Abstract
Previous studies note specific FOXO3 single-nucleotide polymorphisms (SNPs) associated with human longevity. However, it is not clear if these SNPs influence mortality risk beyond the oldest 1 percentile of survival. Using data from four longevity studies (total n = 8,266, age range 96–119 years for cases), we tested gene-wide association between 107 SNPs and survival to at least the oldest 1 percentile of survival for the 1900 birth cohort (≥96, white males; ≥100 white females). This analysis replicated 17 previously published variants, several of which are significant expression quantitative trait loci of FOXO3; rs6911407 and rs2253310 have the most significant effect on FOXO3 expressions in brain tissue. We then performed a survival analysis to determine if any of these 107 SNPs impact upon mortality risk beyond the oldest 1 percentile. While none of the 17 published variants was significantly associated with mortality risk beyond this extreme age, an uncommon homozygote genotype of rs9384680 exhibited the strongest association with mortality risk (p = 2.68E−04) in only 11 females, a heretofore unreported association. These analyses replicate the previous association of common variants of FOXO3 with older age but these common variants do not modify risk for mortality at ages beyond the oldest 1 percentile age of survival.
Keywords: Extreme longevity, Single-nucleotide polymorphisms, Genetic association
Forkhead box O3 (FOXO3) is a constituent of the insulin–IGF1 signaling (IIS) pathway (1), an evolutionarily conserved pathway that has been shown to have a major impact on life span in several model organisms including nematodes (2), fruit flies (3), and mice (4). These results suggested that genes in this pathway may affect human aging and possibly life span, and thus, Willcox and colleagues used a candidate gene approach in demonstrating single-nucleotide polymorphisms (SNPs) in FOXO3 associated with human longevity (5). Following the original work, a total of 17 SNPs in FOXO3 have been associated with what most studies call longevity and what one study calls “extreme longevity” (EL) and some of these SNPs were replicated in multiple cohorts (see Supplementary Table S1 for details). But an important problem that plagues these and other studies is the lack of specificity for the term “longevity” (6). The majority of the studies are of nonagenarians (7–15) (Supplementary Table S2). For whites, according to the U.S. Social Security 1900 birth cohort Life Table, men and women surviving to age 90 years are, respectively, within the 5th and 15th percentiles of survival. In this article, we address this issue of what phenotype of aging (percentile of aging) these FOXO3 variants are actually associated with.
The aim of this work was to systematically examine the association between extreme human longevity and FOXO3 polymorphisms using consistent definitions of cases and controls as suggested in (6), in four studies: the Long Life Family Study (LLFS) (16), the New England Centenarian Study (NECS) (17,18), the Southern Italian Centenarian Study (SICS) (7), and the Longevity Genes Project (LGP) (19). To maintain a consistent definition of extreme longevity and level of selectivity, subjects from each study were included only if they lived past the age at which less than 1% of individuals from the 1900 birth year cohort survived (according to sex and ethnicity). In each cohort, we performed a genetic association study of extreme longevity with 107 SNPs within FOXO3 and combined the results in a meta-analysis. We then used data from the Genotype-Tissue Expression (GTEx) portal (20) to discover if any of the positively associated SNPs were associated with significant changes in FOXO3 expression. Finally, to better understand if SNPs of FOXO3 also affect risk of death in those individuals who survived past the 1 percentile survival age, we conducted a survival analysis restricted to only the EL cases.
Materials and Methods
Definition of Phenotype
The literature regarding human longevity has long been plagued by inconsistent definitions of this phenotype. Different names are used, e.g., “longevity,” “extreme longevity,” “oldest old,” “life span,” etc. and different age cutoffs are used ranging from the 80s to 90s to 100s. These different age criteria represent very different levels of population selection and again very different phenotypes. For example, a large proportion of female nonagenarians (eg, 15th to 10th percentile for the 1900 birth cohort) have long histories of chronic illnesses while a much smaller proportion of females surviving to the oldest 1 percentile (about age 100 years, again for the 1900 birth cohort) do so (18). These very different phenotypes must also have different associated genotypes as well, thus the critical importance, just like other broadly described phenotypes like diabetes, hypertension, heart disease, etc. to be as specific as possible about the phenotype definition. Other investigators have agreed with us and have since adopted this use of birth cohort-specific percentiles of survival to much better define the phenotype they are investigating (21,22). Furthermore, we and other groups have noted the increasing genetic influence with age beyond 95 years upon survival age with older and older ages of survival. Thus, again, it is very important to define the phenotype in terms of the degree of selective survival or how rare the ages of the subjects are (eg, birth year cohort specific percentile of survival).
Following the recommendations in (6,23), extreme longevity was defined as living past the age at which less than 1% of individuals from the 1900 U.S. birth year cohort survived. This definition of cases corresponded to white males aged 96 years and older and white females aged 100 years and older based on birth year cohort life tables from the Social Security Administration (24). Note that these two ages also represent the one percentile survival age in the Danish participants of the LLFS, based on the human mortality database (25). This selection of the one percentile survival age was based on the analysis of the relative risks of extreme survival of sibling of centenarians (6) and is consistent with the definition of longevity used in ref. (5). Controls were defined as either male study participants who died at age less than 90 years, or female study participants who died at age less than 95 years, or study-specific controls (Table 1). We showed in ref. (26) that choosing controls with age at death too different from the minimum age of cases can introduce a bias in the estimation of the genetic effects. Note that with this definition, blood relatives of cases who died at younger ages may be controls.
Table 1.
Characteristics of Subjects in the LLFS, NECS, SICS, and LGP
| Study | Cases (median age, range) | Controls* (median age, range) |
|---|---|---|
| SICS | 126 (99, 96–108) | 540 (41, 18–70) |
| LLFS | 550 (100, 96–111) | 1,210 (73, 43–95) + 1,000 Illumina controls (35, 0–75) |
| LGP | 308 (102, 96–113) | 601 (75, 43–95) |
| NECS | 1,088 (104, 96–119) | 315 (81, 55–95) + 2,593 Illumina controls (35, 0–75) |
| Total | 2,072 | 6,194 |
Note: LGP = Longevity Genes Project; LLFS = Long Life Family Study; NECS = New England Centenarian Study; SICS = Southern Italian Centenarian Study.
*Controls are study-specific controls and/or publically available controls provided by Illumina. Age for the Illumina controls is age at blood draw, though given the rarity of survival to or beyond the oldest 1 percentile of survival, we anticipated that few if any of the controls attained such survival.
Participant Studies
Note that we were careful to include subjects from approximately the same birth cohort, that is, individuals born within 10 years of one another.
LLFS
LLFS is the study of nearly 5,000 individuals from 583 multigeneration families living in the United States and Denmark demonstrating familial longevity. Study eligibility criteria have been described in detail elsewhere (16,27). Generation-specific spouses are referent controls in the study. The study contributed 550 cases and 1,210 controls, including spouses of proband and offspring generation participants.
NECS
NECS is the study of extreme human longevity that has enrolled more than 2,500 centenarians and some of their siblings, about 500 offspring and about 350 controls since 1994 (17). The study contributed 1,088 cases and 315 controls. Controls were spouses of offspring or participants born to at least one parent from the same birth cohort as the centenarian but who died at age 73 years.
SICS
SICS is the study of 410 nonagenarians and centenarians and 553 geographically matched controls from an isolated region of southern Italy east of Naples, with a high prevalence of longevity (28). The study contributed 126 cases and 540 controls. Controls were geographically matched individuals.
LGP
LGP is the study of centenarians, some siblings, offspring, and spousal controls all of Ashkenazi Jewish descent (19). The study contributed 308 cases and 601 controls. Controls were spouses of offspring of centenarians or unrelated individuals of Ashkenazi Jewish descent.
