Abstract
The special design of the Long Life Family Study provides a unique opportunity to investigate the genetics of human longevity by analyzing data on exceptional lifespans in families. In this article, we performed two series of genome wide association studies of human longevity which differed with respect to whether missing lifespan data were predicted or not predicted. We showed that the use of predicted lifespan is most beneficial when the follow-up period is relatively short. In addition to detection of strong associations of SNPs in APOE, TOMM40, NECTIN2, and APOC1 genes with longevity, we also detected a strong new association with longevity of rs1927465, located between the CYP26A1 and MYOF genes on chromosome 10. The association was confirmed using data from the Health and Retirement Study. We discuss the biological relevance of the detected SNPs to human longevity.
Keywords: Lifespan prediction, GWAS of human longevity, CYP26A1, MYOF, rs1927465
Human lifespan is a complex phenotypic trait with many genetic and non-genetic factors contributing to its variability. This trait is affected by individual aging processes, histories of exposures to external conditions, ontogenetic changes, and individual genetic factors. Although each group of factors makes their contributions to the biological mechanisms involved in the regulation of human longevity, many recent analyses have focused solely on the genetic influences on this trait. This focus can be justified, in part, by the abundance of genetic information on individuals for whom data on lifespan, health-related events, and other variables are available in longitudinal or cross-sectional databases. The fact that non-genetic exposure-related factors influence human longevity by activating appropriate genetic mechanisms has also stimulated analyses of the genetic factors involved in this process.
To clarify the roles of genes in human longevity using large amounts of genetic data on common single nucleotide polymorphisms (SNPs), genome wide association studies (GWAS) of this trait have been conducted. These studies have detected a number of genetic variants strongly associated with longevity that have been also replicated in independent analyses. At the same time, the measured associations of variants from many other genes whose importance for longevity has been established in experimental or molecular biological studies have not reached the level of genome-wide statistical significance. To improve the quality of genetic estimates (ie, increase the likelihood of detecting valid genetic associations), researchers typically increase the sample size, perform meta-analysis of data obtained from several independent studies, use special study designs that increase the per-participant information about genetic influences on longevity, and develop special methods for analyzing incomplete lifespan data.
The most common type of incompleteness in lifespan data is caused by right censoring at the latest observation time in an ongoing longitudinal study. Right censoring also occurs when individuals unexpectedly drop out of an ongoing longitudinal study after participating in the initial wave(s) of the study. As a result of right censoring, it will be known that an individual’s attained lifespan is above a certain age but the exact value will be unknown because the individual is still alive or is not being tracked. Typically, some study subjects are alive while others have dropped out at the time of analysis.
Several alternative approaches can be used to perform genetic analyses of incomplete data on lifespan. One approach deals with methods of survival analyses (eg, Cox regression model) that were specifically developed for incomplete survival data; this approach has been studied extensively. An alternative approach is to predict the final attained lifespan for individuals with right censored data and perform genetic analyses using the predicted data together with the available (non-censored) data on attained lifespans for known decedents. The benefits and limitations of this approach for genetic analysis of human longevity are unclear, however; they have yet to be evaluated.
In this article, we use data on white individuals from the Long Life Family Study (LLFS) to evaluate the use of predicted lifespan data in genetic analyses of longevity. The LLFS is a multi-center longitudinal study designed to investigate environmental and genetic factors that contribute to familial clustering of exceptional longevity and to facilitate detection of genetic factors responsible for human longevity by exploiting the fact that longevity is familial. The study participants resided in the United States and Denmark; eligible families had to demonstrate exceptional longevity based on the Family Longevity Selection Score (FLoSS) (1). The first wave of the LLFS (2006–2009) collected genetic and non-genetic data on two generations of living persons in these families. In the present study, we will use genetic data collected for subjects interviewed in the first wave.
Mortality follow-up after the baseline interview has continued, on average, for about 8 years, and is currently ongoing. The LLFS lifespan data are necessarily incomplete as they are right-censored because the period of follow-up is limited to the current time and death may not yet have occurred. The data are also left-truncated on the age dimension because individuals had different ages at the time of the baseline interview. The age dimension is relevant because the lifespan data we seek are the individual ages at death, or equivalently the maximum attained ages for the study participants. The data collection procedures for recording the complete sets of attained lifespans for both generations of the LLFS participants will take several decades. That is why the use of methods for analyzing incomplete data may provide useful insights about the genetics of human longevity today.
The main aims of this article are to investigate: (a) how to predict censored lifespan data for LLFS participants and to use such predictions; (b) whether the use of predicted data on lifespan in GWAS of human longevity results in more significant estimates of genetic associations compared to analyses of incomplete data without such predictions; (c) which statistical models used in GWAS are most appropriate for such analyses; (d) how the numbers and strengths of detected genetic associations depend on the durations of the follow-up periods; and (e) what biological mechanisms regulating human longevity are represented by the detected genes.
The lifespan prediction model was restricted to study subjects aged 80 years or above at the first wave of the LLFS. This range was chosen because the observed number of deaths below age 80 was too small to support reliable estimation. The lifespan prediction model was used to predict the final attained lifespans for study subjects who were still alive at the end of the 8-year longitudinal follow-up and were at least 80 years old at that time. A series of case–control GWAS were performed by applying various longevity thresholds to two alternate forms of LLFS longevity data—one employing predicted lifespan data to replace incomplete lifespan data; the other—using incomplete (observed) lifespan data. The strongest genetic associations with longevity were obtained when the case group was defined as those who lived to age 96 years or beyond.
Methods
Data
The LLFS is a family-based study of healthy aging and longevity that recruited 583 families and 4,900 family members selected for exceptional familial longevity (1). Participants were enrolled during 2006–2009 at three U.S. field centers (Boston, Pittsburgh and New York) and a European field center in Denmark. Potential probands were recruited based on older age, capacity to understand the study, and their Family Longevity Selection Score (FLoSS). The FLoSS score quantifies familial longevity as well as living sibship size using sex and birth-year cohort survival probabilities of each member of the proband generation and their siblings (1). Sibships were eligible for the study if their FLoSS score was greater than 7—a cutpoint corresponding to the top 0.2% of FLoSS sibships in the Framingham Heart Study—and they had at least one living sibling and at least one offspring willing to be enrolled in the study. Sociodemographics, medical history, current medical conditions/medications, physical/cognitive functioning, and blood samples were collected via in-person visits and phone questionnaires for all subjects at the time of enrollment, as described elsewhere (2). Participants are continuing to be followed-up annually to track vital and health status. The ages of the oldest participants were validated against external data (3). Genotyping has been performed by the Center for Inherited Disease Research (CIDR) using SNP Chips manufactured by Illumina (Human Omni 2.5 v1 BeadChip array). Written informed consent was obtained from all subjects following protocols approved by the respective field center’s IRB. Table 1 shows the numbers of individuals and numbers of deaths, for total and genotyped participants by generation (proband, offspring), country (USA, Denmark, combined), and sex (males, females, total). Other details of study design and protocols are described in (2,4).
Table 1.
Study Population in LLFS
| Generation | Country | Females | Males | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | D | NG | DG | N | D | NG | DG | N | D | NG | DG | ||
| Probands | Denmark | 166 | 125 | 157 | 119 | 95 | 80 | 93 | 78 | 261 | 205 | 250 | 197 |
| Probands | USA | 784 | 460 | 711 | 415 | 659 | 455 | 615 | 429 | 1443 | 915 | 1326 | 844 |
| Probands | Combined | 950 | 585 | 868 | 534 | 754 | 535 | 708 | 507 | 1704 | 1120 | 1576 | 1041 |
| Offspring | Denmark | 525 | 26 | 513 | 26 | 483 | 44 | 473 | 43 | 1008 | 70 | 986 | 69 |
| Offspring | USA | 1308 | 47 | 1199 | 42 | 1003 | 57 | 931 | 54 | 2311 | 104 | 2130 | 96 |
| Offspring | Combined | 1833 | 73 | 1712 | 68 | 1486 | 101 | 1404 | 97 | 3319 | 174 | 3116 | 165 |
| Combined | Denmark | 691 | 151 | 670 | 145 | 578 | 124 | 566 | 121 | 1269 | 275 | 1236 | 266 |
| Combined | USA | 2092 | 507 | 1910 | 457 | 1662 | 512 | 1546 | 483 | 3754 | 1019 | 3456 | 940 |
| Combined | Combined | 2783 | 658 | 2580 | 602 | 2240 | 636 | 2112 | 604 | 5023 | 1294 | 4692 | 1206 |
Note: Number of individuals and number of deaths in total sample (N, D) and genotyped (NG, DG) subsample, by generation, country, and sex in LLFS. The numbers exclude individuals with missing lifespan information which were not included in the analyses. D = number of deaths; DG = number of deaths among genotyped; N = sample size; NG = number of genotyped.
