Abstract
Accurate and physiologically meaningful biomarkers for human aging are key to assessing antiaging therapies. Given ethnic differences in health, diet, lifestyle, behavior, environmental exposures, and even average rate of biological aging, it stands to reason that aging clocks trained on datasets obtained from specific ethnic populations are more likely to account for these potential confounding factors, resulting in an enhanced capacity to predict chronological age and quantify biological age. Here, we present a deep learning-based hematological aging clock modeled using the large combined dataset of Canadian, South Korean, and Eastern European population blood samples that show increased predictive accuracy in individual populations compared to population specific hematologic aging clocks. The performance of models was also evaluated on publicly available samples of the American population from the National Health and Nutrition Examination Survey (NHANES). In addition, we explored the association between age predicted by both population specific and combined hematological clocks and all-cause mortality. Overall, this study suggests (a) the population specificity of aging patterns and (b) hematologic clocks predicts all-cause mortality. The proposed models were added to the freely-available Aging.AI system expanding the range of tools for analysis of human aging.
Keywords: Biochemistry aging clocks, Biological age, Deep Learning, Deep Neural Networks, Machine Learning
According to the World Health Organization, from 2000 to 2015 the global life expectancy experienced its fastest growth since the 1990s, increasing by 5 years within this period (1). Life expectancy, however, varies across countries (1) and even across different regions within one country (2). The underlying factors driving this variability often unclear and research into subpopulation specific life expectancies could facilitate the identification of more comprehensive country and region specific aging patterns. Toward this end, we are investigating the populations of three diversely aging including Canada, South Korea, and Eastern Europe. Aging-dependent health care and social costs are rapidly increasing in each of these three (3). Increased life expectancy, even when coupled with decreasing birth rate, is outpacing economic growth (4). Assessing aging is the first step towards interventions to reduce the illness, social, and economic-burden associated with aging (5).
Growing pre-clinical experimental evidence suggests the feasibility of finding interventions for extending human health span (6). Evaluating such interventions might require long follow-up periods and entails the possibility of failing to translate preclinical success into improved clinical outcomes (7). The search for effective geroprotectors (i.e., anti-aging molecules) shown few positive results in humans (8,9). Biomarkers of aging, or aging clocks, are promising tools with the potential to provide a quantitative foundation upon which to evaluate the therapeutic efficacy of clinical healthspan-extending interventions (10). Much progress has been made in measuring aging biomarkers using easily obtained data such as blood DNA methylation (11,12), transcriptomics (13), and metabolomics (14). Data of different modalities provide different levels of precision regarding the magnitude and rate of age-related changes, and biomarkers vary according to their resolving ability. While methylation-based aging clocks provide the most accurate representation of chronological age to date, epigenetic information is relatively stable (15). As such, epigenetic aging clocks appear to be comparatively less effective in quantifying the effect of behavioral, lifestyle, environmental, and interventional factors upon the rate of biological aging (16). Furthermore, epigenetic clocks are not as practically measurable as markers quantifying transcriptional (13,17) and the standardized accredited assays biochemical (14) markers. Nonetheless, the most accurate methods of calculating biological age is a subject of ongoing debate, and recent studies suggest that a suite of biomarkers, rather than any individual biomarker, constitute the most effective means of assessing the health status of a patient (18).
Here, we present several deep learning-based predictors of biological age trained upon population-specific blood biochemistry and hematological cell count datasets. Previously, we showed that blood biochemistry could be used to assess the biological age of a patient (14), an approach that has several advantages compared to other aging clocks, including strong correlation with chronological age (coefficient of determination is greater than 0.8), constancy across the entire adult age range, lack of influence of sex, and ease of assessment compared to methylation-based aging clocks. However, such aging clocks seem to be population-specific (19), therefore, robust aging clocks should be trained upon population-specific data. While current research appears to validate the importance of ethnicity upon life expectancy and mortality rates (20–22), the effect of ethnicity on blood biochemistry-based biomarkers of aging remains unclear. In the present study we decided to focus on emerging machine learning (ML) techniques, such as deep neural networks (DNNs), in the construction of our aging clocks. DNNs are perceived as game-changing methods in data analysis due to their capacity to capture hidden underlying features and learn complex representations of highly multidimensional data (23).
Materials and Methods
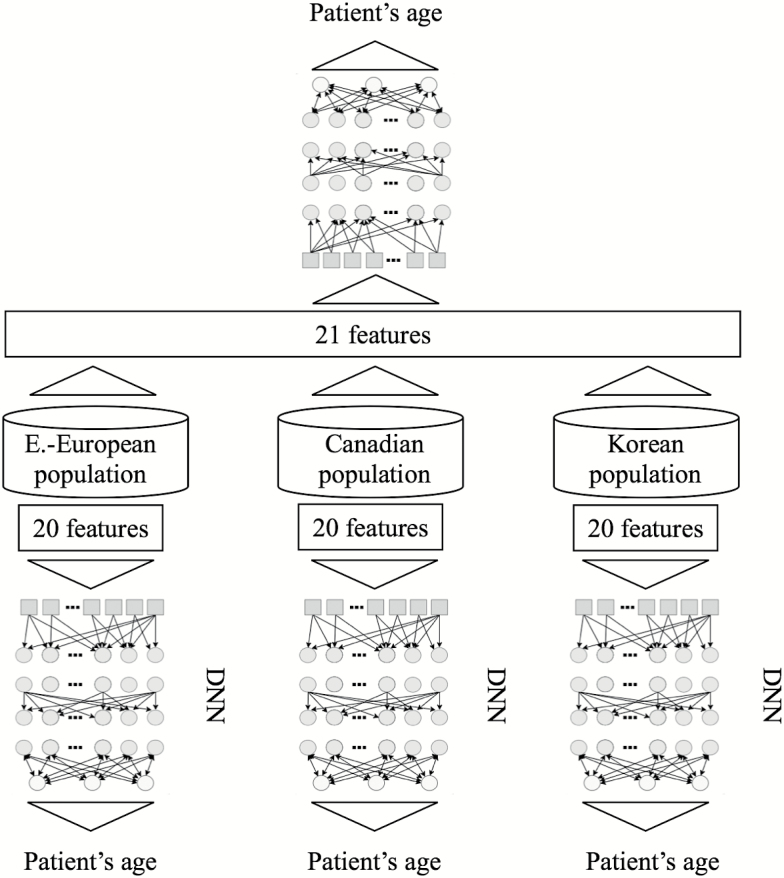
To perform this study, we trained a series of DNNs on anonymized blood tests for patients from three distinct ethnic populations: Korean, Canadian, and Eastern European. We compared the predictive accuracy of our deep learning models first when trained using population specific data, and then when using a combined and ethnically diverse dataset that includes patients from all three patient populations (see Figure 1). We used the same feature space of 20 blood biochemistry markers, cell counts, and sex to train three separate deep networks on three specific ethnic populations. Here, we excluded samples from patients younger than 20 years of age given the high error rate in this age group and the different biochemistry reference ranges associated with adolescents and children.