Additional controls
To improve the statistical power of the current study, an additional set of approximately 3,500 controls from the Illumina control database were added to the control sets of the LLFS and NECS. These controls were of European ethnicity as validated by genome-wide principal component analysis. Details concerning these additional controls are described elsewhere (29).
Genotyping and Imputation
Genotype data of the 107 SNPs were a combination of genotyped and imputed data. Illumina-based genome-wide genotype data were available for LLFS as described in ref. (30), NECS, as described in ref. (29), SICS (28) and LGP participants (31). Genotype data for 1324 SNPs were imputed in each study to the 1,000 Genomes Haplotypes Phase I integrated variant set release (in the National Center for Biotechnology Information build 37 coordinates) as the reference panel using IMPUTE2 (32). Imputation was preceded by prephasing with the ShapeIT program (33). Only SNPs with Hardy–Weinberg Equilibrium p-value > 10−6, imputation quality score > 0.9 in at least one study, and minor allele frequency > 5% in at least one study were retained. An additional filter was applied to remove any SNPs for which the difference in the coded allele frequency between the study controls and Illumina controls was greater than the difference in the coded allele frequency between the study controls and cases. After the quality control procedures, 107 SNPs in FOXO3 remained available for analysis.
Statistical Analysis
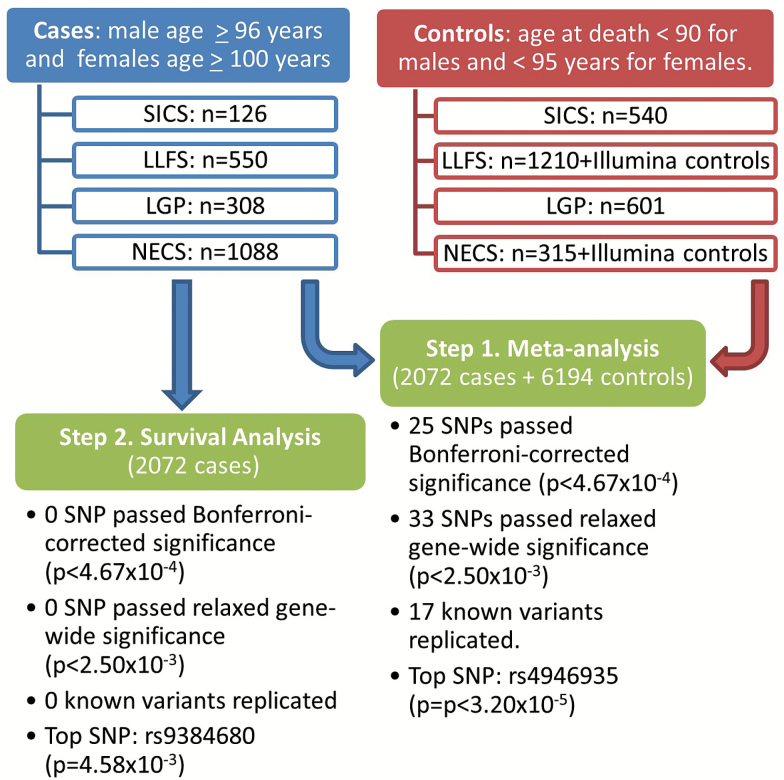
Genome-wide principal component analysis taking into account the family structures was conducted as described in ref. (34). Each study was analyzed independently using hierarchical Bayesian logistic regression, adjusted for sex, principal component 1 (PC1), PC2, PC3, and PC4 using the SNP dosages under additive genetic model (eg, linear effect of the number of coded allele). The top four PCs were included in the model, because they were significantly associated with the outcome variable. Random effects on the log-odds scale were used to model the within-family correlations for related participants in the LLFS, NECS, and LGP, while random effects were not introduced in the SICS data since subjects were independent (35). Marginal estimates of genetic effects were computed using Markov Chain Monte Carlo, with 2,000 iterations after an initial burns-in of 500 iterations, in OpenBUGS. Standard estimates of log-odds ratios and standard errors from Bayesian logistic regression in each study were meta-analyzed using inverse variance weighting implemented in METAL (36). Stringent gene-wide statistical significance was based on Bonferroni correction using the actual number of tested SNPs (0.05/107 = 4.67E−04), and the estimated number of independent loci that was determined to be 20 using the method in (37) leading to gene-wide significance level of 0.05/20 = 2.50E−03. Analyses were also conducted separately in males and females to search for sex-specific genetic effects. Significant expression quantitative trait loci (eQTLs) were discovered using the GTEx portal (20). To investigate how the FOXO3 alleles associate with age at death, survival analysis using a Cox regression model was conducted in the cases only and all study participants, using data aggregated from the four studies. The analysis was adjusted for the top four PCs, it was stratified by sex, and variance correction (robust estimator) was used to account for within family relatedness. The analyses were conducted using the survival package in R v.3, and Kaplan–Meier curves were generated using the survminer package. The study flow of analyses is depicted and summarized in Figure 1.
Figure 1.
Study flow of analyses. In the first step, meta-analysis of case–control data was performed. In the second step, survival analysis in the restricted set of cases only was performed.
Results
Case–Control Meta-analysis
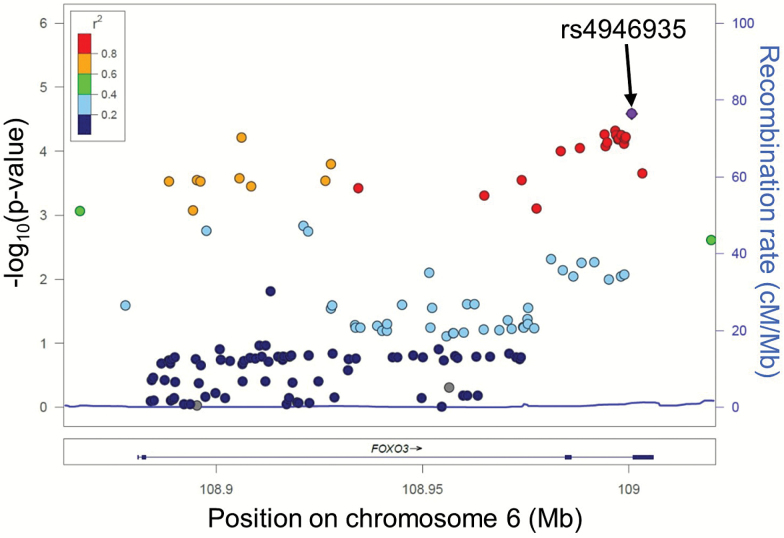
Summary characteristics of participants included in the analysis are depicted in Table 1. Overall, this study included 2,072 cases and 6,194 controls. Supplementary Table S3 shows the results of the association of the 107 SNPs in each individual study. No single SNP in any one study reached significance following Bonferroni correction (p < 4.67E−04). Eleven SNPs in the LLFS and 1 SNP in the SICS reached gene-wide significance (p < 2.50E−03). After combining the results of the studies in the meta-analysis, 25 SNPs out of 107 passed the Bonferroni corrected significance threshold (Table 2), and 33 SNPs passed the gene-wide threshold. The SNP with the lowest p-value was rs4946935 (OR = 1.20, p = 3.20E−05), which was previously shown to be associated with longevity (9). 10 of the 17 published variants (Table 2) surpassed the Bonferroni corrected significance level, and all of the 17 variants were statistically significant at α level of 0.05 with consistent effect directions across the four studies. The effect directions also matched the reported effect directions in publications. The regional plot in Figure 2 shows that the SNPs associated with extreme longevity are all in strong LD and are spread over the whole gene, making it difficult to identify target SNPs for additional studies of the effect of FOXO3 on exceptional longevity.