Data Availability
The LLFS data are available in dbGaP and can be obtained using standard procedure, described at dbGaP website. DbGaP Study Accession: phs000397.v1.p1. The details of genotyping and quality control procedures are described in (5).
Survival Models for Predicting Censored Lifespans
The success of the genetic analyses of predicted lifespan data depends on the quality of the predictions. One would think that demographic life tables for the birth cohorts in the United States and Denmark that correspond to the populations containing the LLFS participants can serve as appropriate predictive models. Indeed, the life table data from the Social Security Administration (SSA) for the United States, and from the Human Mortality Database (HMD) (for Denmark) provide us with robust and reliable estimates of survival probabilities because they are constructed using data on hundreds of thousands or millions of people. The use of such demographic life table models would be completely justified for any population-based study. However, the LLFS participants were recruited using special selection criteria that recruited a cohort that was not population-representative by design (1). This means that one must test whether the survival rates in the LLFS sample can be described by the corresponding demographic life tables. For this purpose, we estimated survival functions from the incomplete LLFS follow-up data and compared them with those calculated from demographic life tables. Because the LLFS participants in the proband generation were older than those in the offspring generation at the baseline interview, and hence subject to higher mortality rates, the proband generation provided lifespan data in the follow-up period that was more complete than the lifespan data for the offspring generation. Overall, about half of the members of the proband generation died during the 8-year follow-up period. For the other half, the lifespan data were right censored. For the purpose of generating the lifespan prediction model, we focused on data from the proband generation with lifespan ≥80 years.
The choice of appropriate life-span prediction model was based on comparison of survival functions constructed from demographic life tables with those obtained from survival analyses of the LLFS data. To represent the impact of heterogeneity in individual lifespans within each combination of country, cohort, and sex, we introduced the “index of cumulative deficits” (DI) (6,7)—a composite index (also known as the “frailty index” (8)—as an additional individual-specific covariate in the Cox regression model (see Supplementary Materials for details). The DI is an established indicator of aging that summarizes the effects on lifespan of a large number of variables spanning multiple health-related domains. The DI was verified and intensively used in a number of recent studies of human aging, health, and longevity (8). For the LLFS data, the DI was constructed using 85 variables measured at baseline (9). To make life-span predictions for individual study participants, we first estimated the best fitting Cox regression model using two covariates—country (United States, Denmark) and birth cohort—stratified by sex with attained age as the time-to-event variable.
We combined the data from the three U.S. field centers because the estimated field-center effects on lifespan were similar. We coded the Danish field center using a separate category because its effects on lifespan were significantly different from the U.S. field centers. Then, using Cox regression models with and without DI, we calculated the mean residual lifespan for each individual in the study sample who was censored according to the data collection protocol, conditional on the attained age at the time of censoring. The mean residual lifespan is the minimum-mean-squared-error (MMSE) estimator of the unobserved yet-to-be attained residual lifespans under the assumption that the respective Cox regression models are correct. The mean residual lifespan estimates were added to the ages at censoring for each censored age at death for use as individual-specific predicted lifespans. In this way, the models with and without DI were used for lifespan prediction for censored individuals. Finally, with both sets of lifespan predictions in hand, we employed corresponding logistic regression models with and without DI to quantify the impact of DI on the survival probabilities for the approximately 8-year follow-up period using Receiver Operating Characteristic (ROC) curves and Area Under the Curve (AUC) techniques.
Quality Control for GWAS (QC)
The QC protocol described in (10) was used with the following settings. QC for study participants: call rate ≥95%; (mean − 3SD) < heterozygosity rate < (mean + 3SD). QC for SNPs: call rate ≥95%, minor allele frequency (MAF) ≥ 1%, (Hardy-Weinberg Equilibrium HWE) p-value ≥1E−10. The number of genotyped study participants before QC: 4,693 (2,112 males, 2,581 females). The number of study participants retained after QC: 4,608 (2,072 males, 2,536 females). The number of autosomal SNPs before QC: 2,225,478. The number of autosomal SNPs after QC: 1,464,314.
GWAS
The generalized linear mixed model association test (GMMAT) logistic regression model (11) was used to conduct the GWAS of human longevity. This model adjusts for relatedness among LLFS family members. The trait “PLS” was defined either as predicted lifespan for individuals with censored ages at death or as observed lifespan for deceased individuals. Study participants were classified as cases or controls, or excluded from the analysis, as follows. Cases comprised males and females from the proband generation including spouses with PLS ≥ 96 years (N = 877). Controls comprised males and females from the offspring generation including spouses with attained lifespan (for deceased persons) or censored lifespan (for currently living persons) <75 years (2,462 individuals). Altogether 3,339 study participants were included in the case and control groups. To control for possible population stratification, we used genomic control (GC) and method based on calculation of principal components using genetic data (12). The primary statistical models used in GWAS included country of origin (United States and Denmark) principal components, and sex as covariates in the analyses. We used the country of origin instead of the individual field centers because the analyses showed non-significant differences in the effects of field center on lifespan in the United States but significant difference between the United States and Denmark. In all cases, we calculated the parameter λ for GC and corrected for possible population stratification. The results were compared with those obtained in corresponding GWAS analyses of observed (non-predicted) data and with analyses of lifespan data predicted without using DI as an observed covariate. The GWAS procedures using the GMMAT model that used country of origin and sex as covariates were repeated for different durations of follow-up from the baseline visits in 2006–2009 until 2010, 2011, 2013, and 2015. Again, we emphasize that these follow-up periods were used solely to resolve the problem of lifespan censoring (for persons currently alive in 2010, 2011, 2013, or 2015) by generating individual-specific predictions of the mean residual lifespans that could be added to the ages at censoring for each censored age at death, yielding the individual-specific predicted lifespans. Once these calculations were done, the GWAS of human longevity using GMMAT logistic regression analyses were performed using the retrospective case–control design.
We carefully investigated the role of thresholds defining the case (longevity) group on the results of genetic analyses of lifespan data. For these purposes, we performed a series of GWAS of human longevity using ages 90, 92, 94, 96, 98, and 100 as longevity defining thresholds for the case group and compared results of these analyses. The lowest p-values for genetic genome-wide significant associations with human longevity were obtained for SNPs on chromosomes 10 and 19 when the case group was defined by setting the longevity threshold to 96 years.
The fact that SNPs and the corresponding genes detected on chromosome 19 are well known genetic determinants of human longevity indicates that the “case” group defined by the 96-year longevity threshold includes individuals whose longevity was strongly affected by genetic factors. The pleiotropic effects and biological functions of the detected genes are discussed in (13).