Figure 1.
Study design. First, blood samples of three populations (Canadian, Korean and Eastern European) with 21 the most relevant features with maximum samples available were used to train three population specific predictors. Afterwards, the resulting dataset consisting of samples from all three populations was used to train and test DNNs for predicting patient age.
Data Overview
Anonymized blood test records were kindly provided by Alberta Health (with IRB approval), Gachon University Gil Medical Center, and an independent laboratory, Invitro. Patient data were fully anonymized. In total, four datasets containing 20,699 samples for the Canadian population, 65,760 samples for the South Korean population, and 55,920 samples for the Eastern European population. Population dataset characteristics are described in Supplementary Table 1.
To investigate the predictive power of trained models on the publicly available data and to assess the predictive value of hematologic clocks to predict all-cause mortality, we analyzed the National Health and Nutrition Examination Survey (NHANES) dataset. We obtained retrospective laboratory data and demographics data for 1996–2016 years of and mortality data for 1996–2011 years using the National Center for Health Statistics website (https://www.cdc.gov/nchs/index.htm). The collected NHANES dataset contained 55,751 samples with blood test values, which were used to predict the age of patients. The blood test dataset was merged with mortality dataset by anonymized patient unique ids resulting in a table of 2,768 samples.
For mortality analysis, we explored Canadian population dataset (with known 340 deaths) and NHANES dataset (with known 873 deaths).
Training and Testing Set Design
Models were trained on 19 blood test features (in this paper, we refer to features implying input data or variables), 15 of which are biochemistry markers, including Albumin, Glucose, Hemoglobin, Cholesterol, Sodium, Urea, LDL Cholesterol, Triglycerides, Hematocrit, HDL Cholesterol, Total Protein, Calcium, Creatinine, Potassium, and Total Bilirubin, and four are cell count markers, including Erythrocytes, and Platelet count. While these markers are common parameters measured for South Korean and Eastern European blood tests, several of these markers were not present in the full Canadian dataset. Therefore, in order to obtain the same feature space for all three population specific datasets, values of Urea, Total Protein, Calcium, and total bilirubin for part of the Canadian dataset were reconstructed via regression analysis using samples with all 19 parameters (Supplementary Table 2). Values of blood tests were treated as continuous values while sex and population labels were treated as binary and dummy variables, respectively. We split the data into the training and testing sets at an 80/20 ratio.
DNNs Implementations
We treated age prediction as a regression task, that is, the model takes a vector of blood test values and returns a single value of patient age. Here we decided to use a deep model with multiple layers, which allows fitting data with high-level of dependencies between input features (blood biochemistry and cell counts) and the output feature (age).
We used multilayer feed-forward neural networks as deep models (ie, having more than three layers) and the Python implementation of the Keras (https://keras.io/) library with Theano (http://deeplearning.net/software/theano/) backend to build and train the neural networks. Grid search over a space of model parameters was used for optimization in order to find the best performing network architecture. We minimized the mean absolute error (MAE) loss function using a back propagation algorithm. We used the Leaky ReLU activation (24) function after each layer, EVE (25) as an optimizer of the cost function, and a dropout (26) with 35% probability after each layer for the purposes of regularization. We trained the networks with fivefold cross validation to compensate for overfitting and to receive more robust performance metrics. All experiments were conducted using an NVIDIA Titan X (Maxwell) graphics processing unit.
We also compared DNNs with a set of conventional ML algorithms, including Elastic Net, Random Forest, Partial Least squares, Gradient Boosting Machines, and Principal component Analysis. All models were implemented using the Scikit-learn library.
Feature Importance Analysis
To address the interpretability problem of DNNs and yield more insight into the data, we have applied permutation feature importance (PFI) analysis to rank input blood markers according to their importance in terms of age prediction. We applied PFI for the best performing models on each dataset. PFI is a wrapper method, which assigns the relative importance to input features based on the level of decreased age prediction accuracy after each feature random reshuffling. The larger the decrease in the accuracy of age prediction, the more important the input feature is. We applied the same technique for the age prediction in the present study (14), with some modification. Sex and population binary vectors were wrapped randomly, but systematically replaced with the opposite values.
Statistical Analysis
The following metrics were used to evaluate the accuracy of the age prediction models:
1) Pearson correlation coefficient
where is chronological age value and is the mean of yi is predicted age value and is the mean of , N is number of samples. r shows the strength of a linear association between predicted and actual age.
2) Coefficient of determination: where is the real value, is the predicted value and is the mean of . R2 shows the percentage of variance explained by the regression between predicted and actual age.
3) Mean absolute error: ; where is a predicted age, yi is age value and N is a number of samples. MAE demonstrates average disagreement between the chronological age and the predicted age.