Table 2.
Meta-analysis of FOXO3 SNPs That Exhibited Statistically Significant Association With Longevity in the Four Populations Studied
| SNP | NCA/ CA | Meta-analysis | SICS | LLFS | LGP | NECS | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | SE | p | CAF1 | CAF0 | β | SE | p | CAF1 | CAF0 | β | SE | p | CAF1 | CAF0 | β | SE | p | CAF1 | CAF0 | β | SE | p | ||
| rs4946935* | G/A | 0.18 | 0.043 | 3.20E−05 | 0.43 | 0.35 | 0.36 | 0.15 | 1.34E−02 | 0.33 | 0.28 | 0.22 | 0.078 | 4.60E−03 | 0.25 | 0.22 | 0.15 | 0.13 | 2.49E−01 | 0.30 | 0.29 | 0.13 | 0.061 | 3.50E−02 |
| rs3800228 | G/T | 0.17 | 0.043 | 4.90E−05 | 0.43 | 0.35 | 0.36 | 0.15 | 1.36E−02 | 0.32 | 0.28 | 0.21 | 0.079 | 8.09E−03 | 0.25 | 0.22 | 0.17 | 0.13 | 1.84E−01 | 0.30 | 0.28 | 0.12 | 0.059 | 3.90E−02 |
| rs3800227 | G/A | 0.18 | 0.045 | 5.60E−05 | 0.4 | 0.32 | 0.37 | 0.15 | 1.36E−02 | 0.30 | 0.26 | 0.22 | 0.082 | 6.28E−03 | 0.22 | 0.20 | 0.13 | 0.13 | 3.27E−01 | 0.27 | 0.26 | 0.13 | 0.063 | 3.80E−02 |
| rs1935951 | A/G | 0.18 | 0.044 | 6.20E−05 | 0.43 | 0.35 | 0.36 | 0.15 | 1.32E−02 | 0.33 | 0.28 | 0.22 | 0.080 | 6.29E−03 | 0.25 | 0.22 | 0.15 | 0.12 | 2.31E−01 | 0.30 | 0.29 | 0.12 | 0.063 | 5.30E−02 |
| rs3800231* | G/A | 0.18 | 0.044 | 6.20E−05 | 0.43 | 0.35 | 0.36 | 0.15 | 1.32E−02 | 0.33 | 0.28 | 0.22 | 0.080 | 6.29E−03 | 0.25 | 0.22 | 0.15 | 0.12 | 2.31E−01 | 0.30 | 0.29 | 0.12 | 0.063 | 5.30E−02 |
| rs9400240 | A/G | 0.18 | 0.044 | 6.20E−05 | 0.43 | 0.35 | 0.36 | 0.15 | 1.32E−02 | 0.33 | 0.28 | 0.22 | 0.080 | 6.29E−03 | 0.25 | 0.22 | 0.15 | 0.12 | 2.31E−01 | 0.30 | 0.29 | 0.12 | 0.063 | 5.30E−02 |
| rs3800229 | T/G | 0.18 | 0.044 | 6.30E−05 | 0.43 | 0.35 | 0.36 | 0.15 | 1.32E−02 | 0.33 | 0.28 | 0.22 | 0.080 | 6.29E−03 | 0.25 | 0.22 | 0.15 | 0.12 | 2.31E−01 | 0.30 | 0.29 | 0.12 | 0.063 | 5.30E−02 |
| rs9374040 | A/G | 0.18 | 0.044 | 6.40E−05 | 0.43 | 0.35 | 0.36 | 0.15 | 1.32E−02 | 0.33 | 0.28 | 0.22 | 0.080 | 6.29E−03 | 0.25 | 0.22 | 0.15 | 0.12 | 2.31E−01 | 0.30 | 0.29 | 0.12 | 0.063 | 5.40E−02 |
| rs1935949* | G/A | 0.18 | 0.044 | 6.60E−05 | 0.43 | 0.35 | 0.36 | 0.15 | 1.32E−02 | 0.33 | 0.28 | 0.22 | 0.080 | 6.29E−03 | 0.25 | 0.22 | 0.15 | 0.12 | 2.31E−01 | 0.30 | 0.29 | 0.12 | 0.063 | 5.60E−02 |
| rs9398172 | A/G | 0.17 | 0.043 | 7.10E−05 | 0.43 | 0.35 | 0.37 | 0.15 | 1.21E−02 | 0.33 | 0.28 | 0.21 | 0.078 | 6.56E−03 | 0.25 | 0.22 | 0.15 | 0.12 | 2.31E−01 | 0.30 | 0.29 | 0.12 | 0.062 | 5.90E−02 |
| rs12206094* | C/T | 0.18 | 0.044 | 7.50E−05 | 0.41 | 0.33 | 0.34 | 0.15 | 2.44E−02 | 0.32 | 0.27 | 0.25 | 0.081 | 1.76E−03 | 0.24 | 0.21 | 0.16 | 0.13 | 2.18E−01 | 0.28 | 0.27 | 0.10 | 0.062 | 9.80E−02 |
| rs2153960 | A/G | 0.17 | 0.043 | 7.90E−05 | 0.45 | 0.36 | 0.43 | 0.15 | 3.52E−03 | 0.34 | 0.29 | 0.20 | 0.081 | 1.27E−02 | 0.28 | 0.25 | 0.15 | 0.12 | 1.92E−01 | 0.31 | 0.30 | 0.11 | 0.062 | 6.90E−02 |
| rs1935952 | C/G | 0.17 | 0.042 | 7.90E−05 | 0.43 | 0.35 | 0.36 | 0.15 | 1.32E−02 | 0.33 | 0.28 | 0.21 | 0.077 | 6.84E−03 | 0.25 | 0.22 | 0.13 | 0.13 | 3.15E−01 | 0.30 | 0.29 | 0.12 | 0.060 | 4.70E−02 |
| rs35396874 | T/C | 0.17 | 0.043 | 9.00E−05 | 0.43 | 0.35 | 0.37 | 0.15 | 1.14E−02 | 0.33 | 0.28 | 0.21 | 0.078 | 7.60E−03 | 0.25 | 0.22 | 0.13 | 0.13 | 3.15E−01 | 0.30 | 0.29 | 0.12 | 0.060 | 5.10E−02 |
| rs9398171 | T/C | 0.17 | 0.043 | 1.06E−04 | 0.45 | 0.35 | 0.44 | 0.15 | 3.51E−03 | 0.34 | 0.29 | 0.21 | 0.079 | 8.56E−03 | 0.28 | 0.24 | 0.13 | 0.12 | 2.78E−01 | 0.30 | 0.30 | 0.11 | 0.062 | 8.50E−02 |
| rs2764261 | G/A | 0.15 | 0.039 | 1.77E−04 | 0.55 | 0.47 | 0.34 | 0.15 | 2.06E−02 | 0.42 | 0.38 | 0.12 | 0.072 | 8.87E−02 | 0.43 | 0.37 | 0.20 | 0.11 | 7.02E−02 | 0.39 | 0.38 | 0.12 | 0.056 | 2.90E−02 |
| rs4946936* | C/T | 0.16 | 0.043 | 1.97E−04 | 0.43 | 0.36 | 0.34 | 0.15 | 2.13E−02 | 0.33 | 0.29 | 0.20 | 0.078 | 1.01E−02 | 0.27 | 0.25 | 0.12 | 0.12 | 3.10E−01 | 0.30 | 0.30 | 0.11 | 0.062 | 6.90E−02 |
| rs2253310* | G/C | 0.14 | 0.039 | 2.31E−04 | 0.54 | 0.47 | 0.31 | 0.14 | 3.32E−02 | 0.41 | 0.38 | 0.12 | 0.072 | 1.06E−01 | 0.41 | 0.36 | 0.18 | 0.10 | 7.81E−02 | 0.39 | 0.38 | 0.13 | 0.056 | 2.60E−02 |
| rs2490272 | T/C | 0.14 | 0.040 | 2.80E−04 | 0.54 | 0.47 | 0.32 | 0.14 | 2.57E−02 | 0.41 | 0.38 | 0.11 | 0.075 | 1.45E−01 | 0.42 | 0.36 | 0.18 | 0.11 | 9.54E−02 | 0.39 | 0.38 | 0.13 | 0.056 | 2.20E−02 |
| rs2802288* | G/A | 0.15 | 0.040 | 2.85E−04 | 0.54 | 0.47 | 0.30 | 0.14 | 3.42E−02 | 0.41 | 0.38 | 0.12 | 0.073 | 1.13E−01 | 0.41 | 0.36 | 0.18 | 0.11 | 9.25E−02 | 0.39 | 0.38 | 0.13 | 0.057 | 2.60E−02 |