Results
Exceptional Survival of LLFS Participants From the Probands’ Generation
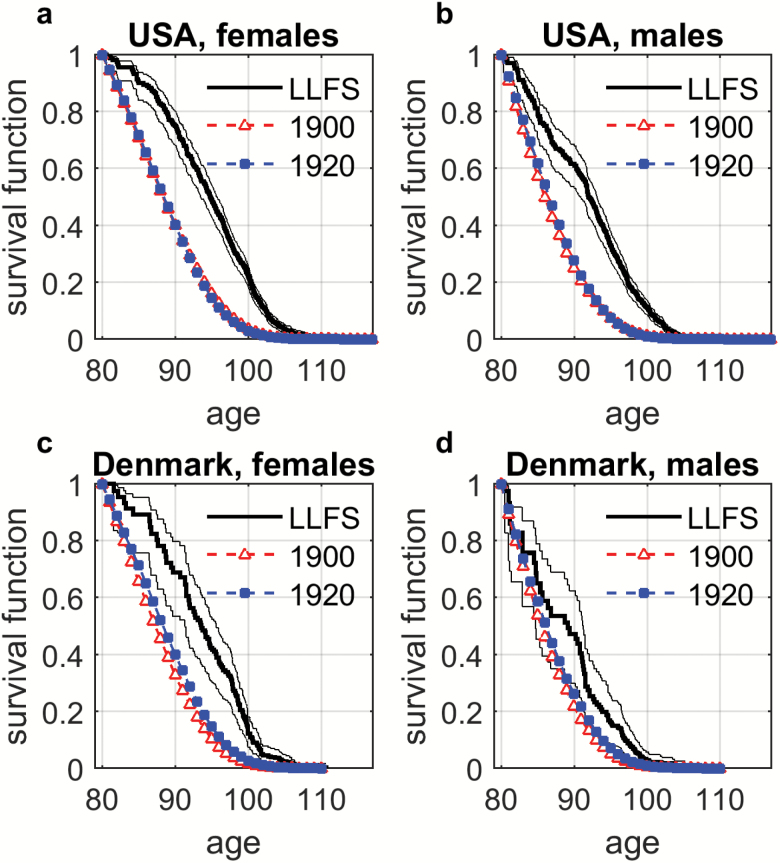
Using incomplete lifespan data collected during about 8 years of follow up since the baseline, we calculated Kaplan–Meier estimates of survival functions for the LLFS’s eldest participants (the probands’ generation) and compared them to survival functions in the 1900 and 1920 U.S. birth cohorts using Social Security Administration (SSA) data (Bell and Miller, 2005). For Denmark, survival functions were calculated using life tables from the HMD. In all cases, survival functions were evaluated for those who survived to age 80. The results are shown in Figure 1.
Figure 1.
Survival functions (conditional on survival to age 80) for the LLFS participants in comparison with 1900 and 1920 birth cohorts: (a) U.S. females; (b) U.S. males; (c) Danish females; (d) Danish males. The LLFS curves are shown by solid thick line with 95% confidence intervals indicated by solid thin lines. Dashed lines display population survival functions from corresponding cohort life tables: Social Security Administration (SSA) for USA and Human Mortality Database (HMD) for Denmark. The numbers after SSA and HMD show the respective birth cohort.
This figure demonstrates that the LLFS participants who survived to age 80 and above have much better survival than their peers represented by the cohort life tables (as expected since the families were selected for exceptional survival). The estimates of survival functions starting below age 80 are not reliable because of the lack of data on deceased individuals in the probands’ generation below this age. The supplementary materials explain why survival of the LLFS participants is much better than the corresponding birth cohorts in the United States and Denmark (see also Supplementary Figure 1).
Lifespan Prediction
Lifespan prediction requires high quality methods for evaluating expected residual lifespans. The simplest prediction is based on demographic information about survival of individuals in the population. This prediction can be improved if additional information on factors affecting human lifespan is used. One such factor is an index of DI whose properties in the LLFS have been evaluated by Kulminski et al. (9). Based on estimated survival functions using the Cox proportional hazard model and values of DI and other covariates (age at baseline, gender, and country), expected residual lifespans were calculated for censored individuals in the LLFS probands’ generation. We then added the estimated residual lifespans to the age at censoring to calculate “predicted” lifespans for the censored cases (see Supplementary Material for further detail). The contribution of the DI to the quality of prediction was estimated using receiver operating characteristic (ROC) curve and area under the curve (AUC) techniques. The results are shown in the Supplementary Figure 2. One can see from this figure that the AUC increased from 0.807 to 0.834 when DI was included as a covariate—about 14% closer to the maximum AUC value of 1.000, strongly supporting the use of DI in our lifespan models.
The lifespan data predicted using the LLFS survival model, combined with the observed ages at death for decedents during the follow-up period, were used in our genetic analyses of human longevity. For simplicity, the combined data are referred to as “predicted lifespan data” and the combined outcome variable is referred to as “predicted lifespan”.
Genetics of Human Longevity From Predicted Data
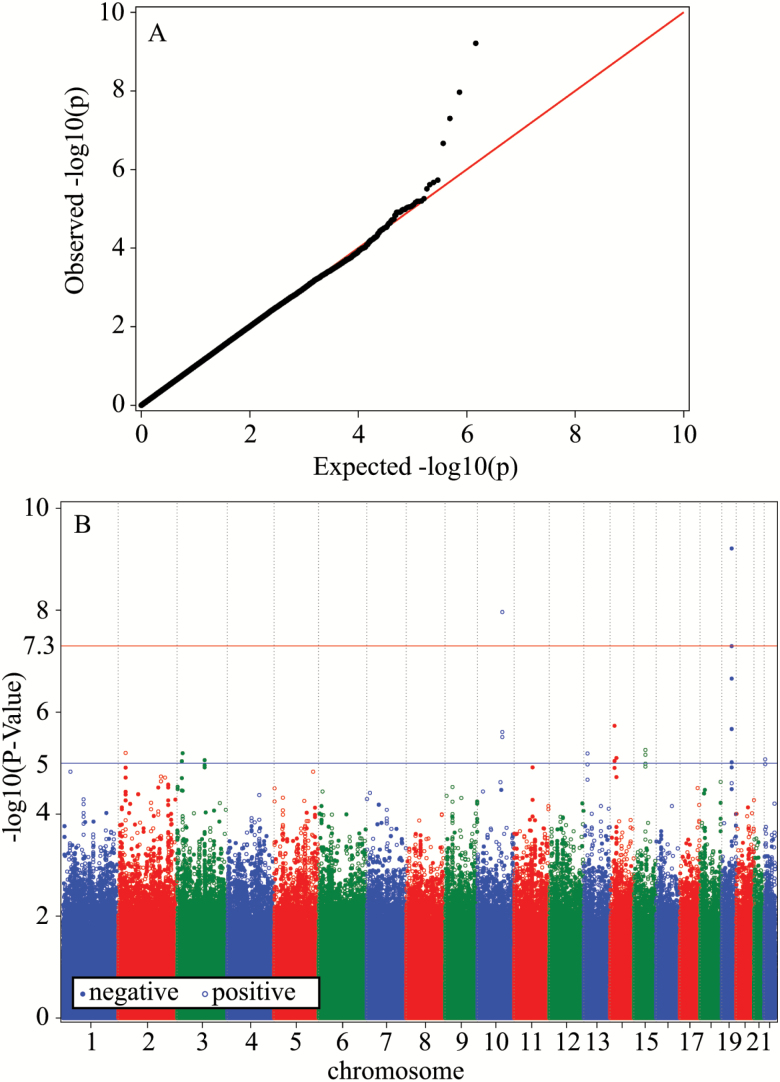
We performed a series of GWAS of predicted data on males and females combined using the GMMAT logistic regression models with different longevity thresholds and different sets of observed covariates, and compared the results. The most significant genetic associations with human longevity were obtained for the case group defined as study subjects survived to age 96 years and beyond with sex and country (DK, US) used as observed covariates. Figure 2A and B display the QQ-plot and Manhattan plot, for this case, respectively.
Figure 2.
(A) QQ-plot and (B) Manhattan plot. The results of GWAS of human longevity using the LLFS data on age at death for deceased individuals and data on predicted lifespan for censored individuals. The logistic regression model for related individuals in the GMMAT software package (11) with sex and country (DK, US) as observed covariates was used. Cases comprised individuals from the proband generation with lifespans (for deceased study subjects) or predicted lifespans (for study subjects with censored lifespans) ≥96 years (877 individuals). Controls comprised individuals from the offspring generation with age at death (for deceased study subjects) or attained age (for study subjects with censored lifespans) <75 years (2,462 individuals).
