ABSTRACT
BACKGROUND AND PURPOSE
Neuroinflammation has been implicated in the pathophysiology of Parkinson's disease (PD), which might be influenced by successful neuroprotective drugs. The uptake of [11C](R)‐PK11195 (PK) is often considered to be a proxy for neuroinflammation, and can be quantified using the Logan graphical method with an image‐derived blood input function, or the Logan reference tissue model using automated reference region extraction. The purposes of this study were (1) to assess whether these noninvasive image analysis methods can discriminate between patients with PD and healthy volunteers (HVs), and (2) to establish the effect size that would be required to distinguish true drug‐induced changes from system variance in longitudinal trials.
METHODS
The sample consisted of 20 participants with PD and 19 HVs. Two independent teams analyzed the data to compare the volume of distribution calculated using image‐derived input functions (IDIFs), and binding potentials calculated using the Logan reference region model.
RESULTS
With all methods, the higher signal‐to‐background in patients resulted in lower variability and better repeatability than in controls. We were able to use noninvasive techniques showing significantly increased uptake of PK in multiple brain regions of participants with PD compared to HVs.
CONCLUSION
Although not necessarily reflecting absolute values, these noninvasive image analysis methods can discriminate between PD patients and HVs. We see a difference of 24% in the substantia nigra between PD and HV with a repeatability coefficient of 13%, showing that it will be possible to estimate responses in longitudinal, within subject trials of novel neuroprotective drugs.
Keywords: Parkinson's, neuroinflammation, positron emission tomography (PET), PK11195, image‐derived input function
Introduction
There is mounting evidence that neuroinflammation contributes to the pathophysiology of Parkinson's disease (PD).1, 2 This suggests that changes in biomarkers of neuroinflammation might be useful in the development of novel neuroprotective drugs,3, 4 regardless of whether the pharmaceuticals have direct effects on neuroinflammation. Several positron emission tomography (PET) biomarkers have been developed to quantify the 18 kD translocator protein (TSPO) on the outer mitochondrial membrane as a proxy of neuroinflammation.5, 6 The Carbon‐11 prototype in this class, [N‐methyl‐11C] (R)‐1‐(2‐chlorophenyl)‐N‐(1‐methylpropyl)‐3‐isoquinolinecarboxamide ([11C]‐PK11195 or simply “PK” in this report), has been used in PET studies for estimating the level of regional cerebral neuroinflammation and activated microglia in the brain.7 Even though it shows low brain uptake and nonspecific binding, PK remains a potentially useful tracer because its binding is not influenced by common allelic variations in the TSPO protein.8, 9 Showing that PK is an adequate response biomarker in clinical trials would suggest that new generation TSPO tracers with more favorable imaging characteristics might be outstanding biomarkers.
The gold standard for analysis of dynamic PET studies using PK is a two‐tissue compartmental model using a metabolite‐corrected arterial input function (AIF).10 The usual way to characterize the AIF is to cannulate a radial artery, separate plasma from red cells, and measure the concentration of the radiotracer in plasma.11 While it is feasible and reasonably safe to perform serial arterial punctures within subjects, tolerance for research procedures can vary in ailing populations.12 Procedures that are challenging for the participants, including repeated arterial punctures, could slow subject accrual or contribute to subject dropout rates in longitudinal studies. The risks of arterial cannulation increase in aging populations, and older patients may require medications that have an effect on the blood clotting cascade. These considerations could make some drug developers pessimistic about the potential for using arterial blood‐based image analysis methods in multicenter, longitudinal trials, even though a few single‐site centers of excellence have shown they can do it.
Several reference tissue models exist to quantify uptake, which seem suited for global trials. However, defining a reference region for PK may be difficult since the pattern of activation in PD is not well established. An alternative, which avoids these issues, is the generation of time‐activity curves using imaging of the vascular compartment to form an image‐derived input function (IDIF).
A number of teams have independently created strategies for utilizing IDIFs.13, 14 Validation of the IDIF‐based outcome measures comes from comparing the results to those using the AIF. Characterizing the precision of the IDIF method should allow estimations of the reproducibility required to detect drug‐induced benefits of novel neuroprotective drugs in longitudinal trials.
Since there are many sources of variance, this study attempted to isolate what is termed “system variance” under “zero‐biological‐change” conditions.15 The repeatability coefficient (RC)16 of PK PET was estimated using a test‐retest study design in which the scans were repeated under nearly identical conditions after a 2‐hour interval.
The goal of this project is to determine if these noninvasive methods will make it possible to distinguish PD patients from HV, and if the reproducibility of an individual between scans is sufficient to estimate responses in longitudinal drug studies.
Methods
All procedures were approved by the Institutional Review Board (IRB) of Weill Cornell Medical College. All subjects gave written informed consent. Participants with PD and healthy volunteers (HVs) were enrolled without regard to race or gender. Primary screening was conducted by a senior neurologist specializing in movement disorders (CH). For subjects with PD, selection was based on the inclusion and exclusion criteria described in the UK Brain Bank Criteria.17 HVs could not have any of the corresponding problems. All subjects were required to be older than 40, have an unremarkable medical history except for benign hypertension or hyperlipidemia, and an unremarkable physical examination. All clinical laboratory studies had to be within normal limits, including a complete blood cell count, and tests of liver and renal function. Urinalysis, urine pregnancy tests when applicable, and urine toxicology screens had to be negative. Exclusion criteria included any condition that could alter brain structure or function, including a history of a drug abuse disorder, as well as any disease associated with inflammation. Participants were required to be drug‐free with the exception of medications to treat PD in PD patients, and drugs for common conditions, such as essential hypertension and hypercholesterolemia, in both groups. Subjects who took anti‐inflammatory drugs chronically or any nonsteroidal anti‐inflammatory drugs during the prior 2 weeks were excluded.
All 19 HVs were studied after a single radial artery cannulation, which served both imaging sessions in the 10 subjects who consented to participate in the test‐retest study. The characteristics of the subject population are shown in Table 1.
Table 1.
Clinical Information for Patients with Parkinson's Disease and Healthy Volunteers
| HV | Gender | Age | PD | Gender | Age |
|---|---|---|---|---|---|
| HV#1 | F | 72 | PD#1 | M | 78 |
| HV#2 | F | 42 | PD#2 | F | 67 |
| HV#3 | M | 73 | PD#3 | M | 72 |
| HV#4 | M | 65 | PD#4 | F | 54 |
| HV#5 | M | 51 | PD#5 | M | 57 |
| HV#6 | M | 55 | PD#6 | F | 60 |
| HV#7 | F | 70 | PD#7 | M | 58 |
| HV#8 | F | 56 | PD#8 | M | 73 |
| HV#9 | M | 53 | PD#9 | F | 54 |
| HV#10 | M | 51 | PD#10 | F | 55 |
| HV#11 | M | 66 | PD#11 | M | 73 |
| HV#12 | F | 67 | PD#12 | M | 70 |
| HV#13 | M | 47 | PD#13 | M | 63 |
| HV#14 | M | 49 | PD#14 | M | 63 |
| HV#15 | F | 46 | PD#15 | F | 59 |
| HV#16 | M | 54 | PD#16 | M | 53 |
| HV#17 | F | 46 | PD#17 | M | 69 |
| HV#18 | M | 57 | PD#18 | M | 64 |
| HV#19 | M | 48 | PD#19 | M | 72 |
| PD#20 | M | 59 | |||
| Average Age | 56.3 | 63.7 | |||
| SD | 9.8 | 7.6 |
Clinical information for healthy volunteers and PD patients examined with [11C](R)‐PK11195 PET. PD = Parkinson's disease; HV = healthy volunteer; M = male; F = female; SD = standard deviation.