4) Log 2 transformed Aging ratio: where is age prediction of the model, yi is a chronological age value, N is a number of samples. Aging ratio is the ratio of predicted age over observed chronological age. Log2Aging ratio of 1 means that the sample is predicted two-fold older than a chronological age and Log2Aging ratio of −1 means sample is predicted half as old
5) ; where and is an age prediction of the model, yi is a true age value. For instance, if epsilon is 5 and the DNN model predicts age of 55 but the real age is 50 or 60, then by epsilon accuracy such sample would be considered as correctly classified.
To evaluate the association of the predicted age acceleration or age slowdown with all-cause mortality, we calculated the hazard ratios. We analyzed survival time data (from the age at blood draw until age at death or last follow-up). For hazard analysis by group, we used “coxph” function from the “survival” R package (27).We adjusted Cox models to chronological age and sex. A delta group was assigned according to the difference between predicted and actual age of the sample. Cases where the samples are predicted older than their chronological age, were compared to norms similarly, cases where , samples predicted young, were compared to norms
Results
Population Specific Biomarkers of Human Aging
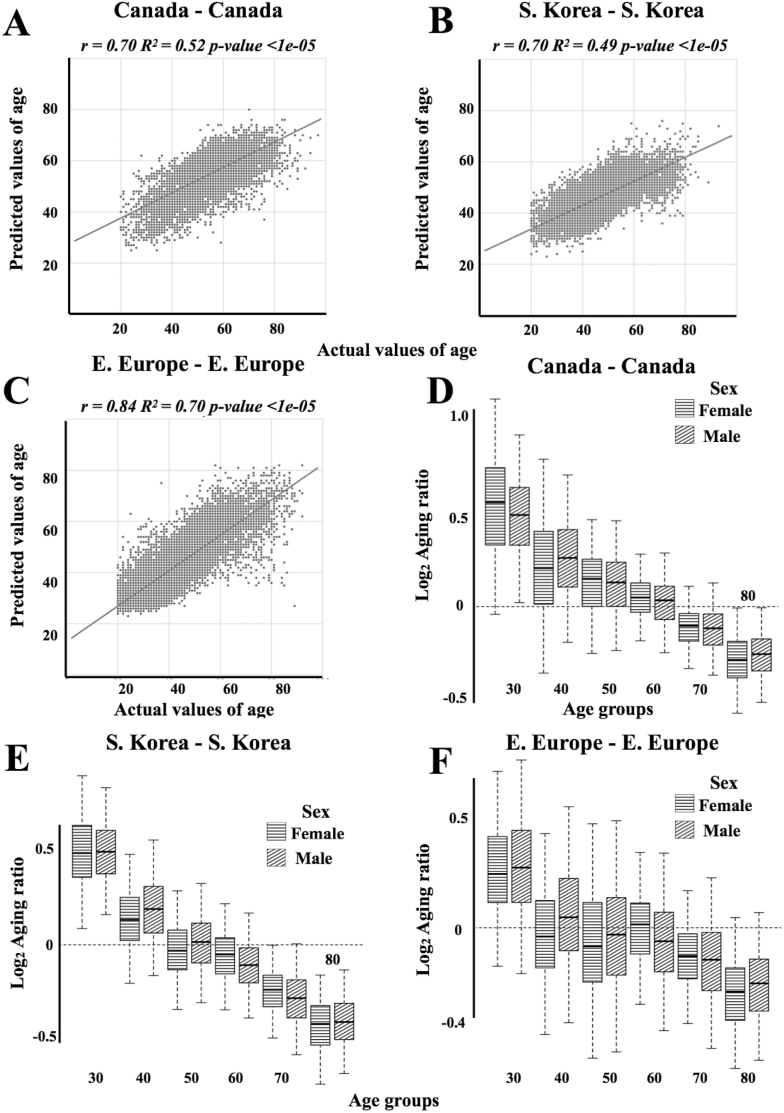
To develop both universal and population specific aging clocks, we trained a series of DNNs on anonymized blood tests for patients from three distinct ethnically diverse populations: South Korean, Canadian, and Eastern-European. The best performing predictor trained upon the Canadian population specific dataset demonstrated an MAE of 6.36 years and an R2 of 0.52 (Figure 2A and D and Table 1). Meanwhile, the best-performing predictor trained upon the South Korean population specific dataset demonstrated an MAE of 5.59 and an R2 of 0.49 (Figure 2B and E and Table 1). Lastly, the best-performing predictor trained upon the Eastern European population specific dataset demonstrated an MAE of 6.25, an R2 of 0.69 (Figure 3C and F and Table 1). The best performing predictor of our previously reported blood biochemistry-based aging clock, Aging.AI, demonstrated R2 = 0.80 and MAE = 6.07 years, while the entire ensemble demonstrated 83.5% epsilon-accuracy R2 = 0.82 and MAE = 5.55 years. Aging.AI was trained on over 56,000 samples of 41 blood biochemistry and cell counts markers of patients from Eastern Europe.
Figure 2.
Actual chronological age vs predicted age for Canadian (A), Korean (B), and European (C) populations of patients. The linear regression line is shown in dark grey. Log2transformed Aging ratio for for Canadian (D), Korean (E), and European (F) population predictions. Log2Aging ratio of 1 means that sample is predicted twice older than a chronological age and Log2Aging ratio of −1 means sample is predicted half as old.
Table 1.