| rs2802290 | A/G | 0.15 | 0.040 | 2.85E−04 | 0.54 | 0.47 | 0.30 | 0.14 | 3.40E−02 | 0.42 | 0.38 | 0.12 | 0.076 | 1.11E−01 | 0.41 | 0.36 | 0.18 | 0.11 | 9.25E−02 | 0.39 | 0.38 | 0.12 | 0.056 | 2.70E−02 |
| rs13217795* | T/C | 0.16 | 0.043 | 2.96E−04 | 0.46 | 0.37 | 0.42 | 0.15 | 4.78E−03 | 0.34 | 0.30 | 0.21 | 0.077 | 5.60E−03 | 0.27 | 0.24 | 0.15 | 0.12 | 2.00E−01 | 0.30 | 0.30 | 0.07 | 0.062 | 2.48E−01 |
| rs2802295 | G/A | 0.14 | 0.040 | 3.30E−04 | 0.54 | 0.47 | 0.32 | 0.14 | 2.75E−02 | 0.42 | 0.38 | 0.11 | 0.075 | 1.37E−01 | 0.42 | 0.36 | 0.20 | 0.11 | 7.28E−02 | 0.39 | 0.38 | 0.12 | 0.055 | 3.00E−02 |
| rs2802292* | T/G | 0.14 | 0.040 | 3.41E−04 | 0.54 | 0.47 | 0.30 | 0.14 | 3.46E−02 | 0.41 | 0.38 | 0.11 | 0.074 | 1.23E−01 | 0.41 | 0.36 | 0.18 | 0.11 | 9.25E−02 | 0.39 | 0.38 | 0.12 | 0.056 | 2.80E−02 |
| rs2764264* | T/C | 0.15 | 0.042 | 4.15E−04 | 0.47 | 0.37 | 0.46 | 0.15 | 1.88E−03 | 0.35 | 0.31 | 0.20 | 0.077 | 1.05E−02 | 0.30 | 0.26 | 0.12 | 0.12 | 3.02E−01 | 0.31 | 0.31 | 0.08 | 0.061 | 2.15E−01 |
| rs10457180* | A/G | 0.15 | 0.043 | 5.29E−04 | 0.47 | 0.37 | 0.44 | 0.15 | 3.04E−03 | 0.34 | 0.30 | 0.21 | 0.078 | 7.72E−03 | 0.28 | 0.25 | 0.13 | 0.12 | 2.68E−01 | 0.30 | 0.30 | 0.07 | 0.060 | 2.70E−01 |
| rs9400239* | C/T | 0.15 | 0.044 | 8.42E−04 | 0.47 | 0.38 | 0.41 | 0.15 | 6.42E−03 | 0.34 | 0.30 | 0.20 | 0.079 | 1.03E−02 | 0.30 | 0.27 | 0.11 | 0.13 | 3.74E−01 | 0.31 | 0.30 | 0.07 | 0.063 | 2.51E−01 |
| rs6911407* | C/A | 0.13 | 0.040 | 1.02E−03 | 0.54 | 0.47 | 0.30 | 0.15 | 4.62E−02 | 0.41 | 0.38 | 0.11 | 0.074 | 1.29E−01 | 0.38 | 0.34 | 0.17 | 0.11 | 1.36E−01 | 0.39 | 0.38 | 0.11 | 0.057 | 5.10E−02 |
| rs479744* | G/T | 0.15 | 0.049 | 2.25E−03 | 0.29 | 0.26 | 0.17 | 0.17 | 3.11E−01 | 0.21 | 0.20 | 0.10 | 0.090 | 2.46E−01 | 0.18 | 0.15 | 0.22 | 0.13 | 9.64E−02 | 0.21 | 0.20 | 0.15 | 0.069 | 2.90E−02 |
| rs13220810* | C/T | 0.11 | 0.047 | 1.55E−02 | 0.85 | 0.8 | 0.33 | 0.20 | 9.28E−02 | 0.77 | 0.75 | 0.10 | 0.084 | 2.23E−01 | 0.79 | 0.74 | 0.30 | 0.12 | 1.41E−02 | 0.76 | 0.76 | 0.04 | 0.066 | 5.44E−01 |
| rs9486902* | C/T | 0.12 | 0.055 | 2.64E−02 | 0.23 | 0.21 | 0.12 | 0.18 | 5.08E−01 | 0.16 | 0.15 | 0.11 | 0.099 | 2.52E−01 | 0.08 | 0.07 | 0.09 | 0.20 | 6.35E−01 | 0.16 | 0.15 | 0.13 | 0.077 | 8.30E−02 |
| rs7762395* | G/A | 0.12 | 0.055 | 2.84E−02 | 0.23 | 0.21 | 0.17 | 0.18 | 3.33E−01 | 0.15 | 0.14 | 0.10 | 0.101 | 3.38E−01 | 0.08 | 0.07 | 0.15 | 0.20 | 4.57E−01 | 0.16 | 0.15 | 0.12 | 0.076 | 1.10E−01 |
Note: CA = Coded allele; CAF1 = Coded allele frequency in cases; CAF0 = Coded allele frequency in controls; LGP = Longevity Genes Project; LLFS = Long Life Family Study; NECS = New England Centenarian Study; NCA = Noncoded allele; SICS = Southern Italian Centenarian Study; SNP = Single-nucleotide polymorphism; β and SE in each study are the mean and standard deviation of the posterior estimates of the log-odds ratio for extreme longevity in carriers of the coded allele.
*Previously published variants.
Figure 2.
Regional association plot of meta-analysis results. The x-axis represents the genomic positions within FOXO3, the y-axis on the left represents –log10 of the p-values from the meta-analysis, and the y-axis on the right represent the recombination rate. The colors show the strength of the linkage disequilibrium between the most strongly associated single-nucleotide polymorphism (SNP) (rs4946935) and the other FOXO3 SNPs studied.
Based on the past literature that suggests FOXO3 alleles may promote longevity only in males (5), sex-specific meta-analyses were also performed. In males, there were five SNPs that were gene-wide significant. However, no SNP reached gene-wide significance in females. For the 33 SNPs that reached gene-wide significance threshold in the sex-combined meta-analysis, the effect directions were consistent between males and females, but the magnitude of the effect (on the log odds scale) decreased in females by up to 50%. For example, rs2802292 has an estimated Beta effect of -0.18 in males (OR = 0.84, p = 3.15E−03), but only −0.10 in females (OR = 0.90, p = 4.58E−02). This SNP is the first FOXO3 variant that was shown to be associated with human longevity in male nonagenarians (5). The full results of sex-specific meta-analysis can be found in the Supplementary Table S3.