The radiopharmaceutical, (R)‐[N‐methyl‐11C]PK11195 (PK), was prepared by modifying previously reported synthetic procedures.18, 19 The average specific activity at the end of bombardment (EOB) was ∼1,500 GBq/μmol (40.6 ± 24.5 Ci/μmol) and the synthesis time was 42 ± 2 minutes. All doses were diluted in saline to produce a final volume of 10 mL. The net injected dose averaged 376 ± 61 MBq (10.2 ± 1.6 mCi) for the test condition, and 388 ± 25 MBq (10.5 ± .7 mCi) for the retest condition. The dose was infused over 60 seconds with an automated pump (Graseby 3400, Smith Medical Ltd., UK) in a “slow bolus” paradigm. There were no significant differences in injected mass between test and retest in either HV or PD subjects.
All PET images were acquired with the same lutetium oxyorthosilicate (LSO) time‐of‐flight whole body PET scanner (mCT, Siemens/CTI, Knoxville, TN). The axial field of view (FOV) was 16.2 cm. PET data were reconstructed in a 400 × 400 matrix with a voxel size of 1.082 × 1.082 × 2.025 mm3 using a zoom of 2.0 and an iterative‐plus‐time of flight (+ TOF) list‐mode reconstruction algorithm provided by the manufacturer using OSEM methods with four iterations and 21 ordered subsets. Brain tissue concentrations were estimated by reconstructing the list mode data into 22 frames (four frames of 15 seconds each, then 4 × 30 seconds, 3 × 60 seconds, 2 × 120 seconds, 8 × 300 seconds, and 1 × 600 seconds) for a total scan time of 60 minutes. The carotid arteries were identified using a summed image of the first 90 seconds after radiotracer injection, and the peak of the curve was determined by dividing the first 180 seconds into frames of 15 seconds each.
Magnetic resonance imaging (MRI) sequences included a magnetization‐prepared rapid gradient‐echo (MPRAGE) acquisition protocol to provide optimal gray matter (GM)/white matter contrast using a 32‐channel head coil on a 3.0 Tesla TIM TRIO MRI system (Siemens Healthcare, Cary, NC). A spatial resolution of 1.0 mm × 1.0 mm × 1.0 mm was used with a 256 × 256 matrix, 160 slices, and 2,170 milliseconds/4.3 milliseconds/1,100 milliseconds repetition time (TR)/echo time (TE)/inversion time (TI) with an acceleration factor of 2 and a flip angle of 7°.
Radial artery angiocatheters were placed in all HVs. Participants in the PD group were not given the opportunity to consent for radial artery catheterization based on concerns that it would contribute to dropout rates in future studies that hoped to include the same subjects. An automated fraction collector was used to continuously drip ∼1 mL of whole blood from the radial artery at 15 second intervals for about the first 10 minutes, followed by 2 mL postpurge samples obtained manually at 15, 20, 40, and 60 minutes.
Analysis of [11C](R)‐PK11195 administration for metabolites was performed as described by Roivainen et al.20 The amount of radiotracer bound to plasma protein was assumed constant for each subject over the 2‐hour period of these scans since there was no intervention and the subjects acted as their own controls.
In the primary analysis, the brain image was segmented based on MPRAGE MRI data using FreeSurfer v8.0 (Martinos Center for Biomedical Imaging, Charlestown, MA). Summed PET images were coregistered to their corresponding MRI scans using PMOD® by rigid registration with mutual information (PMOD Technologies Ltd., Zurich, Switzerland). The VOIs included the caudate, anterior putamen, posterior putamen, substantia nigra (SN), thalamus, hippocampus, amygdala, GM, cerebellum, white matter, and CSF bilaterally. Circular region of interest (ROIs) were placed around the carotid arteries and nearby background regions as shown in Figure 1. For the carotid artery, circular ROIs of 4 mm in diameter placed on summed transaxial PET images acquired between 0 and 90 seconds, and then projected onto each dynamic frame to obtain time activity curves (TACs) expressed as % injected dose/cm3 (decay corrected) versus time. Circular ROIs of 8 mm in diameter were placed about 2 cm from the arteries to assess any background spill in. The ROIs were checked on every PET frame using landmarks in the images to ensure that any head movement did not result in misplacement.
Figure 1.
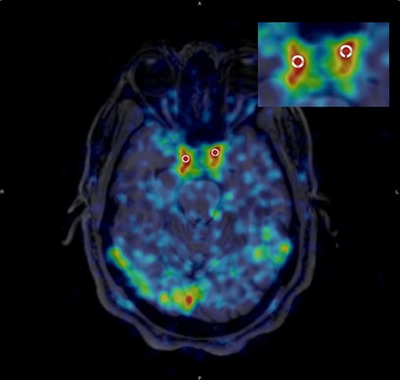
A typical image during the first minute after injection of PK coregistered with MRI. The insert shows the region of interest used to determine the image derived input function.
In a secondary data analysis performed by an independent group in Boston (BOS), motion correction was applied based on rigid registration of each individual frame to the mean of the dynamic PET image. A similar mutual information‐based rigid registration was computed to register the mean PET image to its corresponding MR scan. A study‐specific unbiased reference template was generated using an iterative framework of linear and nonlinear21 registrations applied to the MR data using the Insight Toolkit (ITK). All computed transforms (motion correction, MR registration, and common space registration) were combined prior to application to the dynamic PET data.22
To determine the IDIF, The BOS group, using the summed image of the first 36 seconds as a guide, manually positioned large, encompassing VOIs around the C4 portion of the carotid arteries. An automated algorithm was used to identify the voxels within these VOIs that were most representative of an input function,28 while not an imposed constraint, the algorithm tended to return a cluster of 50–200 voxels within a deep portion of the carotid. These voxels were used with the dynamic PET data to generate an IDIF for each scan.
The true concentration of radioactivity in whole blood (partial volume correction [PVC]) was estimated from images of the internal carotid arteries and surrounding regions following the method of Chen et al26 using Equation (1).
| (1) |
where C measured(t) is the measured level of radioactivity in the blood obtained from the carotid PET image, C carotid(t) is the true concentration of radioactivity in the blood, and C surrounding(t) is the concentration of radioactivity in the tissue near the artery. PV and SF are the experimentally determined partial volume and spillover fraction correction terms required to account for the resolution of the PET scanner and the reconstruction methodology. These terms are calculated as a function of the internal diameter (ID) of each subject's vessel as measured on the corresponding MRI that varied from 4 mm to a maximum of 5.7 mm, with most subjects at about 4.5 mm. The relationship as a function of artery size was determined by fitting the IDIF to the AIF. The IDIF without this PVC was also used for comparison purposes (noPVC‐IDIF).
In the primary analysis done by the NY group, the concentrations of radioactivity in each prespecified brain VOI were determined and then the Logan graphical analysis23 method was used to calculate the total distribution volume (VT) with both the IDIF and AIF in controls and with IDIF in patients using t * = 20 minutes, with a constant weighting factor. The data from each group were expressed as a mean ± SD. The concentration in whole blood as a function of time was analyzed as a linear interpolation of the concentration before the peak and a three‐component exponential fit of concentrations after peak. Kinetic analyses were performed using PMOD 3.5. The Logan reference tissue model24 was applied for calculating nondisplaceable binding potentials (BPND) by the NY group in these studies using the automated reference region extraction.25, 26
The independent BOS group used a fully automated analysis to fit the PET data to a two‐tissue compartment model (2TCM), and to the Logan graphical method. Data were fit using both IDIF and AIF for HV and IDIF for PD for each method. The 2TCM was implemented using a basis function approach.27 Both methods were implemented in MATLAB R2015b (MathWorks, Natick, MA). No weighting was used in either 2TCM or Logan fitting. Logan analysis was performed using t* = 20 minutes.