The performance of models trained on the three populations separately and the combined populations. DNNs were trained and tested on four datasets: the South Korean, Canadian, Eastern European, and the combined dataset (All) with 19, 20, and 21 input features.
| Testing Set | Training Set | Features | r (all [f; m]) | R 2 (all [f;m]) | MAE (years) (All [f; m]) |
ε-Accuracy | Std of Δ Between Predicted and Actual Age |
|---|---|---|---|---|---|---|---|
| Canada | S. Korea | 20 | 0.57 [0.55; 0.59] | 0.24 [0.24; 0.24] | 7.87 [7.66; 8.12] | 0.70 [0.73; 0.69] | 6.19 [6.66; 5.69] |
| Canada | 20 | 0.70 [0.70; 0.70] | 0.52 [0.52; 0.47] | 6.36 [6.46; 6.28] | 0.80 [0.79; 0.81] | 5.24 [5.43; 5.05] | |
| 19 (wout sex) | 0.69 [0.69; 0.69] | 0.47 [0.47; 0.46] | 6.48 [6.51; 6.44] | 0.81 [0.79; 0.82] | 5.35 [5.55; 5.12] | ||
| E. Europe | 20 | 0.52 [0.52; 0.52] | 0.24 [0.27; 0.22] | 9.68 [9.98; 9.2] | 0.62 [0.58; 0.70] | 7.83 [8.34; 7.27] | |
| All | 21 | 0.72 [0.73; 0.71] | 0.52 [0.53; 0.50] | 6.17 [6.19; 6.17] | 0.70 [0.69; 0.72] | 5.12 [5.2; 5.02] | |
| 21 eq pop. Densities | 0.72 [0.73; 0.71] | 0.51 [0.52; 0.50] | 5.98 [6.02; 5.97] | 0.83 [0.82; 0.84] | 5.01 [5.09; 4.96] | ||
| 20 (wout pop.) | 0.72 [0.72; 0.71] | 0.51 [0.52; 0.50] | 6.05 [6.08; 6.03] | 0.83 [0.82; 0.84] | 5.03 [5.09; 5.01] | ||
| 20 (wout sex) | 0.69 [0.69; 0.68] | 0.44 [0.45; 0.44] | 6.62 [6.65; 6.59] | 0.80 [0.80; 0.81] | 5.21 [5.3; 5.06] | ||
| S. Korea | S. Korea | 20 | 0.70 [0.73; 0.66] | 0.49 [0.53; 0.43] | 5.59 [5.45; 5.77] | 0.85 [0.86, 0.84] | 4.76 [4.76; 4.76] |
| 20 (wout sex) | 0.69 [0.72; 0.66] | 0.48 [0.52; 0.44] | 5.64 [5.41; 5.81] | 0.87 [0.88; 0.86] | 4.8 [4.81; 4.8] | ||
| Canada | 20 | 0.52 [0.52; 0,47] | 0.24 [0.27; 0.22] | 7.1 [7.66; 8.12] | 0.72 [0.72; 0.71] | 6.32 [6.52; 6.03] | |
| E. Europe | 20 | 0.54 [0.56; 0.52] | 0.29 [0.27; 0.31] | 9.77 [10.21; 9.02] | 0.65 [0.62; 0.68] | 7.34 [7.49; 7.1] | |
| All | 21 | 0.70 [0.73; 0.66] | 0.49 [0.53; 0.43] | 5.60 [5.46; 5.78] | 0.75 [0.74; 0.76] | 4.82 [4.82; 4.8] | |
| 21 eq pop. Densities | 0.67 [0.7; 0.64] | 0.45 [0.49; 0.4] | 6.49 [6.4; 6.57] | 0.81 [0.8; 0.82] | 5.19 [5.20; 5.19] | ||
| 20 (wout pop.) | 0.69 [0.72; 0.65] | 0.46 [0.49; 0.41] | 5.78 [5.51; 5.91] | 0.86 [0.86; 0.86] | 4.85 [4.84; 4.86] | ||
| 20 (wout sex) | 0.68 [0.72; 0.64] | 0.46 [0.49; 0.41] | 5.77 [5.5; 5.91] | 0.86 [0.85; 0.86] | 4.85 [4.85; 4.85] | ||
| E. Europe | Korea | 20 | 0.76 [0.77; 0.75] | 0.34 [0.36; 0.33] | 8.52 [8.89; 7.93] | 0.31 [0.28; 0.35] | 6.47 [6.73; 5.96] |
| Canada | 20 | 0.68 [0.68; 0.72] | 0.27 [0.24; 0.27] | 9.25 [10.09; 7.91] | 0.38 [0.37; 0.38] | 7.28 [7.78; 6.17] | |
| E. Europe | 20 | 0.84 [0.85; 0.82] | 0.69 [0.72; 0.67] | 6.25 [6.24; 6.28] | 0.82 [0.83; 0.80] | 5.34 [5.51; 5.04] | |
| 19 (wout sex) | 0.83 [0.82; 0.84] | 0.70 [0.73; 0.68] | 6.29 [6.28; 6.31] | 0.83 [0.84; 0.80] | 5.39 [5.61; 5.12] | ||
| All | 21 | 0.84 [0.85; 0.82] | 0.70 [0.72; 0.67] | 6.22 [6.15; 6.33] | 0.81 [0.81; 0.81] | 5.28 [5.4; 5] | |
| 21 eq pop. Densities | 0.75 [0.75; 0.74] | 0.56 [0.57; 0.56] | 5.97 [5.94; 6.02] | 0.84 [0.84; 0.84] | 5.07 [5.29; 4.88] | ||
| 20 (wout pop.) | 0.82 [0.82; 0.82] | 0.68 [0.69; 0.68] | 6.57 [6.55; 6.61] | 0.81 [0.81; 0.81] | 5.37 [5.51; 5.09] | ||
| 20 (wout sex) | 0.82 [0.83; 0.82] | 0.67 [0.68; 0.67] | 6.29 [6.27; 6.32] | 0.81 [0.81; 0.81] | 5.3 [5.45; 5.06] | ||
| All | S. Korea | 20 | 0.73 [0.72; 0.73] | 0.44 [0.52; 0.53] | 7.01 [7.16; 6.82] | 0.85 [0.86; 0.84] | 5.85 [6.13; 5.46] |
| Canada | 20 | 0.65 [0.64; 0.66] | 0.36 [0.37; 0.33] | 8.71 [8.25; 7.54] | 0.88 [0.88; 0.88] | 6.49 [6.93; 5.87] | |
| E. Europe | 20 | 0.68 [0.69; 0.66] | 0.46 [0.48; 0.44] | 8.37 [8.78; 8.31] | 0.82 [0.83; 0.80] | 7.08 [7.31; 6.77] | |
| All | 21 | 0.80 [0.82; 0.78] | 0.65 [0.67; 0.61] | 5.94 [5.86; 6.04] | 0.83 [0.84; 0.82] | 5.04 [5.12; 4.93] | |
