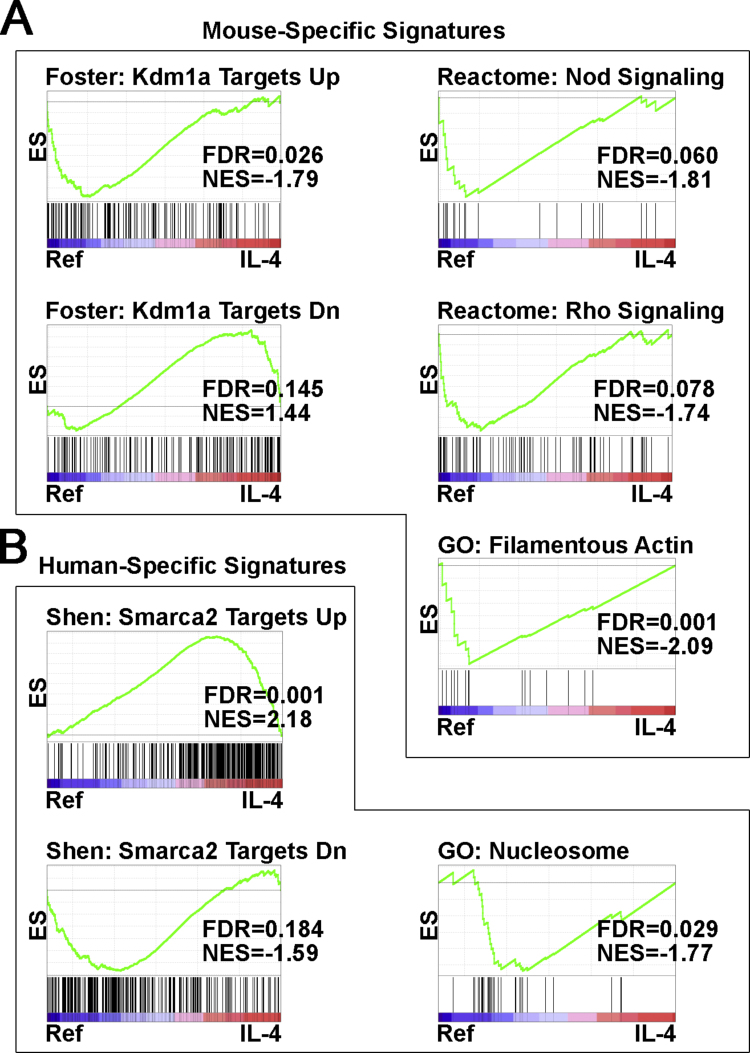
Interesting relationships specifically found in mSBC included the upregulated and downregulated targets of the lysine demethylase Kdm1a, associated to Ref and IL-4, respectively (Fig. 4A), suggesting that this component of histone deacetylase complexes is inhibited by IL-4 in mSBC. In addition, Nod signaling, Rho signaling, and filamentous actin were downregulated by IL-4 in mSBC. On the other hand, the upregulated and downregulated targets of Smarca2 were associated to Ref and IL-4, respectively, specifically in mPBBC (Fig. 4B), suggesting that this component of the SWI/SNF chromatin remodeling complex is activated by IL-4 in hPBBC. Consistent with this finding, the nucleosome was downregulated by IL-4 in hPBBC.
Fig. 4.
Specific pathways and cellular components regulated by IL-4 in mSBC (A) and mPBBC (B) according to GSEA. Histograms depict results of comparison IL-4 vs Ref. The headers of some histograms are shortened designations for the following gene sets: Reactome: Nod1/2 signaling pathway, Reactome: Signaling by Rho GTPases, and GO: Nuclear nucleosome. ES, enrichment score; FDR, false discovery rate; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score.
In summary, the present study dissected and compared the transcriptomic responses of mouse and human B cells to IL-4. Our study is unique in that baseline values were measured, which increased the ability to filter out false positives. In addition, we used GSEA and show for the first time the major axes of the transcriptomic response of B cells to IL-4, which have broad common features in mouse and human but also specific features, such as the choice of pathways that regulate access to chromatin. We conclude that the human and mouse transcriptomic responses of B cells to IL-4 significantly overlap, but mouse and human specific responses are also significant, in contrast to the small overlap between B and T cell responses. IL-4 activated both in mouse and human B cells central processes such as transcription and translation, respiratory chain, cell cycle, survival, and specific pathways such as Wnt signaling, but diverged in others such as histone demethylation, Nod, and Rho signaling. Discrimination between responses of murine and human B cells is important in the interpretation of studies on the function of IL-4, and has implications for its possible therapeutic application for the treatment of allergic diseases, helminth infections and B cell malignancies.