Descriptive statistics of precision and bias included means, SD, and RCs. The RC values were calculated because they quantify the reliability in the same units as the measurement tool.28 The %RC score includes both random and systematic errors, and gives an interval beyond which the absolute differences between two measurements have a higher than .95 probability of representing a true change. The %RC is calculated using the following equation:16
where P is the number of test‐retest pairs.
The intraclass correlation coefficient (ICC) is a measure of reliability, as indicated by the proportion of the variability in the data that is attributed to differences between methods, for instance, between test and retest measurements. The ICC in this study uses the third formulation defined by Shrout and Fleiss29 as shown in Equation (2).
| (2) |
where MSR and MSE are the mean square and mean square error from a one‐way ANOVA, respectively, and k is the number of repeated sessions. ICC ranges from −1 (no reliability) to 1 (maximum reliability). The descriptive statistics and computations for test‐retest analysis were performed using SPSS (IBM Corp. Version 22.0; IBM Corp, Armonk, NY). In all analyses, the statistical significance (alpha level) was set at P < .05.
In the automated analysis by the BOS group, an independent samples t‐test or Wilcoxon rank sum test was used to compare HV and PD subjects at the regional level, according to the outcome of normality and homogeneity of variance assumption testing. Voxel‐level statistics were performed where an independent samples t‐test was used at each voxel. A false discovery rate correction was applied according to Benjamini and Hochberg30 for both region‐ and voxel‐level results.
Results
Most subjects tolerated the study procedures well. The screening MRI scans excluded one potential HV for occult pathology, while three potential subjects with PD withdrew from the study because of claustrophobia. The experience of some pain and anxiety was not uncommon during arterial cannulation, but it was described as mild and resolved without intervention. One HV experienced progressively worsening soreness with activity that persisted for more than 3 months after the study, and was possibly, but not definitely, related to the procedure. Of the 10 HV subjects in the test‐retest paradigm, 4 were eliminated due to technical problems with either the test or the retest scan leaving 6 for analysis. Nine of the 10 PD subjects completed the test‐retest scans with one eliminated due to a technical problem.
High‐performance liquid chromatography (HPLC) analysis of PK11195 metabolites in plasma was carried out in 10 HVs. The unchanged fraction of PK11195 was 56 ± 21% by the end of the measurement time, as shown in Figure 2. As a result of the uncertainly in the measure in the 10 HV and in consideration of the PD group, an average metabolite profile reported in the literature21 was applied to all subjects. The decision not to do arterial sampling and metabolite correction for the PD population was based the fact that PK PET was only one of many research procedures these participants were exposed to and the potential for fatigue was a major concern. VT values were compared before and after applying this correction giving a consistent increase (44 ± 4%) in values for VT in all subjects.
Figure 2.
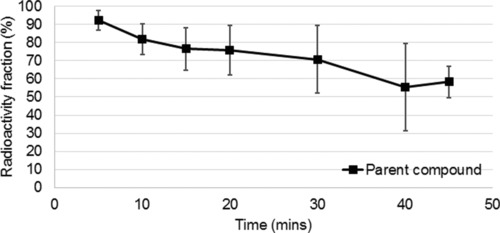
Percentage unchanged tracer as a function of time in healthy volunteers (n = 10).
Assuming the AIF represented the best estimate of the concentration of radioactivity in the blood, the noPVC‐IDIF consistently underestimated the activity at early time points (Fig 3). Correcting for partial volume effects brought the two into close agreement with the area under the curve ratio (AUCR) at .96 ± .18 (see Table 2). In the SN, for example, without PVC, VT was calculated to be .82 compared to .72 calculated using the AIF, but applying the PVC brought the two into close agreement (.70 vs. .72) (see Table 3) Figure 4 shows that the VT estimate using IDIF corrected for partial volume effects in the HV correlates strongly with those calculated with the AIF.
Figure 3.
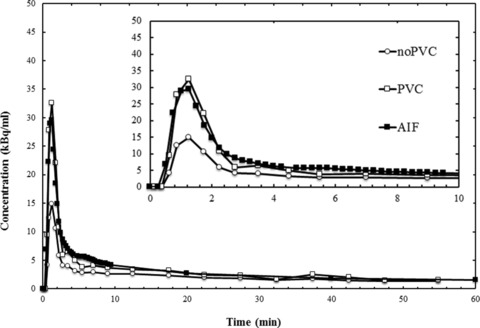
The uncorrected partial volume (noPVC) image‐derived input function (IDIF), the IDIF corrected for partial volume effects (PVC), and the arterial input function (AIF) curves. kBq/mL means kiloBecquerel per milliliter. The insert shows the same data during the first 10 minutes postinjection.
Table 2.
The Ratio of VT Used by Image‐Derived Input Function and Arterial Input Function in Healthy Volunteers
| n = 19 | Region | CAU | PU | TH | SN | CB | GM | HIPP | AMY | WM | CSF |
|---|---|---|---|---|---|---|---|---|---|---|---|
| noPVC | Mean | 1.16 | 1.09 | 1.11 | 1.15 | 1.07 | 1.07 | 1.16 | 1.15 | 1.11 | 1.14 |
| SD | .17 | .28 | .35 | .26 | .27 | .25 | .25 | .28 | .25 | .17 | |
| PVC | Mean | 1.08 | .99 | .99 | .97 | .98 | 1.01 | .96 | .93 | 1.01 | 1.05 |
| SD | .19 | .12 | .14 | .12 | .12 | .13 | .12 | .11 | .14 | .15 |
The ratio of VT values for image‐derived input function divided by values for arterial input function in healthy volunteers with no partial volume correction (noPVC) and with partial volume correction (PVC). CAU = caudate; PU = putamen; TH = thalamus; SN = substatia nigra; CB = cerebellum; GM = gray matter; HIPP = hippocampus; AMY = amygdala; WM = white matter; CSF = cerebrospinal fluid; SD = standard deviation.
Table 3.