Survival Analysis
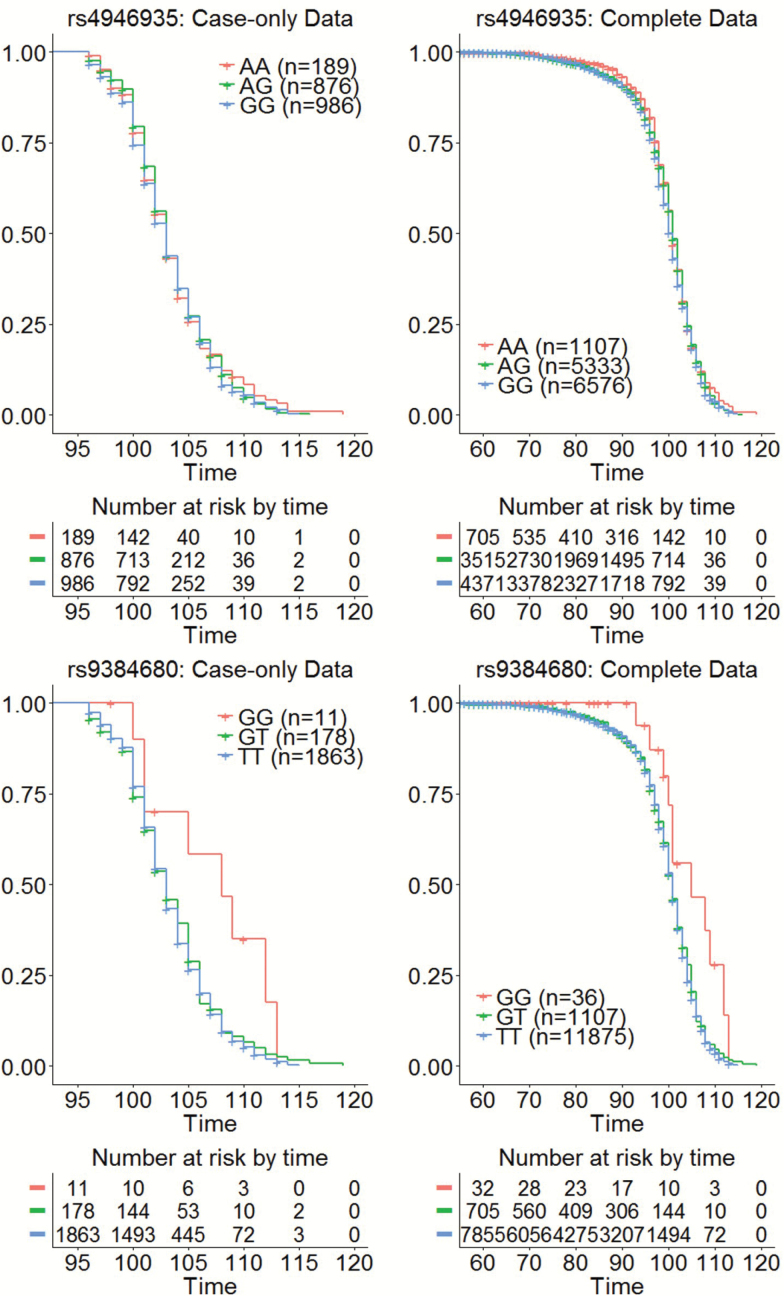
For all of these 33 SNPs that reached at least gene-wide significance, the directions of estimated effects were consistent in the four studies. However, both the magnitude of genetic effects and their frequencies of association with longevity tended to decrease with more extreme ages (Figure 3 and Table 2). We therefore wanted to better understand what degree of survival FOXO3 variants are actually associated with. We hypothesized that if these variants convey an increased risk of living to the 10th percentile of survival, then people who live to the oldest one percentile would also have these variants. The presence of these variants in centenarians does not necessarily mean that the FOXO3 variants influence who survives to the oldest one percentile instead of the 10th percentile. We therefore conducted a series of analyses of age at death using aggregated data from the four studies to better characterize the effect of FOXO3 on survival to extreme old age, conditional on having survived to the 1 percentile survival age. In this analysis, we included only cases, and the estimated hazard ratio for the top SNP in the meta-analysis (rs4946935) was 0.953, but this association was not statistically significant (p = 2.53E−01). The Kaplan–Meier curve (Figure 4) for the three genotypes of rs4946935 clearly shows that there is no discernible difference in survival for the three genotype groups. None of the additional 106 SNPs reached gene-wide significance in the survival analysis after Bonferroni correction for multiple comparisons (Supplementary Table S4). The most significant SNP in the survival analysis in cases was rs9384680 (Hazard ratio = 0.87 and p = 8.76E−02) and this SNP was not significant in the meta-analysis of the four case–control studies (Table 3). The Kaplan–Meier curve (Figure 4, Supplementary Figure S1) shows that homozygotes for the longevity allele (G) may have some survival advantage, but there were only 11 participants with this genotype. Under the recessive genotype model (carrying the recessive genotype versus not), rs9384680 had an estimated hazard ratio of 0.45 (p = 4.58E−03). The frequency of G allele in each study ranged from 4% to 6%, suggesting that rs9384680 was an uncommon SNP. When the same analysis was repeated in males and females separately, the estimated hazard ratio for rs9384680 was 0.37 (p = 2.68E−04) in females, but the hazard ratio was 1.45 (p = 2.66E−01) in males.
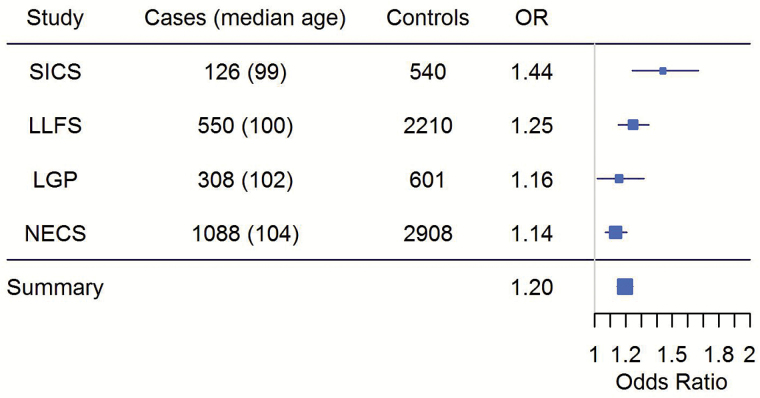
Figure 3.
Forest plot of odds ratios for rs4946935 in the SICS, LLFS, LGP, NECS, and meta-analysis. LGP = Longevity Genes Project; LLFS = Long Life Family Study; NECS = New England Centenarian Study; SICS = Southern Italian Centenarian Study.
Figure 4.
Kaplan–Meier curve of survival stratified by the genotypes of rs4946935 and rs9384690. Blue: carriers of homozygote non-longevity variant. Red: carriers of the heterozygote genotype. Green: carriers of the homozygote longevity variant. Left: analysis in survivors past the age at which less than 1% of the 1900 birth year cohort survived. Right: analysis includes all study participants. Note that the risk table at the bottom of each Kaplan–Meier curve excludes alive subjects at last contact since their age at death is censored. The Kaplan–Meier curves are generated assuming noninformative censoring.
Table 3.