| 21 eq pop. Densities | 0.71 [0.72; 0.70] | 0.51 [0.53; 0.49] | 6.13 [6.05; 6.19] | 0.83 [0.84; 0.82] | 5.19 [5.27; 5.01] | ||
| 20 (wout pop.) | 0.79 [0.80; 0.78] | 0.62 [0.63; 0.61] | 6.09 [6.01; 6.14] | 0.83 [0.84; 0.82] | 5.15 [5.25; 5.01] | ||
| 20 (wout sex) | 0.78 [0.79; 0.78] | 0.61 [0.61; 0.61] | 6.23 [6.21; 6.25] | 0.82 [0.83; 0.81] | 5.32 [5.36; 5.11] | ||
| NHANES | S. Korea | 20 | 0.60 [0.57; 0.68] | 0.36 [0.32; 0.46] | 11.34 [12.53; 9.78] | 0.50 [0.46; 0.56] | 9.89 [10.27; 9.44] |
| Canada | 20 | 0.56 [0.57; 0.59] | 0.31 [0.32; 0.35] | 12.65 [13.84; 11.09] | 0.46 [0.41; 0.51] | 10.93 [10.96; 10.88] | |
| E. Europe | 20 | 0.68 [0.67; 0.70] | 0.46 [0.45; 0.49] | 7.88 [7.23; 8.37] | 0.40 [0.39; 0.42] | 10.00 [10.27; 9.44] | |
| All | 21 | 0.71 [0.71; 0.72] | 0.50 [0.50; 0.52] | 9.93 [10.36; 9.35] | 0.58 [0.57; 0.61] | 10.57 [11.00; 10.09] |
Note: Models trained on the combined dataset more accurately predict age of individual population samples compared to the population specific models. Exclusion of both sex and population features from training datasets resulted in a decrease in the accuracy of the age prediction. r for Pearson correlation coefficient; R2 for coefficient of determination; MAE for mean absolute error, that shows the average disagreement between actual chronological and predicted ages; f for female and m for male; “wout sex” stands for sets without sex as a feature and “wout pop.” stands for sets without population as a feature; ε-accuracy the accuracy of prediction within a period, which was calculated for ε of 10 years; Std for standard deviation; Δ for delta. The performance of best models (in terms of R2) is in bold.
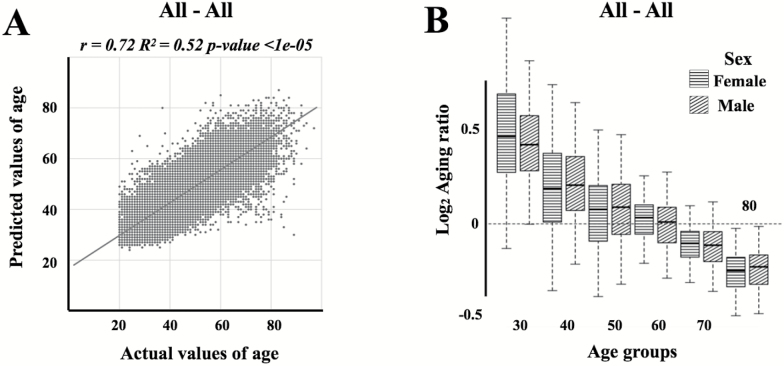
Figure 3.
(A) Actual chronological age vs predicted age for the resulting network trained and tested on the all three populations. The linear regression line is shown in dark grey. (B) Log2 transformed aging ratio. Log2Aging ratio of 1 means that sample is predicted twice older than a chronological age and Log2Aging ratio of −1 means sample is predicted half as old.
To further investigate the importance of population in age prediction, we tested Canadian, South Korean, and Eastern European population of samples on the networks trained on each single population, the results of which are summarized in Table 1. Eastern European samples demonstrated the greatest increase in MAE (9.25 years compared to 6.25 years) and R2 (0.27 compared to 0.69) when tested on the networks trained on the Canadian population. Indeed, almost the same increase in MAE (8.52 compared to 6.25 years) and R2 (0.34 compared to 0.69) were observed when the Eastern European samples were tested on the networks trained on the South Korean population. A similar situation is observed for Canadian and Korean populations tested on the Eastern European network (MAE of 9.68 years compared to 6.36 years and R2 of 0.24 compared to 0.52 for the Canadian population, and MAE of 9.77 years compared to 5.59 years and R2 of 0.29 compared to 0.49 for Korean population). In contrast, the network trained on South Koreans appear to perform almost as well as it does on Canadians, and vica versa.
Next, we trained the age predictor on the combined datasets with population type included as a feature. The best-performing network achieved an R2 of 0.65 and MAE of 5.94 years for the combined population dataset (Figure 3A and B; Table 1), an R2 of 0.52 and MAE of 6.17 years when tested on the Canadian population, an R2 of 0.49 and MAE of 5.60 years when tested on the South Korean population and, an R2 of 0.70 and MAE of 6.22 years when tested on the Eastern European population (Figure 4; Table 1). Using the datasets of all three populations to train the network resulted in an increase in the accuracy of age prediction for both the combined population as well as for each population individually (Table 1 and Supplementary Figure 2).