Comparison of Test‐Retest Results for Healthy Volunteers
| HV: noPVC | HV: PVC | HV: AIF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Test | Retest | RC(%) | ICC | Test | Retest | RC(%) | ICC | Test | Retest | RC(%) | ICC | |
| CAU‐L | .61±.15 | .64±.20 | 18.7 | .92 | .52±.14 | .52±.16 | 20.7 | .94 | .54±.14 | .54±.17 | 17.4 | .96 |
| CAU‐R | .62±.16 | .66±.17 | 19.7 | .91 | .53±.14 | .54±.15 | 22.9 | .9 | .56±.14 | .56±.15 | 18.5 | .96 |
| PU‐L | .77±.18 | .82±.20 | 17.4 | .91 | .65±.18 | .66±.16 | 16.2 | .95 | .68±.14 | .69±.16 | 10.9 | .97 |
| PU‐R | .78±.18 | .85±.20 | 19.5 | .89 | .66±.18 | .69±.17 | 18.2 | .95 | .70±.14 | .71±.17 | 12.1 | .94 |
| PUA‐L | .78±.19 | .83±.21 | 18.5 | .91 | .65±.19 | .66±.17 | 19.8 | .94 | .69±.14 | .70±.17 | 13.6 | .95 |
| PUA‐R | .79±.19 | .85±.21 | 19.4 | .9 | .66±.18 | .68±.16 | 21.1 | .93 | .71±.14 | .71±.17 | 12.1 | .95 |
| PUP‐L | .80±.18 | .84±.21 | 16.7 | .91 | .67±.20 | .68±.18 | 15.3 | .95 | .72±.14 | .72±.17 | 13.3 | .95 |
| PUP‐R | .80±.20 | .87±.23 | 21.8 | .89 | .67±.21 | .70±.20 | 17.8 | .97 | .73±.14 | .74±.19 | 10.1 | .97 |
| TH‐L | .83±.20 | .89±.23 | 18.6 | .91 | .70±.20 | .71±.18 | 17.2 | .95 | .75±.14 | .75±.19 | 11.4 | .97 |
| TH‐R | .84±.19 | .92±.22 | 19.7 | .89 | .72±.19 | .74±.18 | 17.7 | .94 | .76±.14 | .77±.19 | 13.0 | .96 |
| SN | .82±.21 | .86±.21 | 13.6 | .95 | .70±.21 | .71±.17 | 20.5 | .94 | .73±.14 | .72±.16 | 11.0 | .97 |
| CB‐L | .73±.16 | .79±.18 | 17.8 | .89 | .63±.16 | .65±.16 | 15.7 | .95 | .66±.14 | .67±.15 | 12.6 | .96 |
| CB‐R | .73±.16 | .80±.18 | 18.8 | .89 | .63±.16 | .65±.16 | 17.3 | .94 | .67±.14 | .68±.15 | 12.5 | .97 |
| GM‐L | .70±.14 | .75±.16 | 16.9 | .9 | .61±.13 | .61±.13 | 11.2 | .95 | .63±.14 | .63±.13 | 8.8 | .97 |
| GM‐R | .71±.14 | .75±.16 | 16.4 | .9 | .61±.13 | .61±.13 | 13.6 | .94 | .63±.14 | .64±.13 | 9.9 | .96 |
| HIPP‐L | .72±.17 | .76±.18 | 17.1 | .91 | .60±.17 | .62±.14 | 18.1 | .96 | .64±.14 | .64±.15 | 9.5 | .98 |
| HIPP‐R | .72±.16 | .78±.17 | 20.9 | .88 | .62±.14 | .63±.14 | 13.6 | .95 | .65±.14 | .66±.14 | 10.2 | .97 |
| AMY‐L | .69±.17 | .75±.17 | 21.3 | .87 | .58±.15 | .61±.14 | 23.9 | .9 | .62±.14 | .64±.14 | 15.3 | .96 |
| AMY‐R | .70±.15 | .77±.16 | 23.2 | .83 | .59±.15 | .62±.14 | 25.6 | .89 | .64±.14 | .65±.14 | 18.3 | .89 |
| WM‐L | .68±.15 | .72±.17 | 16.5 | .92 | .60±.15 | .60±.15 | 11.1 | .97 | .62±.14 | .62±.14 | 10.1 | .95 |
| WM‐R | .68±.16 | .73±.17 | 16.2 | .92 | .60±.15 | .60±.15 | 12.1 | .97 | .63±.14 | .63±.14 | 10.1 | .97 |
Logan‐VT values of test and retest repeatability in six healthy volunteers (HVs) using the image‐derived input function (IDIF) with (PVC) and without (noPVC) partial volume correction and using the arterial input function (AIF) are listed. RC(%) is the repeatability coefficient, and ICC is the intraclass correlation coefficient. VOI = volume of interest; CAU = caudate; PU = putamen; PUA = putamen, anterior; PUP = putamen, posterior; WM = white matter; GM = gray matter; CB = cerebellum; TH = thalamus; HIPP = hippocampus; AMY = amygdala; SN = substantia nigra; R = right side; L = left side.
Figure 4.
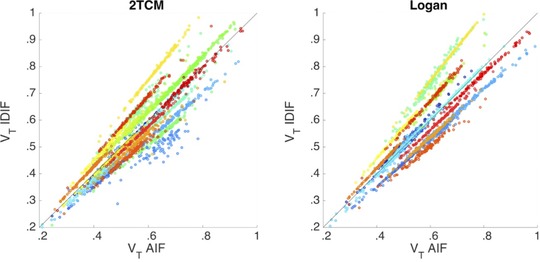
Comparison of distribution volume (VT) values estimated using the image‐derived input function (IDIF) corrected for partial volume effects versus the arterial input function (AIF) for two tissue compartment model (TCM) (left) and Logan (right) methods. Each color represents a unique imaging session and circles represent brain regions. Results are shown for n = 25 healthy volunteers (19 test scans and six retest scans). The solid line is the line of identity.
Table 3 shows the test‐retest variability in VT for the HV (n = 6) comparing noPVC‐IDIF, PVC‐IDIF, and AIF. In PD patients for noPVC‐IDIF, the average RC(%) value was 15 ± 3%, which was significantly more repeatable than the HVs, which was 18 ± 2% (P < .0001), as shown in Table 4. Among the six HVs, calculation of ICCs showed that larger regions exhibited better correlation than smaller regions. For both HV and PD, the RC(%) for most regions and with different input functions was less than 20% indicating good test‐retest repeatability. The correlation between test and retest using the different methods is shown in Figure 5.
Table 4.
The Repeatability of Patients with Parkinson's Disease with/without Corrected Image
| PD: noPVC | PD: PVC | |||||||
|---|---|---|---|---|---|---|---|---|
| VOI | TEST | RETEST | RC(%) | ICC | TEST | RETEST | RC(%) | ICC |
| CAU‐L | .73±.15 | .74±.14 | 22.0 | .87 | .60±.17 | .58±.17 | 22.9 | .93 |
| CAU‐R | .72±.15 | .75±.14 | 17.4 | .93 | .60±.17 | .59±.18 | 17.9 | .94 |
| PU‐L | .98±.13 | .99±.16 | 14.5 | .87 | .81±.21 | .79±.24 | 12.7 | .96 |
| PU‐R | 1.01±.13 | 1.03±.14 | 13.5 | .86 | .83±.20 | .81±.22 | 12.0 | .96 |
| PUA‐L | 1.00±.14 | 1.00±.17 | 14.5 | .88 | .82±.23 | .79±.24 | 13.7 | .97 |
| PUA‐R | 1.01±.13 | 1.02±.17 | 16.2 | .85 | .83±.21 | .81±.24 | 11.7 | .97 |
| PUP‐L | 1.00±.15 | 1.01±.19 | 15.3 | .88 | .83±.23 | .81±.26 | 12.0 | .97 |
| PUP‐R | 1.04±.15 | 1.05±.15 | 14.7 | .86 | .86±.22 | .83±.23 | 13.3 | .96 |
| TH‐L | 1.04±.18 | 1.06±.18 | 16.3 | .88 | .86±.23 | .84±.25 | 15.1 | .96 |
| TH‐R | 1.05±.18 | 1.07±.18 | 16.3 | .9 | .86±.24 | .84±.26 | 14.0 | .96 |
| SN | 1.09±.18 | 1.10±.21 | 11.0 | .82 | .99±.25 | .87±.28 | 16.3 | .95 |
| CB‐L | .90±.14 | 1.83±2.68 | 11.6 | .93 | .77±.21 | .73±.23 | 14.0 | .97 |
| CB‐R | .90±.15 | .91±.15 | 12.5 | .92 | .78±.22 | .75±.23 | 13.6 | .97 |
| GM‐L | .88±.13 | .89±.14 | 11.9 | .91 | .73±.19 | .70±.20 | 13.3 | .96 |
| GM‐R | .89±.13 | .89±.14 | 12.2 | .9 | .74±.19 | .70±.20 | 13.2 | .96 |
| HIPP‐L | .86±.14 | .87±.13 | 18.5 | .84 | .71±.17 | .69±.19 | 18.5 | .93 |
| HIPP‐R | .87±.16 | .89± .14 | 17.3 | .89 | .73±.19 | .70±.20 | 14.7 | .95 |
| AMY‐L | .87±.13 | .90±.14 | 17.3 | .87 | .72±.17 | .71±.20 | 13.8 | .95 |
| AMY‐R | .90±.16 | .91±.14 | 19.0 | .85 | .75±.20 | .72±.20 | 17.1 | .94 |
| WM‐L | .84±.14 | .85±.16 | 13.4 | .92 | .72±.21 | .68±.22 | 13.6 | .97 |
| WM‐R | .85±.15 | .86±.16 | 13.3 | .92 | .72±.21 | .69±.22 | 13.5 | .97 |
Test‐retest repeatability in nine PD subjects using image‐derived input function with and without partial volume correction (PVC). RC(%) is the repeatability coefficient, and ICC is the intraclass correlation coefficient. VOI = volume of interest; PD = Parkinson's disease; CAU = caudate; PU = putamen; PUA = putamen, anterior; PUP = putamen, posterior; WM = white matter; GM = gray matter; CB = cerebellum; TH = thalamus; HIPP = hippocampus; AMY = amygdala; SN = substantia nigra; R = right side; L = left side.