Acknowledgements
We thank Silvia Sebastian Ruiz and María José Alcaraz García for excellent technical assistance, and the animal facility of the University of Murcia. This work was supported by grants from Plan Nacional de I+D+I 2008–2011, Acción Estratégica en Salud, Instituto de Salud Carlos III (PI10/01226 to A.P.) (co-financed by the European Regional Development Fund, “Una manera de hacer Europa”), and II PCTRM 2007–2010, Fundación Séneca, Agencia de Ciencia y Tecnología de la Región de Murcia (08721/PI/08 to A.P.).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.09.007.
Appendix A. Transparency document
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.LeBien T.W., Tedder T.F. B lymphocytes: how they develop and function. Blood. 2008;112:1570–1580. doi: 10.1182/blood-2008-02-078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corcoran L.M., Tarlinton D.M. Regulation of germinal center responses, memory B cells and plasma cell formation–an update. Curr. Opin. Immunol. 2016;39:59–67. doi: 10.1016/j.coi.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman W., Lakkis F.G., Chalasani G. B cells, antibodies, and more. Clin. J. Am. Soc. Nephrol. 2016;11:137–154. doi: 10.2215/CJN.09430915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada H., Banchereau J., Lotze M.T. Interleukin-4. In: Thompson A.W., Lotze M.T., editors. Vol. I. Academic Press; London: 2003. pp. 227–262. (The Cytokine Handbook). [Google Scholar]
- 5.Wojciechowski W., Harris D.P., Sprague F. Cytokine-producing effector B cells regulate type 2 immunity to H. polygyrus. Immunity. 2009;30:421–433. doi: 10.1016/j.immuni.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo B., Rothstein T.L. IL-4 upregulates Igα and Igβ protein, resulting in augmented IgM maturation and B cell receptor–triggered B cell activation. J. Immunol. 2013;191:670–677. doi: 10.4049/jimmunol.1203211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messner B., Stütz A.M., Albrecht B. Cooperation of binding sites for STAT6 and NF kappa B/rel in the IL-4-induced up-regulation of the human IgE germline promoter. J. Immunol. 1997;159:3330–3337. [PubMed] [Google Scholar]
- 8.Shen C.H., Stavnezer J. Interaction of stat6 and NF-kappaB: direct association and synergistic activation of interleukin-4-induced transcription. Mol. Cell. Biol. 1998;18:3395–3404. doi: 10.1128/mcb.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zamorano J., Mora A.L., Boothby M., Keegan A.D. NF-kappa B activation plays an important role in the IL-4-induced protection from apoptosis. Int. Immunol. 2001;13:1479–1487. doi: 10.1093/intimm/13.12.1479. [DOI] [PubMed] [Google Scholar]
- 10.Thieu V.T., Nguyen E.T., McCarthy B.P. IL-4-stimulated NF-kappaB activity is required for Stat6 DNA binding. J. Leukoc. Biol. 2007;82:370–379. doi: 10.1189/jlb.1106707. [DOI] [PubMed] [Google Scholar]
- 11.Schroder A.J., Pavlidis P., Arimura A. Cutting edge: STAT6 serves as a positive and negative regulator of gene expression in IL-4-stimulated B lymphocytes. J. Immunol. 2002;168:996–1000. doi: 10.4049/jimmunol.168.3.996. [DOI] [PubMed] [Google Scholar]
- 12.Lu X., Nechushtan H., Ding F. Distinct IL-4-induced gene expression, proliferation, and intracellular signaling in germinal center B-cell-like and activated B-cell-like diffuse large-cell lymphomas. Blood. 2005;105:2924–2932. doi: 10.1182/blood-2004-10-3820. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz-Lafuente N., Alcaraz-García M.J., Sebastián-Ruiz S. The gene expression response of chronic lymphocytic leukemia to IL-4 is specific, depends on ZAP-70 status and is differentially affected by an NFkB inhibitor. PLoS One. 2014;9:e109533. doi: 10.1371/journal.pone.0109533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue D., Kaufman G.N., Dembele M. Semaphorin 4C protects against allergic inflammation: requirement of regulatory CD138+ plasma cells. J. Immunol. 2017;198:71–81. doi: 10.4049/jimmunol.1600831. [DOI] [PubMed] [Google Scholar]