One can see from Figure 2B and Table 2 that association with longevity reached genome-wide levels of significance for two genetic variants located on chromosome 19 (rs769449 in APOE, and rs2075650 in TOMM40) and for one variant on chromosome 10 (rs1927465 near MYOF gene). Visualizations of genetic regions for these three SNPs and nearby genes are shown in Supplementary Figure 7. There were also promising genetic signals with p-values less than or around 10–5 on chromosome 19 and elsewhere. Table 2 and Supplementary Table 1 provide details about the top SNPs and related genes.
Table 2.
Top-Ranked SNPs and Their Characteristics
| SNP | Chr | A1 | A2 | P-val (GC) | MAF Case | MAF Control | Closest Gene | Gene Region | Regulatory Region |
|---|---|---|---|---|---|---|---|---|---|
| rs769449 | 19 | A | G | 6.19E−10 | 0.039 | 0.096 | APOE | Intron | eQTL, enhancer |
| rs1927465 | 10 | A | G | 1.09E−08 | 0.222 | 0.15 | MYOF | 70kb 3′ of | enhancer |
| rs2075650 | 19 | G | A | 5.05E−08 | 0.062 | 0.117 | TOMM40 | Intron | eQTL, enhancer |
| rs71352238 | 19 | C | T | 2.18E−07 | 0.065 | 0.119 | TOMM40 | 140bp 5′ of | eQTL |
| rs17102226 | 14 | T | C | 1.86E−06 | 0.074 | 0.119 | LOC102724945 | enhancer | |
| rs56131196 | 19 | A | G | 2.15E−06 | 0.098 | 0.151 | APOC1 | 239bp 3′ of | eQTL, enhancer |
| rs6765409 | 3 | T | C | 6.39E−06 | 0.28 | 0.342 | FBLN2 | Intron | eQTL, enhancer |
| rs7140186 | 14 | T | C | 9.06E−06 | 0.088 | 0.134 | LOC102724945 | ||
| rs34095326 | 19 | A | G | 9.60E−06 | 0.051 | 0.087 | TOMM40 | Intron | eQTL, enhancer |
| rs157582 | 19 | A | G | 1.21E−05 | 0.139 | 0.195 | TOMM40 | Intron | eQTL, enhancer |
| rs61981596 | 14 | G | T | 1.24E−05 | 0.095 | 0.142 | LOC102724945 | enhancer | |
| rs75736662 | 1 | G | A | 1.48E−05 | 0.03 | 0.013 | MAN1C1 | Intron | enhancer |
| rs73052307 | 19 | C | T | 2.49E−05 | 0.189 | 0.1472 | NECTIN2 | Intron | eQTL, enhancer |
| rs56161136 | 4 | G | A | 4.24E−05 | 0.163 | 0.118 | LOC105377441 | Intergenic | |
| rs34558922 | 9 | A | G | 5.66E−05 | 0.063 | 0.098 | ABCA2 | Intron | eQTL, enhancer |
| rs35590326 | 9 | G | T | 5.77E−05 | 0.065 | 0.1 | ABCA2 | Synonym | eQTL, enhancer |
| rs7778004 | 7 | C | T | 1.93E−04 | 0.352 | 0.415 | IQUB | 100kb 3′ of | enhancer |
| rs35902749 | 9 | A | G | 2.21E−04 | 0.055 | 0.086 | ABCA2 | Intron | eQTL, enhancer |
Note: Top-ranked SNPs (and respective genes) that showed most significant associations with predicted lifespan in GWAS of human longevity using GMMAT. A1 = minor allele; A2 = major allele; Chr = chromosome number; Closest Gene = GENCODE gene name; Gene Region = SNP location in gene, or distance to closest gene; MAF Case = minor allele frequency for case; MAF Contr = minor allele frequency for control; p-val (GC) = p-value after genomic control; Regulatory Region = SNP location in eQTL or Enhancer region of genome; SNP = rs-number.
Among the three top significant SNPs associated with longevity, the rs1927465 was a new finding (the associated allele: A; MAF in cases = 0.22; MAF in controls = 0.15; beta = 0.42, SE = 0.07; GMMAT p = 1.09E−08; GLIMMIX p = 5.50E−09).
Comparing Results of GWAS Obtained Using Predicted and Observed Data
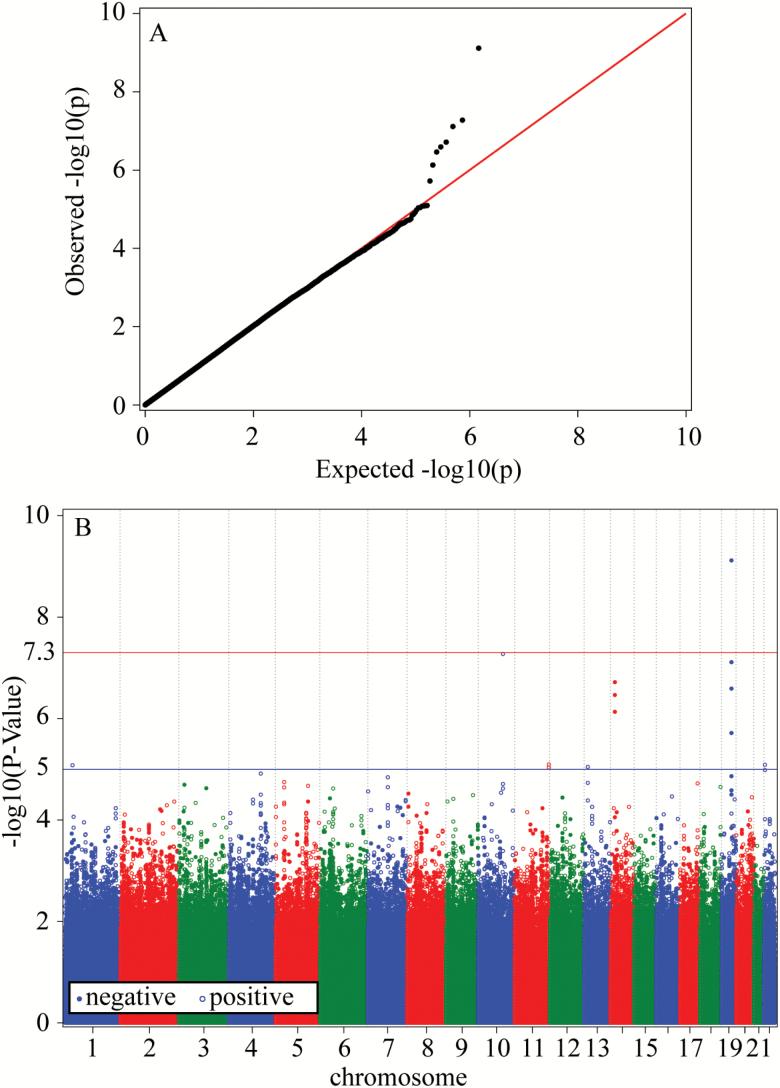
To better understand the benefits of conducting GWAS using the predicted lifespan, these results were compared with those obtained in GWAS of observed (non-predicted) longevity, as shown in Figure 3. One can see from this figure and Table 2 that two variants located on chromosomes 19 (rs769449) and 10 (rs1927465) still reached genome-wide levels of significance.
Figure 3.
(A) QQ-plot; (B) Manhattan plot. The results of GWAS of human longevity using observed LLFS data on lifespan. The logistic regression model for related individuals in the GMMAT software package (11) with sex and country (DK, US) as observed covariates was used. Cases comprised individuals from the proband generation with lifespans (for deceased subjects) or censored lifespans (for living study subjects, or for those who dropped out from the study) ≥96 years (723 individuals). Controls comprised individuals from the offspring generation whose age at death (for deceased study subjects) or attained age (for study subjects with censored lifespans) <75 years (2,462 individuals).
The comparisons of Figures 2 and 3 indicated that after about 8 years of follow-up from baseline, the use of predicted lifespan data provides slightly lower p-values for the estimated associations of genetic variants with human longevity for most of genetic variants with the highest levels of statistical significance. The comparisons of the annotation files obtained in the GWAS of the predicted and observed data indicated that, despite the slight difference in the p-values, the top 100 genetic variants were essentially the same. These may indicate that the about 8-year follow-up period provided enough information to reliably detect genetic variants associated with longevity in the probands’ generation.