Figure 5.
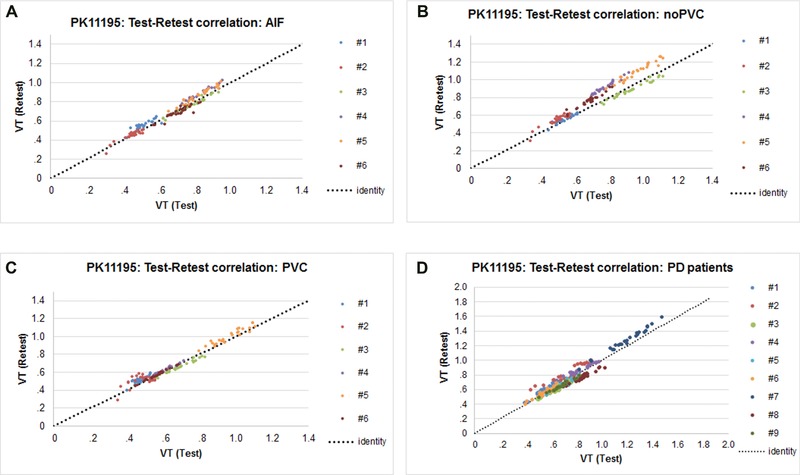
Graph shows correlations between test‐retest with AIF (A), noPVC IDIF (B), PVC IDIF (C), and noPVC IDIF with Parkinson's disease (PD) patients (D). These correlations include the six healthy volunteers and nine PD patients. AIF = arterial input function; IDIF = image‐derived input function; PVC = corrected for partial volume; noPVC = not corrected for partial volume; VT = distribution volume.
A comparison of the average IDIFs for HVs and Parkinson's patients does not indicate any systematic difference in the time activity curves of the blood as shown in Figure 6. The Logan analysis carried out by the NY group using either the IDIF with or without PVC showed widespread and significantly increased VT values in the PD patient group compared to HVs (see Table 5). The uptake was greater by about 19 ± 3%, and similar across all of the brain regions. The mean VT for the 10 regions in each hemisphere was .72 ± .1 in HVs compared to .85 ± .1 in the PD patients (two‐way ANOVA P = .008 for group difference).
Figure 6.
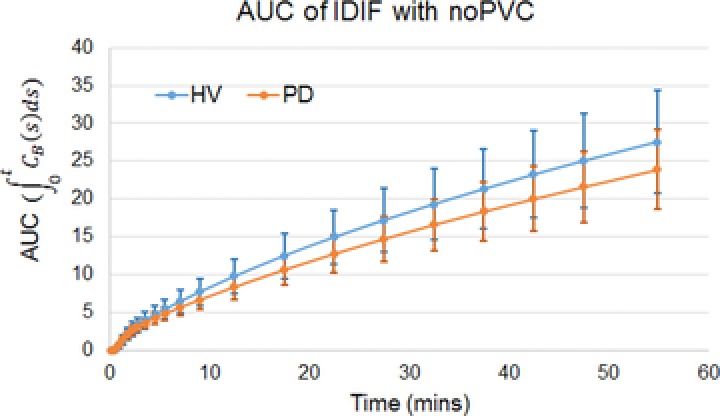
The area under the curve (AUC) of the image‐derived input function (IDIF) without partial volume correction (noPVC) showing no statistically significant difference between healthy volunteers (HVs) and Parkinson's disease (PD) patients.
Table 5.
Values of VT in Patients with Parkinson's Disease and Healthy Volunteers
| HV (n = 19) | PD (n = 20) | |||
|---|---|---|---|---|
| Region | Logan‐VT (mL/cm−3) | Logan‐VT (mL/cm−3) | % Increase in PD | P‐Value |
| CAU | .59±.16 | .68±.18 | 16 | .08 |
| PU | .78±.20 | .93±.17 | 20 | .01 |
| PU‐L | .76±.20 | .92±.17 | 20 | .01 |
| PU‐R | .79±.21 | .95±.17 | 20 | .02 |
| PUA‐L | .77±.20 | .93±.17 | 21 | .02 |
| PUA‐R | .79±.21 | .95±.17 | 20 | .02 |
| PUP‐L | .79±.21 | .94± .18 | 19 | .02 |
| PUP‐R | .80±.22 | .96±.19 | 20 | .02 |
| TH | .84±.22 | 1.00±.20 | 18 | .03 |
| SN | .81±.30 | 1.04±.21 | 28 | .01 |
| CB | .73±.19 | .86±.15 | 18 | .02 |
| GM | .70±.18 | .84±.14 | 19 | .01 |
| HIPP | .72±.19 | .82±.17 | 15 | .07 |
| AMY | .70±.18 | .84±.17 | 20 | .02 |
| WM | .68±.19 | .79±.14 | 17 | .05 |
Calculated P‐values of Logan‐VT between HVs and PD patients using an image‐derived input function without partial volume correction. The % increase values reflect the effect size between the two groups. PD = Parkinson's disease; HV = healthy volunteer; CAU = caudate; PU = putamen; PUA = putamen, anterior; PUP = putamen, posterior; WM = white matter; GM = gray matter; CB = cerebellum; TH = thalamus; HIPP = hippocampus; AMY = amygdala; SN = substantia nigra; R = right side; L = left side.
The independent automated analysis by the BOS group showed both the 2TCM and Logan methods produced similar VT estimates for each subject. VT estimates were globally greater in PD subjects relative to HV subjects by about 21%, while K 1 values (estimated by 2TCM) were not different between groups (Fig 7). Regions that had the greatest percent differences included several areas of the frontal lobe, cingulum, portions of the temporal lobe, and nucleus accumbens. This difference was most statistically significant in the temporal and frontal regions of the brain.
Figure 7.
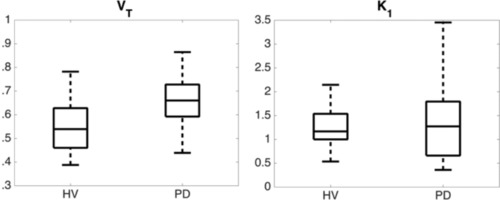
Box and whisker plots of distribution volume (VT) values (left panel) and influx rate (K 1) values (right panel) estimated using two tissue compartment model and partial volume corrected image‐derived input function showing the difference between healthy volunteers (HVs) and patients with Parkinson's disease (PD) indicating no significant difference in influx, but a significant difference in VT values. (left, n = 19 HV and n = 18 PD, P = .0031; right, n = 19 HV and n = 18 PD, P = .60).