- 15.Mokada-Gopal L., Boeser A., Lehmann C.H.K. Identification of novel STAT6-regulated proteins in mouse B cells by comparative transcriptome and proteome analysis. J. Immunol. 2017;198:3737–3745. doi: 10.4049/jimmunol.1601838. [DOI] [PubMed] [Google Scholar]
- 16.García-Serna A.M., Alcaraz-García M.J., Ruiz-Lafuente N. Dock10 regulates CD23 expression and sustains B-cell lymphopoiesis in secondary lymphoid tissue. Immunobiology. 2016;221:1343–1350. doi: 10.1016/j.imbio.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Madan B., Walker M.P., Young R. USP6 oncogene promotes Wnt signaling by deubiquitylating Frizzleds. Proc. Natl. Acad. Sci. USA. 2016;113:E2945–E2954. doi: 10.1073/pnas.1605691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klinkert K., Echard A. Rab35 GTPase: a central regulator of phosphoinositides and F-actin in endocytic recycling and beyond. Traffic. 2016;17:1063–1077. doi: 10.1111/tra.12422. [DOI] [PubMed] [Google Scholar]
- 19.Ishibashi K., Kanno E., Itoh T., Fukuda M. Identification and characterization of a novel Tre-2/Bub2/Cdc16 (TBC) protein that possesses Rab3A-GAP activity. Genes Cells. 2009;14:41–52. doi: 10.1111/j.1365-2443.2008.01251.x. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe C., Shu G.L., Zheng T.S. Caspase 6 regulates B cell activation and differentiation into plasma cells. J. Immunol. 2008;181:6810–6819. doi: 10.4049/jimmunol.181.10.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krebs D.L., Hilton D.J. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Fear D.J., Willis-Owen S.A. Global gene regulation during activation of immunoglobulin class switching in human B cells. Sci. Rep. 2016;6:37988. doi: 10.1038/srep37988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taubenheim N., Tarlinton D.M., Crawford S. High rate of antibody secretion is not integral to plasma cell differentiation as revealed by XBP-1 deficiency. J. Immunol. 2012;189:3328–3338. doi: 10.4049/jimmunol.1201042. [DOI] [PubMed] [Google Scholar]
- 24.James J., Weaver V., Cantorna M.T. Control of circulating IgE by the vitamin D receptor in vivo involves B cell intrinsic and extrinsic mechanisms. J. Immunol. 2017;198:1164–1171. doi: 10.4049/jimmunol.1601213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bod L., Douguet L., Auffray C. IL-4-induced gene 1: a negative immune checkpoint controlling B cell differentiation and activation. J. Immunol. 2018;200:1027–1038. doi: 10.4049/jimmunol.1601609. [DOI] [PubMed] [Google Scholar]
- 26.Richman T.R., Davies S.M., Shearwood A.M. A bifunctional protein regulates mitochondrial protein synthesis. Nucleic Acids Res. 2014;42:5483–5494. doi: 10.1093/nar/gku179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz-Lafuente N., Alcaraz-García M.J., Sebastián-Ruiz S. IL-4 up-regulates miR-21 and the miRNAs hosted in the CLCN5 gene in chronic lymphocytic leukemia. PLoS One. 2015;10:e0124936. doi: 10.1371/journal.pone.0124936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero-Camarero I., Jiang X., Natkunam Y. Germinal centre protein HGAL promotes lymphoid hyperplasia and amyloidosis via BCR-mediated Syk activation. Nat. Commun. 2013;4:1338. doi: 10.1038/ncomms2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X., Lu X., McNamara G. HGAL, a germinal center specific protein, decreases lymphoma cell motility by modulation of the RhoA signaling pathway. Blood. 2010;116:5217–5227. doi: 10.1182/blood-2010-04-281568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hipp N., Symington H., Pastoret C. IL-2 imprints human naive B cell fate towards plasma cell through ERK/ELK1-mediated BACH2 repression. Nat. Commun. 2017;8:1443. doi: 10.1038/s41467-017-01475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z., Lund R., Aittokallio T. Identification of novel IL-4/Stat6-regulated genes in T lymphocytes. J. Immunol. 2003;171:. 3627–3635. doi: 10.4049/jimmunol.171.7.3627. [DOI] [PubMed] [Google Scholar]
- 32.Kanduri K., Tripathi S., Larjo A. Identification of global regulators of T-helper cell lineage specification. Genome Med. 2015;7:122. doi: 10.1186/s13073-015-0237-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material