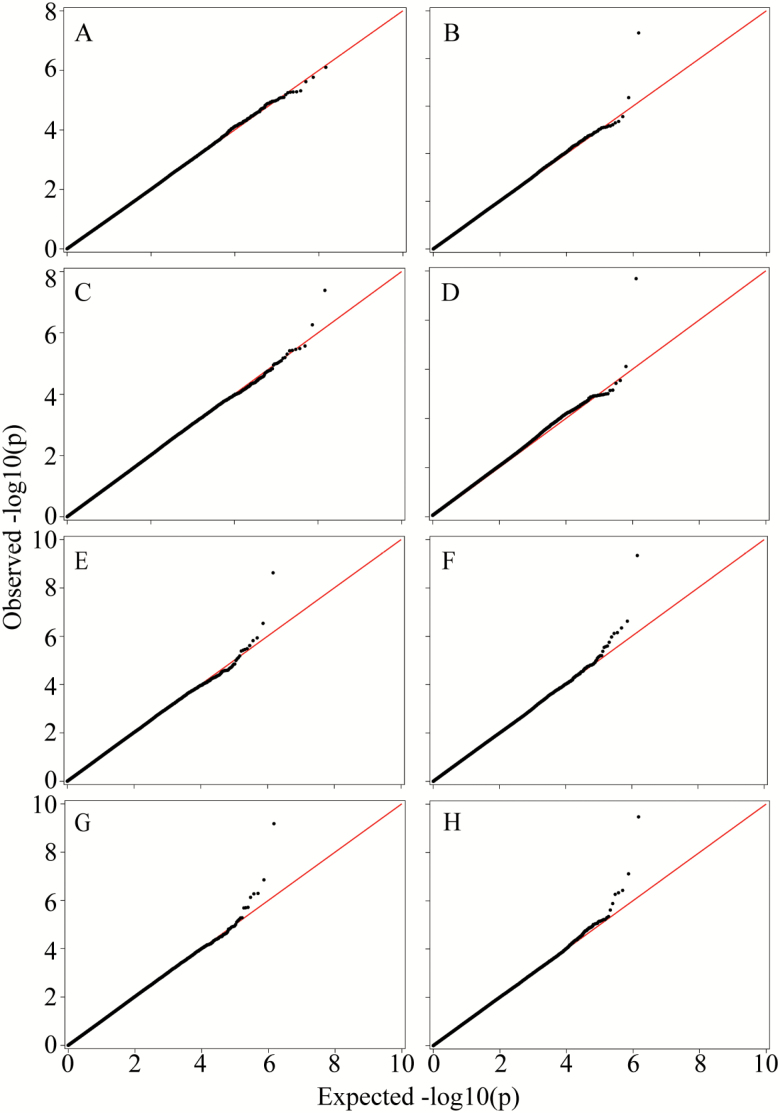
We hypothesized that the benefits of using predicted data in genetic studies of human longevity would be more visible in situations with shorter follow-up periods. To test this hypothesis, we performed further GWAS of human longevity using the LLFS data on males and females combined from the probands’ generation for different durations of follow-up. Figure 4 displays the QQ-plots resulting from these analyses, calculated for follow-up periods from baseline until 2010, 2011, 2013, and 2015, respectively.
Figure 4.
QQ plots corresponding to analyses of predicted and observed data with different periods of follow-up. The results of the GWAS of human longevity obtained in the GMMAT analyses of unpredicted (left panels) and predicted (right panels) LLFS data on males and females combined for different durations of follow-up. The panels from the top to the bottom represent follow-up periods from baseline to 2010, 2011, 2013, and 2015, respectively.
One can see from the QQ-plots on Figure 4 that the benefits of using predicted versus observed data in the GWAS of human longevity are substantial when the durations of follow-up are relatively short. For example, the QQ-plot (top left panels A and B) resulting from the analysis of observed data with durations extending from baseline only until 2010 does not show any visible signals. However, the analyses of predicted data for the same durations (top right panel) exhibit clear genetic signals. When the periods of follow-up were increased, the genetic signals resulting from the analyses of the observed data gradually became more visible and showed a tendency to converge to the results of the analyses of the predicted data (see also Figures 2 and 3). The analyses showed that these genetic signals correspond to SNPs on chromosomes 10 and 19 (Table 2).
Replicating newly discovered longevity SNP, rs1927465
To replicate the novel association of rs1927465 with longevity, we performed additional analysis of the Health and Retirement Study (HRS) data (14), using rs1927465 as candidate longevity SNP. Altogether we had 3,395 carriers and 9,186 non-carriers of the minor allele of this SNP for males and females combined. In the logistic regression model, the case group included participants who survived to age 90 or beyond (892 individuals). The controls included those who died before age 90, or whose current age did not exceed 70 years (4,300 individuals). The results of this analysis showed that the minor allele A of rs1927465 is positively associated with human longevity (OR = 1.19, p = .038; SE = 0.08; Confidence interval = 1.009–1.394). The age trajectory of the minor allele frequency of the rs1927456 is shown in Supplementary Figure 6.
Discussion
LLFS is an Outstanding Resource for Studying Exceptional Longevity
Our analysis confirmed that the special design of the LLFS resulted in selection of individuals with exceptional survival. Recent studies also showed that severe mortality-associated diseases are also less prevalent among LLFS participants (2,15–17). This indicates that the LLFS data is a unique resource for analyzing causes of exceptional health and longevity.
Benefits of Lifespan Prediction
The lifespan prediction model used survival probabilities estimated from the mortality experience of LLFS participants to generate individualized predictions of residual lifespan for older participants still alive at the end of follow-up. More specifically, lifespan predictions were made for individuals with censored data whose age at the end of follow-up was 80 years and older. The 8 years of follow-up produced enough data to reliably estimate the survival probabilities used for lifespan prediction at age 80+. Such probabilities could not be reliably estimated below age 80 because of the insufficiency of mortality data for LLFS participants in the offspring generation. This entire problem would not exist for participants in population-based studies because their survival probabilities, and hence their predicted residual lifespans, could be readily estimated for any age using data from demographic life tables for the selected population.
The results showed that the benefits of lifespan prediction for genetic analyses of human longevity decrease with increasing duration of follow up. In our case, the data on predicted lifespan were most useful for genetic analyses up to 3–4 years of follow-up. This timing depends on the amount of lifespan data accumulated during the follow-up period which in turn depends on the number of individuals at risk in the corresponding age groups. This timing property of predicted data, however, does not lessen the importance of continued follow-up beyond 3–4 years; such follow-up will eventually provide complete lifespan data. Analyses of complete lifespan data will have better power, will yield more accurate estimates of genetic effects, and may lead to discoveries of new associations.
Novel Longevity SNP
This study identified a new SNP (rs1927465) associated with familial longevity (survival ≥ 96 years in probands from long-lived families) with genome-wide significance, and replicated this SNP in the HRS data (p = .038). The rs1927465 SNP is located on chromosome 10 between the MYOF (myoferlin) and CYP26A1 (cytochrome P450 family 26 subfamily A member 1) genes, and is not in LD with SNPs from these genes. It is in closer proximity to MYOF (70 kb 3′ of), which plays a role in membrane repair, focal adhesion, and endocytosis, and has also been associated with cancer invasion (18–20). Since rs1927465 is in a noncoding region, we explored its potential regulatory role using the HaploReg v4.1 tool (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php) (see also (21). The analysis found that rs1927465 is located in a region with marks of an active enhancer in at least 10 tissues, which means that it may potentially influence expression of other genes. This SNP is also in moderate LD (r2 = 0.45 in the European (EUR) sample) with rs11187345, which sits closer to the MYOF gene (59 kb 3′ of) and is an eQTL influencing the expression of the neighboring CYP26A1 gene, whose product is involved in regulation of retinoic acid, cell differentiation, stem cell renewal, and some cancers (22,23).