For both PD and HV groups, reference tissue curves were generated by Super‐PK software26 (Imperial Innovations, UK). In the test‐retest studies, lower uptake regions showed higher values for the RC (60‐150%), while higher uptake region values had lower values (20‐50%) in both the HV and PD groups due to the higher signal to noise ratio. The ICC values for the PD group were similar to those of the HV group in most brain regions (Table 6).
Table 6.
Test‐Retest Repeatability of Nondisplaceable Binding Potential between Parkinson's Disease and Healthy Volunteers
| BPND | HV (n = 6) | PD (n = 9) | ||||
|---|---|---|---|---|---|---|
| VOI | Mean±SD | RC(%) | ICC | Mean±SD | RC(%) | ICC |
| CAU‐L | −.09±.16 | 126.7 | .95 | 0±.15 | 81.9 | .83 |
| CAU‐R | −.04±.13 | 152.6 | .91 | .01±.13 | 159.1 | .85 |
| PU‐L | .21±.14 | 3.0 | .97 | .36±.15 | 48.6 | .79 |
| PU‐R | .25±.14 | 21.0 | .97 | .40±.14 | 28.3 | .88 |
| PUA‐L | .21±.15 | 37.1 | .95 | .37±.16 | 51.4 | .81 |
| PUA‐R | .24±.15 | 24.0 | .95 | .40±.16 | 38.4 | .90 |
| PUP‐L | .26±.15 | 57.3 | .86 | .40±.16 | 54.6 | .76 |
| PUP‐R | .27±.15 | 42.3 | .91 | .45±.16 | 32.6 | .89 |
| TH‐L | .31±.17 | 21.0 | .97 | .44±.16 | 44.8 | .79 |
| TH‐R | .34±.16 | 25.4 | .96 | .45±.13 | 37.1 | .80 |
| SN | .34±.17 | 25.8 | .94 | .52±.14 | 34.1 | .79 |
| CB‐L | .14±.11 | 9.7 | .91 | .18±.12 | 94.2 | .83 |
| CB‐R | .13±.11 | 109.4 | .94 | .19±.12 | 94.8 | .85 |
| GM‐L | .07±.16 | 57.2 | .98 | .22±.08 | 57.3 | .76 |
| GM‐R | .07±.13 | 71.7 | .98 | .23±.08 | 55.6 | .76 |
| Hipp‐L | .15±.14 | 57.2 | .96 | .20±.12 | 178.0 | .67 |
| Hipp‐R | .16±.13 | 33.4 | .96 | .23±.13 | 85.0 | .80 |
| AMY‐L | .10±.13 | 64.1 | .89 | .22±.12 | 82.1 | .70 |
| AMY‐R | .14±.13 | 142.9 | .88 | .26±.13 | 125.8 | .77 |
| WM‐L | .12±.12 | 145.8 | .97 | .20±.09 | 73.5 | .82 |
| WM‐R | .12±.13 | 102.6 | .98 | .21±.09 | 73.6 | .82 |
Binding potential values of test and retest calculated using the Logan reference tissue model with a reference curve generated by Super‐PK software. BPND = nondisplaceable binding potential; PD = Parkinson's disease; HV = healthy volunteer; CAU = caudate; PU = putamen; PUA = putamen, anterior; PUP = putamen, posterior; WM = white matter; GM = gray matter; CB = cerebellum; TH = thalamus; HIPP = hippocampus; AMY = amygdala; SN = substantia nigra; R = right side; L = left side. RC(%) is the repeatability coefficient, and ICC is the intraclass correlation coefficient.
The Super‐PK analysis showed substantial differences in the SN and cortical GM (SN: P = .03, GM: P = .05, Table 7). In this sample, Super‐PK did not show a significant difference between the PD and HV groups (P = .09) in the putamen, even though the pattern of binding between Super‐PK and the other methods was similar (Fig 8). The Super‐PK analysis did show a difference between PD and HV in the reference region as shown in Figure 9, which may indicate widespread inflammation.
Table 7.
Nondisplaceable Binding Potential in Parkinson's Diseaseand Healthy Volunteers
| HV (n = 17) | PD (n = 19) | |||
|---|---|---|---|---|
| Region | LRTM BPND | LRTM BPND | % Increase in PD | P‐Value |
| CAU | −.04±.18 | −.03 ±.19 | 25 | .76 |
| PU | .27±.18 | .35±.16 | 30 | .09 |
| PU‐L | .26±.14 | .33±.16 | 27 | .07 |
| PU‐R | .29±.16 | .36±.16 | 24 | .11 |
| PUA‐L | .24±.15 | .32±.17 | 30 | .08 |
| PUA‐R | .26±.16 | .36±.17 | 38 | .07 |
| PUP‐L | .26±.18 | .36±.15 | 38 | .24 |
| PUP‐R | .29±.17 | .38±.17 | 31 | .13 |
| TH | .35±.15 | .42±.17 | 20 | .35 |
| SN | .36±.16 | .49±.16 | 36 | .03 |
| CB | .23±.14 | .20±.15 | −13 | .26 |
| GM | .13±.17 | .21±.1 | 61 | .05 |
| HIPP | .16±.14 | .19±.16 | 18 | .75 |
| AMY | .13 ±.17 | .21±.16 | 61 | .18 |
| WM | .15±.14 | .18±.11 | 20 | .41 |
BPND with P‐values for Logan reference tissue model (LRTM) using reference tissue curves generated by Super‐PK with P‐values. The % increase values reflect the effect size between the two groups. BPND = nondisplaceable binding potential; CAU = caudate; PU = putamen; PUA = putamen, anterior; PUP = putamen, posterior; WM = white matter; GM = gray matter; CB = cerebellum; TH = thalamus; HIPP = hippocampus; AMY = amygdala; SN = substantia nigra; R = right side; L = left side; PD = Parkinson's disease; HV = healthy volunteer.
Figure 8.
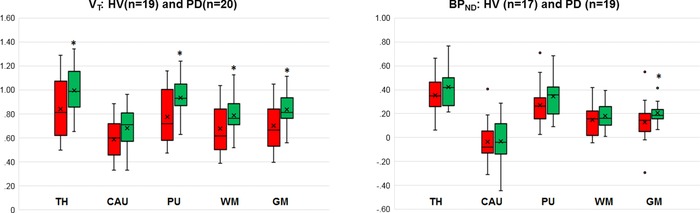
The plot on the left gives the distribution volume (VT) comparison between two groups using image‐derived input function with no partial volume correction (noPVC‐IDIF) and the plot on the right compares nondisplaceable binding potential (BPND) using Super‐PK reference curves. Even though both measures give the same result (patients with Parkinson's disease [PD] have higher binding than healthy volunteers [HVs]), the VT values showed a greater difference in all regions. TH = thalamus; CAU = caudate; PU = putamen; WM = white matter; GM = gray matter. * represents P‐value is less than 0.05 (significant difference).
Figure 9.
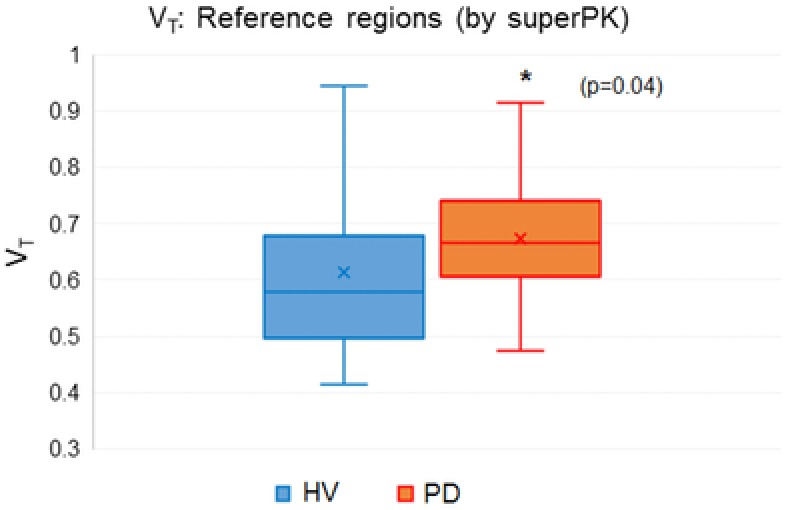
Box and whisker plot of distribution volume (VT) values calculated using the image‐derived input function without partial volume correction (noPVC‐IDIF) at the reference regions defined by the Super‐PK algorithm between the healthy volunteers (HVs) and the Parkinson's patients (PDs). This suggests that there is a general increase in areas assumed to represent nonspecific binding.