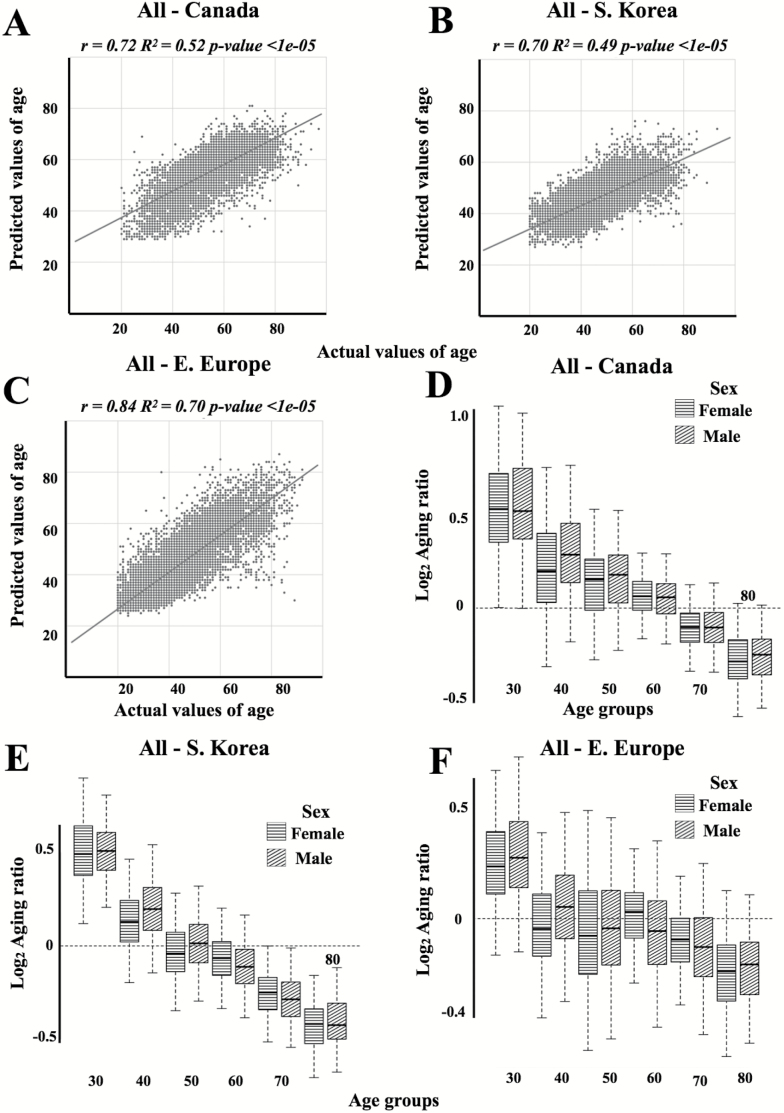
Figure 4.
Actual chronological age vs predicted age for (A) Canadian, (B) Korean, and (C) European patient populations tested on the network trained on all population samples. Linear regression lines are shown in dark grey. Log2, transformed aging ratio for (D) Canadian, (E) Korean, and (F) European populations tested on the network trained on all population samples. Log2Aging ratio of 1 means that sample is predicted twice older than a chronological age and Log2Aging ratio of −1 means sample is predicted half as old.
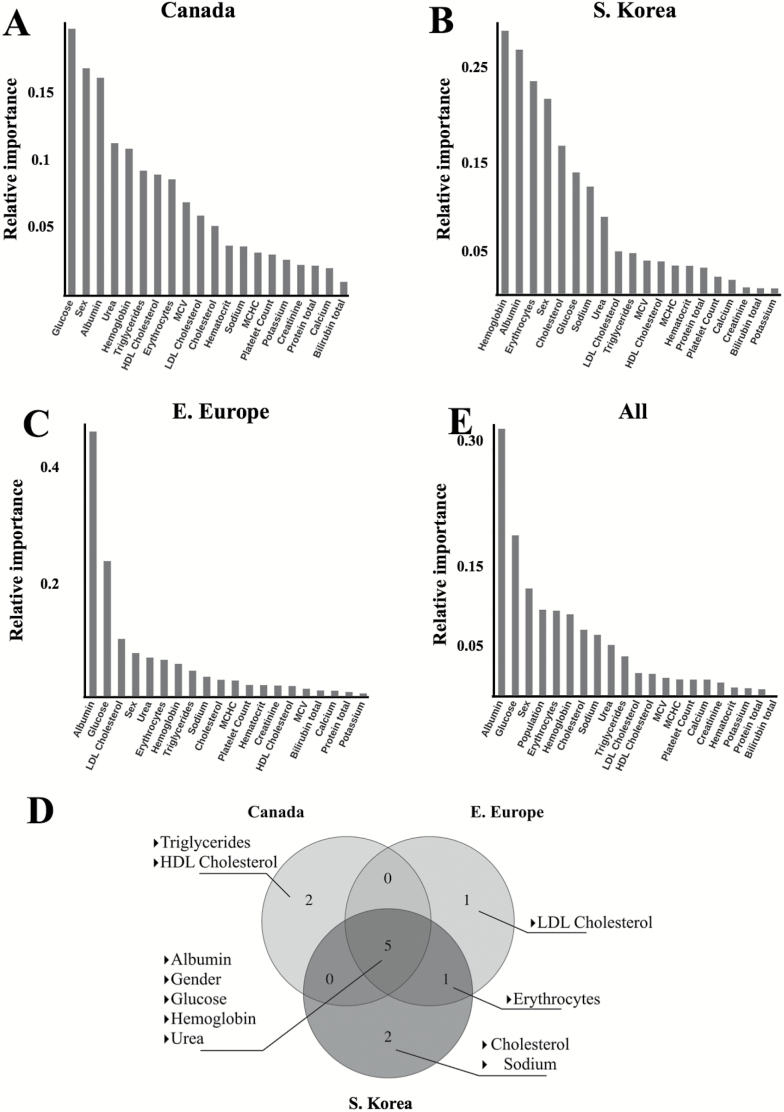
To determine which markers contributed to the predictive accuracy of the network the most and to evaluate possible differences in biological aging between each distinct population, we performed PFI analysis as described in Putin et al. (13). Given that the age distribution and sample size of each population specific dataset differed (Supplementary Figure 1), we equalized the sample size and age distribution of each set, so that an equal number of samples from each age-group of each population specific dataset were included in the final dataset, in order to avoid incorrect ranking of population type during PFI. Additionally, we trained a separate model on a combined dataset excluding population type as a feature to evaluate the change in the predictive accuracy of the model.