Top SNPs in the Survival Analysis in the Case-Only Data and Complete Data.
| a) Case-Only Data | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | NCA | CA | Survival Analysis in Case-Only Data | Case–Control Meta-analysis | SICS | LLFS | LGP | NECS | ||||||
| HR | p | OR | p | OR | p | OR | p | OR | p | OR | p | |||
| rs9384680 | T | G | 0.87 | 8.76E−02 | 1.08 | 4.20E−01 | 1.32 | 4.32E−01 | 0.82 | 2.79E−01 | 1.25 | 3.19E−01 | 1.14 | 3.13E−01 |
| rs9384681 | A | G | 0.87 | 9.00E−02 | 1.07 | 4.40E−01 | 1.31 | 4.50E−01 | 0.82 | 2.93E−01 | 1.25 | 3.19E−01 | 1.13 | 3.38E−01 |
| rs57099886 | G | A | 0.87 | 9.01E−02 | 1.07 | 4.62E−01 | 1.30 | 4.63E−01 | 0.81 | 2.58E−01 | 1.25 | 3.19E−01 | 1.13 | 3.38E−01 |
| rs72940674 | C | T | 0.87 | 9.02E−02 | 1.08 | 4.08E−01 | 1.32 | 4.42E−01 | 0.83 | 3.01E−01 | 1.25 | 3.19E−01 | 1.14 | 2.97E−01 |
| rs9372188 | C | T | 0.87 | 9.02E−02 | 1.08 | 3.76E−01 | 1.33 | 4.26E−01 | 0.84 | 3.29E−01 | 1.25 | 3.19E−01 | 1.14 | 2.96E−01 |
| b) Complete Data | ||||||||||||||
| SNP | NCA | CA | Survival Analysis in Complete Data | Case–Control Meta-analysis | SICS | LLFS | LGP | NECS | ||||||
| HR | p | OR | p | OR | P | OR | p | OR | p | OR | p | |||
| rs6911407* | C | A | 0.93 | 7.92E−03 | 1.14 | 1.02E−03 | 1.34 | 4.62E−02 | 1.12 | 1.29E−01 | 1.18 | 1.36E−01 | 1.12 | 5.08E−02 |
| rs479744* | G | T | 0.92 | 2.72E−02 | 1.16 | 2.25E−03 | 1.19 | 3.11E−01 | 1.11 | 2.46E−01 | 1.25 | 9.64E−02 | 1.16 | 2.91E−02 |
| rs2802290 | A | G | 0.94 | 3.27E−02 | 1.16 | 2.85E−04 | 1.36 | 3.40E−02 | 1.13 | 1.11E−01 | 1.20 | 9.25E−02 | 1.13 | 2.68E−02 |
| rs2253310* | G | C | 0.94 | 3.30E−02 | 1.16 | 2.31E−04 | 1.36 | 3.32E−02 | 1.12 | 1.06E−01 | 1.20 | 7.81E−02 | 1.13 | 2.58E−02 |
| rs2764265 | T | C | 0.94 | 3.31E−02 | 1.15 | 1.01E−03 | 1.37 | 2.92E−02 | 1.10 | 1.94E−01 | 1.19 | 1.15E−01 | 1.12 | 4.89E−02 |
| rs2802292* | T | G | 0.94 | 3.40E−02 | 1.15 | 3.41E−04 | 1.35 | 3.46E−02 | 1.12 | 1.23E−01 | 1.20 | 9.25E−02 | 1.13 | 2.83E−02 |
| rs2490272 | T | C | 0.94 | 3.54E−02 | 1.16 | 2.80E−04 | 1.38 | 2.57E−02 | 1.12 | 1.45E−01 | 1.20 | 9.54E−02 | 1.14 | 2.21E−02 |
| rs2802288* | G | A | 0.94 | 3.97E−02 | 1.16 | 2.85E−04 | 1.36 | 3.42E−02 | 1.12 | 1.13E−01 | 1.20 | 9.25E−02 | 1.14 | 2.63E−02 |
| rs3800228 | G | T | 0.94 | 4.34E−02 | 1.19 | 4.86E−05 | 1.44 | 1.36E−02 | 1.23 | 8.09E−03 | 1.19 | 1.84E−01 | 1.13 | 3.94E−02 |
| rs2764261 | G | A | 0.94 | 4.76E−02 | 1.16 | 1.77E−04 | 1.40 | 2.06E−02 | 1.13 | 8.87E−02 | 1.22 | 7.02E−02 | 1.13 | 2.90E−02 |
| rs2802295 | G | A | 0.94 | 4.84E−02 | 1.15 | 3.30E−04 | 1.38 | 2.75E−02 | 1.12 | 1.37E−01 | 1.22 | 7.28E−02 | 1.13 | 3.01E−02 |
Note: Case-only analysis included only subjects who survived beyond the 1 percentile survival of the 1900 birth year cohort (males: age > 95; females: age > 99) while the complete data analysis included all subjects in the four studies. CA = coded allele; HR = hazard ratio for mortality estimated from aggregated data from the four studies; LGP = Longevity Genes Project; LLFS = Long Life Family Study; NCA = noncoded allele; NECS = New England Centenarian Study; OR is the mean of the posterior estimates of the odds ratio for extreme longevity in carriers of the coded allele; SICS = Southern Italian Centenarian Study; SNP = Single-nucleotide polymorphism.
*Previously published variants.
When the same analysis of the age at death was conducted in the aggregated data from all participants in the four studies (N = 13,062; all individuals for whom we had age information in the four studies and Illumina controls), the estimated hazard ratio for rs9384680 was 0.89 (p = 8.42E−02) under the additive genetic model and 0.46 (p = 9.65E−03) under the recessive genetic model. In females, the estimated hazard ratio was 0.43 (p = 9.05E−03); in males, the estimated hazard ratio was 0.64, but this effect was not statistically significant (p = 2.52E−02). The survival analysis in the complete data set yielded no variant that achieved gene-wide significance. Eleven SNPs achieved nominal statistical significance (p < .05), and five of these SNPs were previously published variants (Table 3). The SNP rs4946935 reached only nominal significance in this survival analysis and the Kaplan–Meier curve in Figure 4 shows that the survival advantage in carriers of the longevity variant is limited to ages less than 90 years, assuming noninformative censoring. Full results of the survival analysis in the case-only and complete data sets are available in the Supplementary Tables S4 and S5. Supplementary Figure S3 shows some sensitivity analysis of the Kaplan–Meier curves.
Functional Annotation
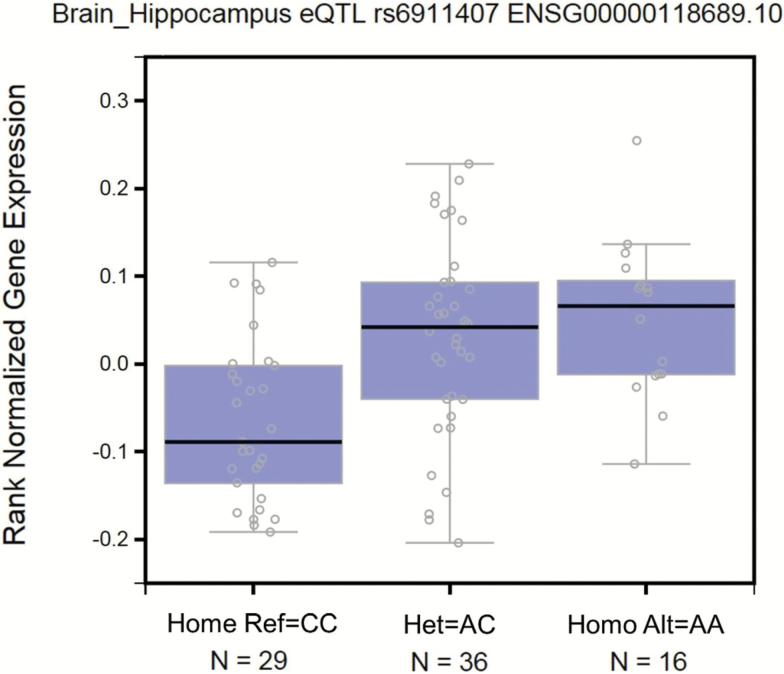
We annotated the 33 variants in Table 3 using eQTL from the GTEx database in all 45 tissues (20). Annotations revealed significant eQTLs in tissues such as brain (hippocampus, cerebellar hemisphere, and cerebellum), pancreas, and transformed fibroblast cells. The most significant eQTL was rs6911407 (p = 3.10E−05) in hippocampus tissue, and the boxplot of expression levels by the three genotypes is shown in Figure 5. This SNP reached gene-wide significance in the meta-analysis but did not reach Bonferroni corrected significance and the longevity allele A was associated with increased expression of FOXO3 in a dominant manner. The second most significant association was for SNP rs2802295 (p = 7.50E−05), also in the hippocampus tissue, which is in strong LD with rs6911407. This SNP showed consistent effects in the four studies and reached Bonferroni corrected significance in the meta-analysis. Additional eQTL SNPs were present in the hippocampus tissue and include rs2253310 and rs2802292 which are also previously published variants. The full results of eQTL annotations are available in Supplementary Table S6. eQTL analysis also showed that the longevity allele of rs9384680 (G) has a significant increase of expression in brain cerebellar hemisphere (p = 2.80E−02).