Discussion
As stated in the Introduction, the goal of this project is to determine if noninvasive methods will make it possible to distinguish PD patients from HV, and if the reproducibility of an individual between scans is sufficient to estimate responses in longitudinal drug studies. We have shown that values of VT using an IDIF with or without PVC in the same subject are repeatable and that we can easily discriminate between PD and HV groups. We have established the effect size that would be required to distinguish true drug‐induced changes from system variance in longitudinal trials using the Logan graphical method with a noninvasive IDIF blood input function. We have also shown that correction for partial volume effects brings the values from the IDIF noninvasive method into close agreement with those derived from AIF.
Among blood‐based methods, the AIF produced the smoothest curves, the least variance between subjects, and the most repeatable results within subjects confirming that the AIF is the standard against which other methods of quantification should be compared. However, the AIF had more technical failures and the discomfort associated with it was not negligible. VT estimates using an IDIF showed excellent correlation with the AIF across brain regions despite reliance on the concentration of radioactivity in whole blood instead of the free fraction in the plasma. This might be explained by the fact that [11C]PK11195 is greater than 98% plasma protein bound, which does not change significantly over the first 60 minutes postinjection.21
Repeatability was measured after a 2‐hour interval that minimizes the impact of normal biological variability and instrumental instability. When the interval between successive scans is longer, it is quite possible that ordinary fluctuations in biological factors could require larger effect sizes. These biological fluctuations might partially explain why the system variability in this study seems substantially less than the variability reported in prior studies of PK that repeated the scans on different days.24, 31
Some studies investigating the extent of neuroinflammation in PD have found elevated PK binding in PD patients, while others have not. The results from both the NY and BOS groups in this study showed that both the BPND derived from the Super‐PK analysis as shown in Table 6 and the VT based on the IDIF as shown in Table 5 were higher in patients than controls. There were statistically significant differences between PD and HV in more regions than some studies,32, 33 but not in as many as the other analytical methods.34 There were no differences in K 1 values observed between HV and PD groups at either the global or the regional level (see Fig 7). This supports claims suggesting that the greater VT values seen in PD subjects are more likely due to differences in tracer binding rather than blood flow.
In this sample, the power to discriminate between the two groups was better for VT using the IDIF than for the BPND from Super‐PK. The RC% for the test‐retest in PD using VT with or without PVC ranged from 11% for the SN to about 17% for GM (Table 5). This is similar to the values obtained for HV using either AIF or IDIF with or without PVC as shown in Table 3. This can be compared to the average difference of 19% between the HV and PD subjects and a 25% difference in SN and putamen (Table 4).
Super‐PK produced statistically significant differences in only the SN and the cortex. Perhaps, even more importantly for using TSPO tracers in longitudinal trials, the RCs ranged from 21% to 151% for HV and from 34% to 178% for PD subjects (Table 6). This is substantially larger than the difference in BPND values between the PD and HV subjects shown in Table 7. The difference in BPND between the PD and the HV groups might be somewhat obscured because the clustering algorithm generated contaminated reference regions as shown in Figure 9. This could explain the discordance between our positive findings and studies that found no abnormal microglial activation in patients with PD.
One limitation of this study was the absence of individual metabolite correction for the HV and PD groups. Although this is an important consideration, in consideration of PD patient comfort and the fact that we were testing the ability of a test to be applied in a clinical setting where full metabolite analysis might not be possible, we felt that this approach was justified. The point here is to carry out longitudinal studies where the patient is being compared to themselves and therefore the metabolite profile is likely to be relatively consistent.
The results of this study using IDIF seem particularly encouraging since the difference in HV and PD patients is clear, but IDIF avoids some of the potential pitfalls of AIF such as subject selection biases and technical failures during complex procedures. Correcting for partial volume effects makes VT more accurate compared to the AIF but does not improve the sensitivity to changes in neuroinflammation. This is shown in Table 8 where we compare the percent difference between the VT values for HV and PD populations using the IDIF with and without PVC. Our results also demonstrate that the importance of finding a reference region that is free of any interference caused by specific radiotracer binding when calculating BPND using the Super‐PK method.
Table 8.
Comparison in the Percent Difference in Distribution Volume Values Calculated Using Image‐Derived Input Function with or without Partial Volume Correction
| HV | PD | |||||
|---|---|---|---|---|---|---|
| No PVC | PVC | No PVC | PVC | No PVC % diff. | PVC % diff. | |
| CAU | .61 | .53 | .73 | .60 | 18 | 12 |
| PU | .78 | .66 | 1.00 | .82 | 25 | 22 |
| PU‐L | .77 | .65 | .98 | .81 | 24 | 22 |
| PU‐R | .78 | .66 | 1.01 | .83 | 26 | 23 |
| PUA‐L | .78 | .65 | 1.00 | .82 | 25 | 23 |
| PUA‐R | .79 | .66 | 1.01 | .83 | 24 | 23 |
| PUP‐L | .80 | .67 | 1.00 | .83 | 22 | 21 |
| PUP‐R | .80 | .67 | 1.04 | .86 | 26 | 25 |
| TH | .84 | .71 | 1.05 | .86 | 22 | 19 |
| SN | .82 | .70 | 1.09 | .99 | 28 | 34 |
| CB | .73 | .63 | .90 | .78 | 21 | 21 |
| GM | .70 | .61 | .89 | .74 | 24 | 19 |
| HIPP | .72 | .61 | .87 | .72 | 19 | 17 |
| AMY | .70 | .59 | .89 | .74 | 24 | 23 |
| WM | .68 | .60 | .85 | .72 | 22 | 18 |
Comparison of the difference between calculated VT values of healthy volunteer and Parkinson's disease patient populations using image‐derived input functions with and without partial volume correction. HV = healthy volunteers; PD = Parkinson's disease; PVC = partial volume corrected; no‐PVC = no partial volume correction; CAU = caudate; PU = putamen; PUA = putamen, anterior; PUP = putamen, posterior; WM = white matter; GM = gray matter; CB = cerebellum; TH = thalamus; HIPP = hippocampus; AMY = amygdala; SN = substantia nigra; R = right side; L = left side.
It seems likely that an IDIF‐based analysis of PK PET with or without PVC can be a viable alternative to an AIF in the context of multicenter neuroprotective drug development. For a repeated study in any 1 patient, the effect size in the posterior basal ganglia will need to be about 14% for an IDIF to be >95% certain that a change in the VT for PK11195 represents a true change in biological state (see Table 4).