Strong Replication of Findings from Earlier Studies
We replicated genome-wide significant associations with human longevity of SNPs in the APOE and TOMM40 genes found in earlier GWAS of human longevity that used different data. Specifically, we directly replicated: rs769449 in APOE (14,24); rs2075650 in TOMM40 (25,26); rs71352238 in TOMM40 (27). Using the HaploReg 4.1 tool (28), we also indirectly replicated SNP rs4420638 previously associated with longevity (29) as the rs56131196 SNP in a region of APOC1, identified in our study, is in high LD (r2 = 1, D′ = 1) with the “longevity SNP” rs4420638.
The study of the Han Chinese population showed that in addition to rs2075650 (TOMM40) and rs405509 (APOE), the rs12978931, rs519825, and rs395908 SNPs (all three from PVRL2 (NECTIN2)) are associated with longevity (30). The associations of SNPs from TOMM40, APOE, and APOC1 genes with longevity were also confirmed in other study of Chinese population (26). The association of the APOE gene with exceptional longevity was also detected by Garatachea et al. (31). In the LLFS, it was found that the chances of carrying “bad” alleles (APOE ε4 allele, or a G allele in rs2075650) among family members of long lived individuals in the offspring generation were lower than among their spouse controls (32). Several recent GWAS of long lived individuals identified rs2075650 in TOMM40 as associated with longevity (29,33,34). Since rs2075650 is in LD with rs429358—the APOE ε4 allele—it was proposed that SNPs from TOMM40 may not have independent effects on longevity. The influence of TOMM40 polymorphisms on human longevity was confirmed by Maruszak et al. (35).
Thus, genes and genetic variants at chromosome 19 detected in our study of human longevity replicated research findings from these earlier studies. The roles of these and other genes associated with human longevity were also discussed in 34,36–43. Other promising associations were found for SNPs in NECTIN2 and APOC1genes, located on chromosome 19 (Tables 2, Supplementary Table 1).
Functional Properties of the Top Significant Genes
One should stress that two of the top three significant SNPs associated with longevity in our study (rs769449 and rs2075650) as well as other SNPs from the TOMM40/APOE/APOC1/NECTIN2 region are also among the top SNPs that have been most consistently associated with late onset Alzheimer’s disease (AD) and cognitive decline in multiple studies (44–50), including our recent analysis of the HRS, CHS, FHS, and LOADFS data (13). Both SNPs have also been significantly associated with LDL cholesterol levels. The rs769449 SNP in APOE is in strong LD (r2 = 0.82, D′ = 1) with rs429358 representing the APOE e2/e3/e4 polymorphism, a major genetic risk factor for AD. The rs769449 SNP is in moderate LD (r2 = 0.6, D′ = 0.8) with rs2075650 in TOMM40, another SNP robustly associated with AD across many datasets, including in our analyses (13). The rs2075650 SNP is in strong LD (R2 ≥ 0.92; D ≥ 0.98) with several SNPs in the PVRL2 (NECTIN2) gene which is involved in adherens junctions and host resistance to viral infection. It is also eQTL and may influence expression levels of both TOMM40 (regulating protein precursors’ transport into mitochondria) and NECTIN2. It is important to note that these SNPs and corresponding genes are physically connected by chromatin contacts and therefore may be functionally connected. The common functions of the top SNPs associated with longevity indicate that cholesterol transport may potentially play central roles in both cognitive decline and response to infection, and through this in longevity.
We explored the functional effects of the 100+ top-ranked significant SNPs that influenced lifespan with p-value ≤ 10–4, and related genes, to get insights into potential biological mechanisms of their effects on longevity. For this, we gathered the information about SNPs, genes, and regulatory genomic regions from multiple established online resources, such as NCBI (PubMed, Entrez Gene, dbSNP, OMIM, and others), 1000Genomes, GO, ENCODE, Roadmap Epigenomics, GTEx consortium, Ensembl, GRASP, HGRI-EBI Catalog of published GWAS, and others. We also used the Haploreg v4.1 online tool (21) for comparative assessment of the prospective regulatory effects of the selected SNPs (eg, SNP location in eQTL and enhancer regions of the genome), as well as a commercial tool for enrichment analysis and pathway exploration (MetaCore, by Thomson Reuters). We conducted the enrichment analysis for genes corresponding to the top 103 SNPs that influenced lifespan with p-value ≤ 10–4, using MetaCore. The analysis detected significant enrichment by GO Processes related to lipid transport, lipid synthesis, and lipid metabolism. Overall, about 10% of the detected genes were involved in the respective processes. Examples of relevant genes include APOE, APOC1, APOC4, ABCA2, CPT1A, PLA2G4C, HNF4A, and ERBB4.
We looked more closely at the functions of the 18 most significant SNPs, and the respective genes, that influenced the predicted lifespans (see Supplementary Table 1). The genes that were closest to the most significant SNPs are involved in a number of cellular and tissue processes, including lipid transport, cell junctions, and extracellular matrix (ECM) remodeling. They also have common associations with a number of health related traits including AD, cancer, and viral or bacterial infections. Strong LD between SNPs in TOMM40 and NECTIN2 gene, which is involved in resistance to herpes viruses, indicates that the latter may potentially play an important role in the observed associations.
One hypothetical scenario that integrates this information and addresses common functions of the top SNPs associated with longevity in this study (especially rs769449/rs429358 and rs2075650) could be that the aging-related decline in cholesterol transport is accelerated in the presence of certain genetic variants. This may lead to cholesterol and myelin deficiency in the brain and CNS, which could in turn compromise the repair of neurons and reduce their capacity to recover after various damage, including infection. This may promote neural apoptosis and overall decline in brain capacity. Proper cell junctions are important for controlling the blood–brain barrier (BBB) permeability and protecting the brain from infection. The BBB permeability may increase with aging, and so the infection burden in the brain, which together with compromised brain repair (due to cholesterol/myelin deficiency) could lead to accumulating brain damage over age and eventually to limiting longevity.
Most of the top-ranked SNPs belong to regulatory genomic regions such as eQTLs and enhancers. Such SNPs may influence transcription levels and protein concentrations without changes in protein structure. This indicates that longevity can potentially be extended by modulating patterns of gene expression. Note that such genes, whose expression is modulated, do not necessarily have SNPs associated with human longevity or health-related traits.
The large-sample genome-wide association meta-analysis performed by Deelen et al. (51) detected genome wide significant association of rs2149954, on chromosome 5, with longevity. Although rs2149954 is not available in our genetic data, we found four SNPs on chromosome 5 that are in strong LD with it: rs4704775 (r2 = 0.94, D′ = 1), rs6863179 (r2 = 0.95, D′ = 1), rs7715501 (r2 = 0.87, D′ = 0.94), and rs11960210 (r2 = 0.95, D′ = 1). The genetic analyses of LLFS data on males and females combined showed that all four SNPs are associated with human longevity at the nominal level of statistical significance: rs4704775 (p ≤ .03), rs6863179 (p ≤ .03), rs7715501 (p ≤ .04), and rs11960210 (p ≤ .04). The nominally significant association of this SNP with longevity was also confirmed in recent study (52)
Using the Index of DIs as a Covariate for Better Lifespan Prediction
In addition to lifespan data, the LLFS collected extensive information on other variables measured at baseline among the study participants. A set of 85 such variables was used to construct the index of DIs, called the DI or Frailty Index (FI) (9). Numerous studies have shown that FI (DI) is a good predictor of lifespan (8). The use of the ROC-AUC techniques indicated that the inclusion of this index as a covariate in the predictive survival model improves the prediction of lifespans among censored participants in the LLFS data. Comparisons of the predictive models with and without use of DI are shown in Supplementary Figure 2. These models were used to predict the missing lifespan data for the primary GWAS analyses in this article.
GMMAT and GLIMMIX (SAS) are two programs that can be used in GWAS of complex traits for related individuals. The advantage of GMMAT is that it is faster than GLIMMIX. To test whether the results are consistent we performed analyses of the same data using each program. Supplementary Figure 3 shows that both programs are appropriate tools for GWAS of human longevity using LLFS data on lifespan. This figure indicates that the p-values for highly significant SNPs obtained using these two computer programmes are about the same. The limitation of GMMAT is that the output includes the p-values but not the estimates of regression coefficients and odds ratios. Supplementary Table 1 provides estimates of these statistical characteristics for selected SNPs, obtained using GLIMMIX.