Glucose, albumin, sex, urea, hemoglobin, HDL cholesterol, and triglycerides were ranked as the seven most important markers for the Canadian population. Hemoglobin, albumin, erythrocytes, sex, cholesterol, glucose, and sodium were ranked as the seven most important markers for the South Korean population. Albumin, glucose, LDL cholesterol, gender, urea, and erythrocytes were ranked as the seven most important markers for the Eastern European population. The biochemistry markers found to be most important for the predictive accuracy of all three population specific predictors were Albumin, hemoglobin, urea, and glucose (Figure 5A–D). For the model trained on samples from all three populations, we found that the population-type ranked as one of the most important features for age prediction (Figure 5E). Consistent with this finding, the network trained on a feature space that included population type as a feature demonstrated higher performance than the network that excluded population type as a feature (R2 of 0.65 compared to 0.62 and MAE of 5.94 compared to 6.09 years) (Table 1). Exclusion of sex as a feature also significantly reduced the accuracy of the model trained on all three populations (R2 of 0.61 compared to 0.65 and MAE of 6.23 years compared to 5.94 years), but not for models trained on the individual populations (Table 1). In addition, the age of female samples tends to be predicted more accurately compared to male samples for South Korean and Eastern European populations by both population specific models and the universal model, however, no significant difference is observed. Notably, female samples in the Canadian population were predicted less accurately compared to male samples.
Figure 5.
Feature importance plots of the model trained on (A) Canadian population samples, (B) on Korean population samples, and (C) on Eastern European population samples. Permutation feature importance (PFI) method was used to rank blood markers and sex by their importance in age prediction. (D) The top seven most important features across all predictors trained on different populations. Albumin, sex, hemoglobin, and urea are ranked as the most important markers for age prediction in all three models; (E) the most important markers for the network trained on the three populations. Albumin, glucose, and erythrocyte count were ranked as the most markers for age prediction in this model. PFI method was applied to rank blood markers, sex and population by their importance in age prediction.
Validation of Models
The National Health and Nutrition Examination Survey (NHANES) dataset were used to validate our models. We excluded population as a feature in models trained on the individual Canadian, South Korean and Eastern European datasets, as well as the combined dataset to predict the age of NHANES samples based on their blood biochemistry values.
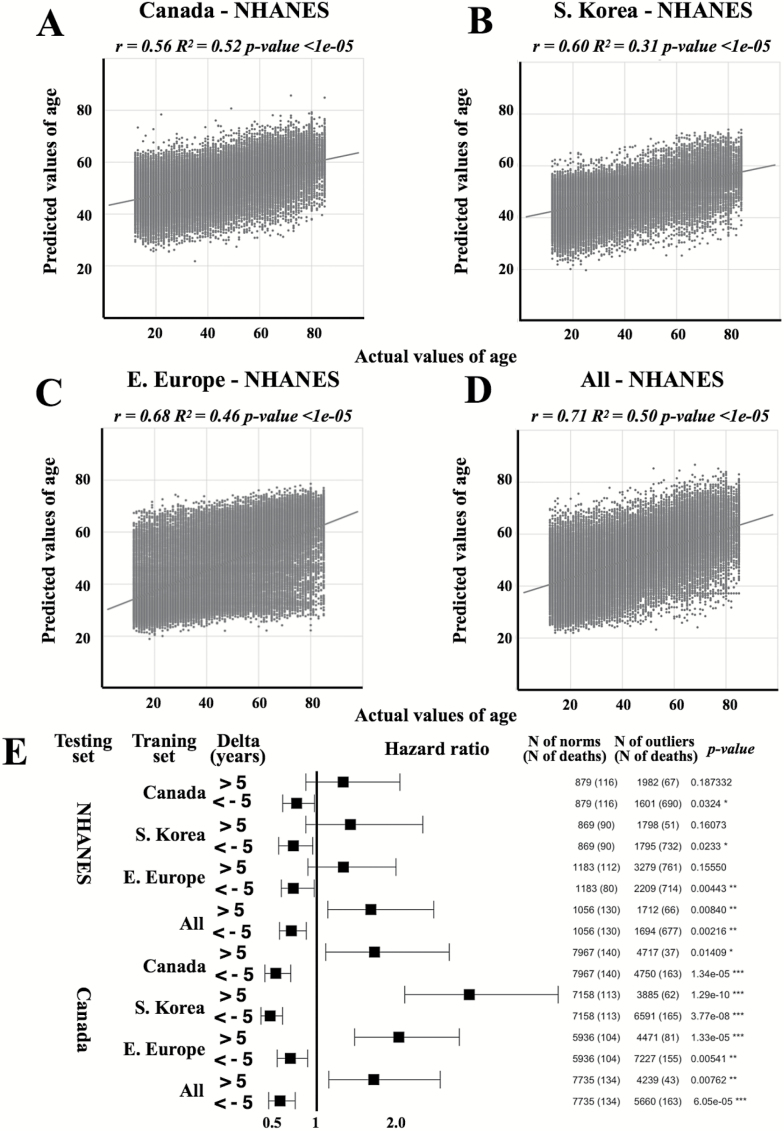
Networks trained on Eastern European and all samples demonstrated higher accuracy in prediction of age of NHANES samples and achieved R2 of 0.46 and MAE of 7.88 and R2 of 0.50 years and MAE of 9.93 years, respectively (Table 1; Figure 6). This performance coincides with the performance of population specific networks tested on other populations. Interestingly, for the NHANES dataset the difference in accuracy of age prediction for male and female samples is higher compared to internal datasets investigated in this study and the age of female samples is predicted less accurately compared to male samples.