Acknowledgments and Disclosure: This study was designed in collaboration with its sponsor, Biogen Idec, Inc., a pharmaceutical company that has no market in imaging per se, but sometimes uses imaging to make decisions about new drug development. The authors declare that they have no conflicts‐of‐interests. The authors are grateful to Robert B. Innis of NIMH, who provided guidance on the design and execution of the study throughout its development. The authors also want to acknowledge the guidance by Gary S. Dorfman, who shepherded the study and actively edited the protocol, its derivative documents, and this manuscript. Elizabeth Monohan ran the FreeSurfer analyses that delivered the regions of interest. Nancy Obuchowski of the Cleveland Clinic guided the application of the repeatability coefficient as a primary study deliverable and helped make it consistent with the endpoints calculated by the Quantitative Imaging Biomarker Alliance of the Radiological Society of North America. Linda M. Gerber and Gulce Askin of Weill Cornell Medicine reviewed the other statistical analyses. This study was conducted by the professional staff at the Citigroup Biomedical Imaging Center of Weill Cornell Medical College, including, in alphabetical order, John W. Babich, Jonathan Dyke, Simon Morim, Amelia Ng, and Nelsie Pastrano‐Redula.
References
- 1. Tansey MG, Goldberg MS. Neuroinflammation in Parkinson's disease: its role in neuronal death and implications for therapeutic intervention. Neurobiol Dis 2010;37:510‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Le W, Wu J, Tang Y. Protective microglia and their regulation in Parkinson's disease. Front Mol Neurosci 2016;21:9‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sarkar S, Raymick J, Iman S. Neuroprotective and therapeutic strategies against Parkinson's disease: recent perspectives. Int J Mol Sci 2016;17:904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Q, Liu Y, Zhou J. Neuroinflammation in Parkinson's disease and its potential as therapeutic target. Transl Neurodegener 2015;12:4‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ransohoff RM. How neuroinflammation contributes to neurodegeneration. Science 2016;353:777‐83. [DOI] [PubMed] [Google Scholar]
- 6. Venneti S, Lopresti BJ, Wiley CA. Molecular imaging of microglia/macrophages in the brain. Glia 2013;61:10‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vivash L, O'Brien TJ. Imaging microglial activation with TSPO PET: lighting up neurologic diseases? J Nucl Med 2016;57:165‐8. [DOI] [PubMed] [Google Scholar]
- 8. Owen DR, Yeo AJ, Gunn RN, et al. An 18‐kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J Cereb Blood Flow Metab 2012;32:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turkheimer FE, Rizzo G, Bloomfield PS, et al. The methodology of TSPO imaging with positron emission tomography. Biochem Soc Trans 2015;43:586‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kropholler MA, Boellaard R, Schuitemaker A, et al. Development of a tracer kinetic plasma input model for (R)‐[11C]PK11195 brain studies. J Cereb Blood Flow Metab 2005;25:842‐51. [DOI] [PubMed] [Google Scholar]
- 11. Zanotti‐Fregonara P, Chen K, Liow J‐S, et al. Image‐derived input function for brain PET studies: many challenges and few opportunities. J Cereb Blood Flow Metab 2011;31:1986‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Everett BA, Oquendo MA, Abi‐Dargham A, et al. Safety of radial arterial catheterization in PET research subjects. J Nucl Med 2009;50:1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zanotti‐Fregonara P, Hines CS, Zoghbi SS, et al. Population‐based input function and image‐derived input function for [11C](R)‐rolipram PET imaging: methodology, validation and application to the study of major depressive disorder. Neuroimage 2012;63:1532‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fung EK, Planeta‐Wilson B, Mulnix T, et al. Multimodal approach to image‐derived input functions for brain PET. IEEE Nucl Sci Symp Conf Rec 2009;2009:2710‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weber WA, Gatsonis CA, Mozley PD, et al. Repeatability of 18F‐FDG PET/CT in advanced non‐small cell lung cancer: prospective assessment in two multicenter trials. J Nucl Med 2015;56:1137‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Obuchowski NA, Buckler A, Kinahan P, et al. Statistical issues in testing conformance with the quantitative imaging biomarker alliance (QIBA) profile claims. Acad Radiol 2016;23:496‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hughes AJ, Daniel SE, Kilford L, et al. Accuracy of clinical diagnosis of idiopathic Parkinson's disease. A clinico‐pathological study of 100 cases. J Neurol Neurosurg Psychiatry 1992;55:181‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashimoto K, Inoue O, Suzuki K, et al. Synthesis and evaluation of 11C‐PK 11195 for in vivo study of peripheral‐type benzodiazepine receptors using positron emission tomography. Ann Nucl Med 1989;3:63‐71. [DOI] [PubMed] [Google Scholar]
- 19. Shah F, Hume SP, Pike VW, et al. Synthesis of the enantiomers of [N‐methyl‐11C]PK 11195 and comparison of their behaviours as radioligands for PK binding sites in rats. Nucl Med Biol 1994;21:573‐81. [DOI] [PubMed] [Google Scholar]
- 20. Roivainen A, Nagren K, Hirvonen J, et al. Whole‐body distribution and metabolism of [N‐methyl‐11C] (R)‐1‐(2‐chlorophenyl)‐N‐(1‐methylpropyl)‐3‐isoquinolinecarboxamide in humans; an imaging agent for in vivo assessment of peripheral benzodiazepine receptor activity with positron emission tomography. Eur J Nucl Med Mol Imaging 2009;36:671‐82. [DOI] [PubMed] [Google Scholar]
- 21. Avants B, Tustison N, Song G. Advanced normalization tools (ANTS). Insight J 2009;2:1‐35. [Google Scholar]
- 22. Klein S, Staring M, Murphy K, et al. elastix: a toolbox for intensity‐based medical image registration. IEEE Trans Med Imaging 2010;29:196‐205. [DOI] [PubMed] [Google Scholar]
- 23. Logan J. Graphical analysis of PET data applied to reversible and irreversible tracers. Nucl Med Biol 2000;27:661‐70. [DOI] [PubMed] [Google Scholar]
- 24. Logan J, Fowler JS, Volkow ND, et al. Distribution volume ratios without blood sampling from graphical analysis of PET data. J Cereb Blood Flow Metab 1996;16:834‐40. [DOI] [PubMed] [Google Scholar]
- 25. Turkheimer FE, Edison P, Pavese N, et al. Reference and target region modeling of [11C]‐(R)‐PK11195 brain studies. J Nucl Med 2007;48:158‐67. [PubMed] [Google Scholar]
- 26. Yaqub M, van Berckel BN, Schuitemaker A, et al. Optimization of supervised cluster analysis for extracting reference tissue input curves in (R)‐[11C]PK11195 brain PET studies. J Cereb Blood Flow Metab 2012;32:1600‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gunn RN, Gunn SR, Turkheimer E, et al. Positron emission tomography compartmental models: a basis pursuit strategy for kinetic modeling. J Cereb Blood Flow Metab 2002;22:1425‐39. [DOI] [PubMed] [Google Scholar]
- 28. Vaz S, Falkmer T, Passmore AE, et al. The case for using the repeatability coefficient when calculating test‐retest reliability. PLoS One 2013;8:e73990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull 1979;86:420‐8. [DOI] [PubMed] [Google Scholar]
- 30. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc 1995;57:289‐300. [Google Scholar]
- 31. Jučaite A, Cselényi Z, Arvidsson A, et al. Kinetic analysis and test‐retest variability of the radioligand [11C](R)‐PK11195 binding to TSPO in the human brain ‐ a PET study in control subjects. EJNMMI Res 2012;2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerhard A. TSPO imaging in parkinsonian disorders. Clin Transl Imaging 2016;4:183‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koshimori Y, Ko JH, Mizrahi R, et al. Imaging striatal microglial activation in patients with Parkinson's disease. PLoS One 2015;10:e0138721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gerhard A, Pavese N, Hotton G, et al. In vivo imaging of microglia activation with [11C](R)‐PK11195 PET in idiopathic Parkinson's disease. Neurobiol Dis 2006;21:404‐12. [DOI] [PubMed] [Google Scholar]