Figure 5.
Boxplot of gene expression levels by genotype of rs6911407 using the genotype-tissue expression database. Reference allele C, alternative allele A.
Discussion
We examined the association between consistently defined extreme human longevity and 107 SNPs in FOXO3 in more than 2,000 individuals who lived past the age at which just 1% of the 1900 birth year cohort survived, and more than 6,000 controls. The case–control analysis detected 25 SNPs that met Bonferroni corrected level of significance, and 33 that met gene-wide significance, and the SNPs had consistent effects in the four participating studies. In addition, we replicated the association of 17 published variants (ie, statistically significant at α level of 0.05 with consistent effect directions across the four studies), although seven of them reached only nominal significance level. All SNPs associated with extreme longevity were in strong LD.
While the analysis confirms the association of FOXO3 and human extreme longevity, some results were largely driven by the LLFS and the SICS, and evidence from the LGP and NECS was weaker or in some cases was absent. Both the LGP and NECS have enrolled mainly centenarians and several subjects from the NECS survived past age 110, thus the case sets in both studies tend to be older than the case sets in the LLFS and SICS (median ages of cases in LGP and NECS was 102 and 104 years respectively, while the median ages of LLFS and SICS cases were 99 and 100 years, Table 1). Genetic effects that decrease with more extreme ages suggest that the effect of FOXO3 becomes weaker as individuals reach exceptionally old ages, which was also suggested in ref. (10). This observation is also supported by the survival analysis. When only the cases were analyzed in the survival analysis, the SNP rs4946935 was not associated with risk for mortality at ages beyond the 1 percentile survival age (Figure 4). In addition, the frequencies of the longevity associated alleles tend to decrease in older subjects. For example, the frequency of the longevity allele of rs4946936 was 43% in SICS, 33% in LLFS, 30% in NECS, and 26% in LGP. The results suggest that while some FOXO3 variants may have an effect in helping individuals survive to their 90s (eg, 10th–15th percentile for women born in 1900, 5th percentile for men), having these variants will not confer a survival advantage at ages more extreme than the oldest 1 percentile of survival. This result is further confirmed by the Kaplan–Meier survival curves stratified by the genotype of rs4946935 in all study participants that shows a survival advantage limited to ages less than 90 years. We note that the decreasing frequency of EL-associated SNPs in FOXO3 is inconsistent with the original results in (5) that showed increasing prevalence of the longevity allele G of rs2802292. All cohorts included in the present study are of European ethnicities, whereas the study by Willcox et al. (5) included only Japanese or Japanese-American participants, and perhaps some ethnicity-specific effects could explain this difference. Replicating the Willcox et al. findings with a survival analysis similar to what we have performed here in a Japanese centenarian sample would be helpful.
Our analysis in extreme old individuals identified an uncommon genotype in FOXO3 that is associated with different rates of survival in a recessive mode of inheritance in females. This variant did not show significant association in the case-control analyses that used an additive genetic model. The fact that different FOXO3 alleles have different effects on human longevity is consistent with our hypothesis that the genetics of aging and extreme longevity are different (6) and only an analysis restricted to individuals who reached extreme ages can characterize longevity variants that promote survival to the oldest 1 percentile.
Annotation by significant eQTLs through the GTEx portal (20) showed several SNPs in Table 2 associated with changes of expression of FOXO3 in hippocampus and pancreas. SNP rs6911407 had the most significant eQTL in hippocampus, and the longevity allele A was associated with increased expression of the gene. Although a full characterization of the biological mechanisms by which FOXO3 affects human aging remains elusive, it has been conjectured that FOXO3 is implicated with resistance to oxidative stress and activation of FOXO3 may increase antioxidant capacities of cells (38). The analysis shown here suggests that different gene alleles may be associated with different expression, and variants of the gene associated with living longer have higher expression in some tissues and are better equipped to respond to oxidative stress. Interestingly, the two SNPs associated with extreme survival in the case-only analysis also had significant eQTLs in brain cortex. The significant eQTL in different tissues is consistent with the hypothesis that mechanisms leading to older age are different from basic aging mechanisms that would impact all tissues equally.
Limitations
One limitation may come from using external controls from the Illumina repository in the analysis of LLFS and NECS participants. Most of the ages in these controls were censored. From this control set, there may be individuals who may meet the case definition of the current study. However, we expect this number to be small as living past the age at which less than 1% of individuals from the 1900 birth year cohort survived is a rare condition. Another limitation would be that cases and controls are not matched for birth year cohort, which may confound the associations due to unmeasured confounders associated with by different birth year cohorts. However, the survival analysis limited to case only was not affected by this limitation. The sample of extreme survivors included in this analysis may not be representative of extreme survivors in the population, since healthy individuals may be more likely to enroll in studies of extreme longevity. However, if the unmeasured “healthy volunteer effect” is independent of the genetic background, it cannot confound the association between SNPs and EL. If there was an association between SNPs and the healthy volunteer effect, then this factor would likely become a mediator on the casual path between SNPs and EL and ignoring this variable would not bias the estimate of the effects of SNPs on EL. In the survival analysis that included all participants, the birth cohort is confounded with the outcome since controls are born on average 20–25 years after the long-lived individuals. Therefore, we conducted a separate survival analysis stratified by sex and birth cohort. However, the estimates of the genetic effects did not change from our initial analysis, suggesting that confounding was not a major issue (see Supplementary Figure S2 for comparisons). Finally, even though eQTL analysis may reveal initial insight about the potential link between a SNP and a biologic mechanism affecting longevity, identified eQTLs do not necessarily lead to directly interpretable mechanisms. In order to fully characterize the mechanisms by which FOXO3 SNPs affect longevity, more functional analyses are required in the future.
Conclusion
Previous studies assert replicated associations between SNP variants of FOXO3 and human longevity. However, definitions of what studies mean by longevity are inconsistent. The majority of studies used controls either alive at young age or who died at most in their 80s and cases surviving to their 90s. So while variants of FOXO3 appear to affect variation to reach old age, the evidence is not compelling for an effect on survival to extreme old age. Along the same lines, with the controls used in the four studies included in our meta-analysis, we found 17 SNPs associated with aging, replicating previous findings, but the effect of these associations decreased with ages beyond the oldest 1 percentile. Still the role of FOXO3 in human aging remains both elusive and interesting. Using the GTEx database to annotate the significant variants noted in the case-control study, we found several SNPs that had specific effects in the hippocampus indicating the need for tissue specific studies when attempting to decipher the roles of some genes in aging. To understand if any of the FOXO3 variants affected survival to extreme old age, we performed a survival analysis just among the cases and we found one novel rare variant that has a significant effect on survival.