Auxiliary analyses conducted using two alternative sets of covariates (1): sex, PC1, and PC2; and (2) sex, country of origin, PC1, and PC2 (DK, US) practically did not affect the significance of the SNPs on chromosome 19 (Supplementary Figures 4 and 5). Although the p-value of the SNP on chromosome 10 was reduced to about 10–7, it remained the most significant SNP other than those detected on chromosome 19. Age trajectories of minor allele frequencies for rs1927465 are shown in Supplementary Figure 6. The LocusZoom regional visualization of genome-wide association scan results are shown in Supplementary Figure 7 for three genome-wide significant SNPs: rs1927465, rs769449, and rs2075650.
Summary
The survival of the LLFS populations is substantially better than that of the same birth cohorts in the United States and Denmark. This indicates that survival probabilities calculated from the LLFS data, not demographic life tables, should be used for predicting lifespan. This better survival also highlights the high potential of the LLFS data for analyzing causes of exceptional longevity. The analyses showed that the benefits of using the predicted lifespan data in GWAS of human longevity are most visible (Figure 4) for relatively short periods of follow up. The p-values of the genetic estimates obtained in the analyses of predicted and non-predicted data tended to converge as the length of follow-up increased, when more data on observed lifespans are available. The GWAS of the two variations of the predicted lifespan data—using survival models with versus without the cumulative DI—produced similar estimates of genetic associations with human longevity when the estimates were based on 8 years of follow-up data. These findings suggest that improvement of the quality of genetic analyses of incomplete lifespan data will require a corresponding improvement in the accuracy of the lifespan predictions. One improvement would be to add more composite indices (eg, the multi-morbidity index, healthy aging index) into the prediction model. Our analyses also showed that the logistic regression model in GWAS of LLFS data yielded stronger genetic associations with human longevity than the Cox regression model. The replication of the effect of the newly detected rs1927465 SNP located between the CYP26A1 and MYOF genes on chromosome 10 on longevity in the HRS data confirms the relevance of this SNP for human longevity for both sexes. The replication of strong associations of genetic variants from the APOE, TOMM40, NECTIN2, and APOC1 genes on chromosome 19 with human longevity indicates that the statistical model used in our analyses is capable of reliably detecting strong genetic associations with this trait. Additional analyses are needed to evaluate sex-specific genetic effects on human longevity in these data. The common functions of the top SNPs associated with longevity in our study (especially rs769449 and rs2075650) indicate that cholesterol transport may potentially play a central role in cognitive decline and response to infection, and through this in longevity.
Supplementary Material
Supplementary data is available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Funding
This work was supported by the National Institute on Aging, National Institutes of Health (NIA/NIH) grant U01 AG023712. The work of A.I.Y., K.A., D.W., O.B., A.K., I.C., M.K., I.Z., E.S., and S.U. was also partly supported by the NIA/NIH grants R01AG046860 and P01AG043352. The Long Life Family Study is funded by U01 AG023749, U01 AG023744, and U01 AG023712 from the National Institute on Aging, National Institutes of Health. The Health and Retirement Study genetic data is sponsored by the National Institute on Aging (grant numbers U01 AG009740, RC2 AG036495, and RC4 AG039029) and was conducted by the University of Michigan. This study used data provided by the database of Genotypes and Phenotypes (dbGaP), dbGaP Study Accession: phs000428.v1.p1.
Acknowledgements
Author contributions: A.I.Y. and coauthors conceptualized the idea of the article in a series of conference calls. A.I.Y. wrote the first draft and the ultimate version of the article. K.G.A. and L.A. performed statistical analyses of survival probabilities and generated predicted data on lifespan; D.W. prepared genetic data and performed series of GWASs of predicted and non-predicted data using different statistical models; O.B. prepared survival data for analyses; S.V.U. performed functional genetic analyses of research findings; A.I.Y., K.G.A., D.W., E.S., A.M.K., I.A., F.F., M.K.W., K.C., A.B.N., R.B., M.A.P., S.T., T.P., A.P., I.E., and S.V.U. discussed each stage of analyses at series of regular conference calls. All coauthors read the manuscript and provided valuable comments and suggestions.
Conflict of interest statement
None declared.
References
- 1. Sebastiani P, Hadley EC, Province M, et al. . A family longevity selection score: ranking sibships by their longevity, size, and availability for study. Am J Epidemiol. 2009;170:1555–1562. doi:10.1093/aje/kwp309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Newman AB, Glynn NW, Taylor CA, et al. . Health and function of participants in the Long Life Family Study: a comparison with other cohorts. Aging (Albany NY). 2011;3:63–76. doi:10.18632/aging.100242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elo IT, Mykyta L, Sebastiani P, Christensen K, Glynn NW, Perls T. Age validation in the long life family study through a linkage to early-life census records. J Gerontol B Psychol Sci Soc Sci. 2013;68:580–585. doi:10.1093/geronb/gbt033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cosentino S, Schupf N, Christensen K, Andersen SL, Newman A, Mayeux R. Reduced prevalence of cognitive impairment in families with exceptional longevity. JAMA Neurol. 2013;70:867–874. doi:10.1001/jamaneurol.2013.1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee JH, Cheng R, Honig LS, et al. . Genome wide association and linkage analyses identified three loci-4q25, 17q23.2, and 10q11.21-associated with variation in leukocyte telomere length: the Long Life Family Study. Front Genet. 2013;4. doi:10.3389/fgene.2013.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kulminski AM, Ukraintseva SV, Akushevich IV, Arbeev KG, Yashin AI. Cumulative index of health deficiencies as a characteristic of long life. J Am Geriatr Soc. 2007;55:935–940. doi:10.1111/j.1532-5415.2007.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yashin AI, Arbeev KG, Kulminski A, Akushevich I, Akushevich L, Ukraintseva SV. Cumulative index of elderly disorders and its dynamic contribution to mortality and longevity. Rejuvenation Res. 2007;10:75–86. doi:10.1089/rej.2006.0500 [DOI] [PubMed] [Google Scholar]
- 8. Mitnitski A, Rockwood K. Aging as a process of deficit accumulation: its utility and origin. Interdiscip Top Gerontol. 2015;40:85–98. doi:10.1159/000364933 [DOI] [PubMed] [Google Scholar]
- 9. Kulminski AM, Arbeev KG, Christensen K, et al. . Do gender, disability, and morbidity affect aging rate in the LLFS? Application of indices of cumulative deficits. Mech Ageing Dev. 2011;132:195–201. doi:10.1016/j.mad.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–73. doi:10.1016/j.mad.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen H, Wang C, Conomos MP, et al. . Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98:653–66. doi:10.1016/j.ajhg.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi:10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 13. Yashin AI, Fang F, Kovtun M, et al. . Hidden heterogeneity in Alzheimer’s disease: insights from genetic association studies and other analyses. Exp Gerontol. 2018;107:148–160. doi:10.1016/j.exger.2017.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang C, Pierce BL. Genetic susceptibility to accelerated cognitive decline in the US health and retirement study. Neurobiol Aging. 2014;35:1512.e11–1512.e18. doi:10.1016/j.neurobiolaging.2013.12.021 [DOI] [PubMed] [Google Scholar]