Supplementary Material
Supplementary data is available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Funding
This work was funded by the National Institute on Aging (T.T.P.: U19-AG023122, U01-AG023755; N.B.: P01-AG027734, P30-AG038072; G.A.: R01-AG042188), The William M. Wood Foundation (T.T.P.), the National Institute of General Medical Sciences (P.S.: T32GM074905), and the Glenn Foundation for the Biology of Aging (N.B.).
Conflict of Interest
None reported.
Supplementary Material
References
- 1. Morris BJ, Willcox DC, Donlon TA, Willcox BJ. FOXO3: a major gene for human longevity–a mini-review. Gerontology. 2015;61:515–525. doi:10.1159/000375235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi:10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- 3. Clancy DJ, Gems D, Harshman LG, et al. . Extension of life-span by loss of CHICO, a drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi:10.1126/science.1057991 [DOI] [PubMed] [Google Scholar]
- 4. Bartke A. Impact of reduced insulin-like growth factor-1/insulin signaling on aging in mammals: novel findings. Aging Cell. 2008;7:285–290. doi:10.1111/j.1474-9726.2008.00387.x [DOI] [PubMed] [Google Scholar]
- 5. Willcox BJ, Donlon TA, He Q, et al. . FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–13992. doi:10.1073/pnas.0801030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sebastiani P, Nussbaum L, Andersen SL, Black MJ, Perls TT. Increasing sibling relative risk of survival to older and older ages and the importance of precise definitions of “aging,” “Life Span,” and “Longevity”. J Gerontol A Biol Sci Med Sci. 2016;71:340–346. doi:10.1093/gerona/glv020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anselmi CV, Malovini A, Roncarati R, et al. . Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi:10.1089/rej.2008.0827 [DOI] [PubMed] [Google Scholar]
- 8. Flachsbart F, Caliebe A, Kleindorp R, et al. . Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci USA. 2009;106:2700–2705. doi:10.1073/pnas.0809594106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y, Wang WJ, Cao H, et al. . Genetic association of FOXO1A and FOXO3A with longevity trait in han chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi:10.1093/hmg/ddp459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soerensen M, Dato S, Christensen K, et al. . Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell. 2010;9:1010–1017. doi:10.1111/j.1474-9726.2010.00627.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pawlikowska L, Hu D, Huntsman S, et al. ; Study of Osteoporotic Fractures. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi:10.1111/j.1474-9726.2009.00493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Broer L, Buchman AS, Deelen J, et al. . GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J Gerontol A Biol Sci Med Sci. 2015;70:110–118. doi:10.1093/gerona/glu166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun L, Hu C, Zheng C, et al. . FOXO3 variants are beneficial for longevity in southern chinese living in the red river basin: a case-control study and meta-analysis. Sci Rep. 2015;5:9852. doi:10.1038/srep09852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bao JM, Song XL, Hong YQ, et al. . Association between FOXO3A gene polymorphisms and human longevity: a meta-analysis. Asian J Androl. 2014;16:446–452. doi:10.4103/1008-682X.123673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He YH, Lu X, Yang LQ, Xu LY, Kong QP. Association of the insulin-like growth factor binding protein 3 (IGFBP-3) polymorphism with longevity in Chinese nonagenarians and centenarians. Aging (Albany NY). 2014;6:944–956. doi:10.18632/aging.100703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Newman AB, Glynn NW, Taylor CA, et al. . Health and function of participants in the long life family study: a comparison with other cohorts. Aging (Albany NY). 2011;3:63–76. doi:10.18632/aging.100242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sebastiani P, Perls TT. The genetics of extreme longevity: lessons from the new England centenarian study. Front Genet. 2012;3:277. doi:10.3389/fgene.2012.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andersen SL, Sebastiani P, Dworkis DA, Feldman L, Perls TT. Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J Gerontol A Biol Sci Med Sci. 2012;67:395–405. doi:10.1093/gerona/glr223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Atzmon G, Schechter C, Greiner W, Davidson D, Rennert G, Barzilai N. Clinical phenotype of families with longevity. J Am Geriatr Soc. 2004;52:274–277. doi:10.1111/j.1532-5415.2004.52068.x [DOI] [PubMed] [Google Scholar]
- 20. GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi:10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brodaty H, Woolf C, Andersen S, et al. . ICC-dementia (International Centenarian Consortium - dementia): an international consortium to determine the prevalence and incidence of dementia in centenarians across diverse ethnoracial and sociocultural groups. BMC Neurol. 2016;16:52. doi:10.1186/s12883-016-0569-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeng Y, Nie C, Min JX, et al. . Novel loci and pathways significantly associated with longevity. Sci Rep. 2016;6:21243. doi:10.1038/srep21243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sebastiani P, Gurinovich A, Bae H, et al. . Four genome-wide association studies identify new extreme longevity variants. J Gerontol A Biol Sci Med Sci. 2017; 72:1453–1464. doi:10.1093/gerona/glx027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bell F, Miller M. Life tables for the united states social security area 1900–2100. Actuarial Study No 116. 2005. Retrieved from https://www.ssa.gov/oact/NOTES/pdf_studies/study120.pdf
- 25. Lindahl-Jacobsen R, Hanson HA, Oksuzyan A, Mineau GP, Christensen K, Smith KR. The male-female health-survival paradox and sex differences in cohort life expectancy in Utah, Denmark, and Sweden 1850-1910. Ann Epidemiol. 2013;23:161–166. doi:10.1016/j.annepidem.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sebastiani P, Bae HT, Gurinovich A, Soerensen M, Puca A, Perls TT. Limitations and risks of meta-analyses of longevity studies. Mech Ageing Dev. 2017. In press. doi:10.1016/j.mad.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sebastiani P, Hadley EC, Province M, et al. . A family longevity selection score: ranking sibships by their longevity, size, and availability for study. Am J Epidemiol. 2009;170:1555–1562. doi:10.1093/aje/kwp309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malovini A, Illario M, Iaccarino G, et al. . Association study on long-living individuals from Southern Italy identifies rs10491334 in the CAMKIV gene that regulates survival proteins. Rejuvenation Res. 2011;14:283–291. doi:10.1089/rej.2010.1114 [DOI] [PubMed] [Google Scholar]
- 29. Sebastiani P, Solovieff N, Dewan AT, et al. . Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi:10.1371/journal.pone.0029848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bae HT, Sebastiani P, Sun JX, et al. . Genome-wide association study of personality traits in the long life family study. Front Genet. 2013;4:65. doi:10.3389/fgene.2013.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eny KM, Lutgers HL, Maynard J, et al. ; LifeLines Cohort Study Group; DCCT/EDIC Research Group. GWAS identifies an NAT2 acetylator status tag single nucleotide polymorphism to be a major locus for skin fluorescence. Diabetologia. 2014;57:1623–1634. doi:10.1007/s00125-014-3286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi:10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi:10.1038/nmeth.1785 [DOI] [PubMed] [Google Scholar]
- 34. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi:10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 35. Li B, Lingsma HF, Steyerberg EW, Lesaffre E. Logistic random effects regression models: a comparison of statistical packages for binary and ordinal outcomes. BMC Med Res Methodol. 2011;11:77. doi:10.1186/1471-2288-11-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi:10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2010;34:100–105. doi:10.1002/gepi.20430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15:196–207. doi:10.1111/acel.12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.