- 15. Barral S, Cosentino S, Costa R, et al. . Exceptional memory performance in the Long Life Family Study. Neurobiol Aging. 2013;34:2445–8. doi:10.1016/j.neurobiolaging.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sebastiani P, Sun FX, Andersen SL, et al. . Families enriched for exceptional longevity also have increased health-span: findings from the Long Life Family Study Front Public Health. 2013;1. doi:10.3389/fpubh.2013.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ash AS, Kroll-Desrosiers AR, Hoaglin DC, Christensen K, Fang H, Perls TT. Are members of long-lived families healthier than their equally long-lived peers? evidence from the long life family study. J Gerontol A Biol Sci Med Sci. 2015;70:971–976. doi:10.1093/gerona/glv015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bernatchez PN, Sharma A, Kodaman P, Sessa WC. Myoferlin is critical for endocytosis in endothelial cells. Am J Physiol Cell Physiol. 2009;297:C484–C492. doi:10.1152/ajpcell.00498.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turtoi A, Blomme A, Bellahcene A, et al. . Myoferlin is a key regulator of EGFR activity in breast cancer. Cancer Res. 2013;73:5438–5448. doi:10.1158/0008-5472.CAN-13–1142 [DOI] [PubMed] [Google Scholar]
- 20. Blackstone BN, Li R, Ackerman WET, Ghadiali SN, Powell HM, Kniss DA. Myoferlin depletion elevates focal adhesion kinase and paxillin phosphorylation and enhances cell-matrix adhesion in breast cancer cells. Am J Physiol Cell Physiol. 2015;308:C642–C649. doi:10.1152/ajpcell.00276.2014 [DOI] [PubMed] [Google Scholar]
- 21. Zhbannikov IY, Arbeev K, Ukraintseva S, Yashin AI. haploR: an R package for querying web-based annotation tools. F1000Research. 2017;6. doi:10.12688/f1000research.10742.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osanai M, Sawada N, Lee GH. Oncogenic and cell survival properties of the retinoic acid metabolizing enzyme, CYP26A1. Oncogene. 2010;29:1135–1144. doi:10.1038/onc.2009.414 [DOI] [PubMed] [Google Scholar]
- 23. Ghiaur G, Yegnasubramanian S, Perkins B, Gucwa JL, Gerber JM, Jones RJ. Regulation of human hematopoietic stem cell self-renewal by the microenvironment’s control of retinoic acid signaling. Proc Natl Acad Sci USA. 2013;110:16121–16126. doi:10.1073/pnas.1305937110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ryu S, Atzmon G, Barzilai N, Raghavachari N, Suh Y. Genetic landscape of APOE in human longevity revealed by high-throughput sequencing. Mech Ageing Dev. 2016;155:7–9. doi:10.1016/j.mad.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shadyab AH, Kooperberg C, Reiner AP, et al. . Replication of genome-wide association study findings of longevity in white, African American, and hispanic women: the Women’s Health Initiative. J Gerontol A Biol Sci Med Sci. 2017; 72:1401–1406. doi:10.1093/gerona/glw198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin R, Zhang Y, Yan D, et al. . Association of common variants in TOMM40/APOE/APOC1 region with human longevity in a Chinese population. J Hum Genet. 2016;61:323–328. doi:10.1038/jhg.2015.150 [DOI] [PubMed] [Google Scholar]
- 27. Zeng Y, Nie C, Min J, et al. . Novel loci and pathways significantly associated with longevity. Sci Rep. 2016;6. doi:10.1038/srep21243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi:10.1093/nar/gkv1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nebel A, Kleindorp R, Caliebe A, et al. . A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech Ageing Dev. 2011;132:324–330. doi:10.1016/j.mad.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 30. Lu F, Guan H, Gong B, et al. . Genetic variants in PVRL2-TOMM40-APOE region are associated with human longevity in a Han Chinese population. PLoS One. 2014;9:e99580. doi:10.1371/journal.pone.0099580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garatachea N, Emanuele E, Calero M, et al. . ApoE gene and exceptional longevity: insights from three independent cohorts. Exp Gerontol. 2014;53. doi:10.1016/j.exger.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 32. Schupf N, Barral S, Perls T, et al. . Apolipoprotein E and familial longevity. Neurobiol Aging. 2013;34:1287–1291. doi:10.1016/j.neurobiolaging.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deelen J, Beekman M, Uh HW, et al. . Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10:686–698. doi:10.1111/j.1474-9726.2011.00705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sebastiani P, Solovieff N, Dewan AT, et al. . Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi:10.1371/journal.pone.0029848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maruszak A, Peplonska B, Safranow K, et al. . TOMM40 rs10524523 polymorphism’s role in late-onset Alzheimer’s disease and in longevity. J. Alzheimers Dis. 2012;28:309–322. doi:10.3233/JAD-2011–110743 [DOI] [PubMed] [Google Scholar]
- 36. Broer L, Buchman AS, Deelen J, et al. . GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 Candidacy. J Gerontol A Biol Sci Med Sci. 2015; 70:110–118. doi:10.1093/gerona/glu166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chung WH, Dao RL, Chen LK, Hung SI. The role of genetic variants in human longevity. Ageing Res Rev. 2010;9(Suppl 1):S67–S78. doi:10.1016/j.arr.2010.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lunetta KL, D’Agostino RB Sr, Karasik D, et al. . Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham Study. BMC Med Genet. 2007;8(Suppl 1S13). doi:10.1186/1471-2350-8-S1-S13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murabito JM, Yuan R, Lunetta KL. The search for longevity and healthy aging genes: insights from epidemiological studies and samples of long-lived individuals. J Gerontol A Biol Sci Med Sci. 2012;67:470–479. doi:10.1093/gerona/gls089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Newman AB, Walter S, Lunetta KL, et al. . A meta-analysis of four genome-wide association studies of survival to age 90 years or older: the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. J Gerontol A Biol Sci Med Sci. 2010;65:478–487. doi:10.1093/gerona/glq028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soerensen M. Genetic variation and human longevity. Dan Med J. 2012;59:B4454. [PubMed] [Google Scholar]
- 42. Walter S, Atzmon G, Demerath EW, et al. . A genome-wide association study of aging. Neurobiol Aging. 2011;32:2109.e15–2109.e28. doi:10.1016/j.neurobiolaging.2011.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Slagboom PE, Heijmans BT, Beekman M, Westendorp RG, Meulenbelt I. Genetics of human aging. The search for genes contributing to human longevity and diseases of the old. Ann N Y Acad Sci. 2000;908. doi:10.1111/j.1749–6632.2000.tb06635.x [DOI] [PubMed] [Google Scholar]
- 44. Bagnoli S, Piaceri I, Tedde A, et al. . TOMM40 polymorphisms in Italian Alzheimer’s disease and frontotemporal dementia patients. Neurol Sci. 2013;34:995–998. doi:10.1007/s10072-013-1425-6 [DOI] [PubMed] [Google Scholar]
- 45. Cruchaga C, Nowotny P, Kauwe JS, et al. . Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. Arch Neurol. 2011;68:1013–1019. doi:10.1001/archneurol.2011.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lyall DM, Harris SE, Bastin ME, et al. . Alzheimer’s disease susceptibility genes APOE and TOMM40, and brain white matter integrity in the Lothian Birth Cohort 1936. Neurobiol Aging. 2014;35:1513.e25–1513.e33. doi:10.1016/j.neurobiolaging.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma XY, Yu JT, Wang W, et al. . Association of TOMM40 polymorphisms with late-onset Alzheimer’s disease in a Northern Han Chinese population. Neuromol Med. 2013;15:279–287. doi:10.1007/s12017-012-8217-7 [DOI] [PubMed] [Google Scholar]
- 48. Seripa D, Bizzarro A, Pilotto A, et al. . TOMM40, APOE, and APOC1 in primary progressive aphasia and frontotemporal dementia. J Alzheimers Dis. 2012;31:731–740. doi:10.3233/JAD-2012–120403 [DOI] [PubMed] [Google Scholar]
- 49. Takei N, Miyashita A, Tsukie T, et al. . Genetic association study on in and around the APOE in late-onset Alzheimer disease in Japanese. Genomics. 2009;93:441–448. doi:10.1016/j.ygeno.2009.01.003 [DOI] [PubMed] [Google Scholar]
- 50. Logue MW, Schu M, Vardarajan BN, et al. . A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol. 2011;68:1569–1579. doi:10.1001/archneurol.2011.646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deelen J, Beekman M, Uh HW, et al. . Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum Mol Genet. 2014;23:4420–4432. doi:10.1093/hmg/ddu139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nygaard M, Thinggaard M, Christensen K, Christiansen L. Investigation of the 5q33.3 longevity locus and age-related phenotypes. Aging (Albany NY). 2017;9:247–255. doi:10.18632/aging.101156 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The LLFS data are available in dbGaP and can be obtained using standard procedure, described at dbGaP website. DbGaP Study Accession: phs000397.v1.p1. The details of genotyping and quality control procedures are described in (5).