Figure 6.
Validation of models. Actual chronological age vs predicted age for NHANES dataset using networks trained on Canadian (A), Korean (B), European (C), and (D) all patient population samples. The linear regression lines are shown in dark grey. Networks trained on both E. European and all patient samples demonstrated the higher accuracy of age prediction of NHANES dataset. (E) Hazard ratios for the NHANES and Canada datasets. A Cox proportional hazards regression model was used to relate survival time to the accelerated aging group (delta >5) and slowed aging group (delta <5). Patients predicted younger their chronological age has a lower mortality risk, while patients predicted older has a higher risk. Each row represents a hazard ratio and 95% confidence interval. Note: “∗∗∗” for p-value of .001; “∗∗” for p-value of .01; “∗” for p-value of .05.
To investigate the predictive ability of deep hematologic aging clocks on mortality, we employed chronological age- and sex-adjusted Cox regression models. Samples predicted younger consistently demonstrated a decrease in the hazard ratio (from 49.2 to 31.5% for the Canadian dataset and from 30.4 to 24% for the NHANES dataset), while samples predicted older demonstrated a significant increase in the hazard ratio (from 69.5 to 185.8% for the Canadian dataset and from 32.6 to 66.2% for the NHANES dataset) (Figure 6E, Supplementary Table 3).
Discussion
Here, we present several novel hematological aging clocks trained using data from several diverse populations. The best-performing predictor achieved an MAE of 5.94 years having greater predictive accuracy than the best-performing predictor of our previously reported aging clock (which achieved an MAE of 6.07 years), despite being trained on a narrower feature space (21 compared to 41 features). Importantly, our deep learned predictors outperformed conventional ML models (Supplementary Figure 3). These results are in line with the hypothesis that ethnically-diverse aging clocks have the potential to predict chronological age and quantify biological age with greater accuracy than generic aging clocks. Furthermore, they have a greater capacity to account for the confounding effect of ethnic, geographic, behavioral, and environmental factors upon the prediction of chronological age and the measurement of biological age.
Albumin, glucose, urea and hemoglobin were among the most important blood biochemistry parameters for all three population specific predictors. Albumin is the most prevalent protein in blood and its primary function is the regulation of oncotic pressure, which is critical for transcapillary fluid dynamics, and deviations in serum albumin levels is associated with a number of pathophysiological conditions (28). Hypoalbuminemia (i.e. abnormally low levels of albumin) is often associated with malnutrition, liver disease, injury, chronic inflammation (29) and the aging process (30). Blood glucose levels, on the contrary, tend to increase with age, and glucose is able to modify proteins via irreversible glycosylation, a feature that is directly associated with the aging process (31). In this regard, low-calorie and low-glucose diets are considered to be one of the most effective antiaging interventions, as well as metformin, a biguanide that reduces glucose levels. Levels of serum urea also increase with age, which is associated with age-related decrease in muscle mass (32). Age-related decrease in hemoglobin is common in the elderly (33), a condition that increases the risk of cardiovascular disease, cognitive decline and an overall decline in quality of life (34). Our hematological clock is consistent with what is already known about the biology and pathophysiology of aging. While the blood parameters are not accurate biomarkers of aging by themselves, when analyzed in combination they can be used to reasonably accurately predict chronological and biological age.
Other groups have reported previously upon sex and ethnicity specific differences in the rate of biological aging as for example quantified by epigenetic aging clocks (35). Here, we report similar findings obtained through the use of hematologic aging clocks. The results of our PFI analysis also found that sex was ranked as one of the important features for age prediction by our system, which is consistent with sex-specific differences in the rate of aging as reported by many other groups; indeed, the phenomenon of sex-dependent differences in aging rates has been widely investigated in model organisms and humans with fairly consistent results (35,36). However, the model trained on both males and females predicts age equally well for both sexes, despite the fact that blood biochemistry values vary between males and females and that such parameters as creatinine, hemoglobin, or HDL Cholesterol have sex-specific normal reference ranges.
The performance of population specific networks is better on the population dataset they were trained on. At the same time, the inclusion of a population into the training set increases the accuracy of age prediction for this population. Accordingly, the network trained on all populations demonstrates almost the same level of performance compared to population specific networks tested on population specific data. Population type was also ranked as one of the most important markers for age quantification perhaps indicating that different populations show variability in their aging phenotype. Indeed, our PFI analysis determined the population type to be one of the most important features in our model, and our population specific predictors achieved greater predictive accuracy than our previously reported DL-based age predictor, despite being trained on a narrower feature space. However, we should emphasize that, while the tests used are completely standardized it is possible that slight biases between batches and labs in different regions could introduce subtle changes in the results. This could perhaps contribute to some of the differences observed between populations.
Previously, Cohen et al. showed age prediction for four blood test datasets obtained from multiple different patient populations of non-Eastern European descent, using a freely available validated blood-based aging clock (www.Aging.AI) (14). Authors highlighted that the lower accuracy reported in their study was partly the result of using a much lower number of input samples [ie, 100 samples compared to 6,242 in our previous study (14)], the limited age range of the samples, and a minimum allowed number of input markers (ie, 10 markers compared to 41 in our previous study (14)). Our current results for the NHANES dataset correspond with the results presented by Cohen and colleagues, DNNs trained upon one population show lower accuracy when predicting age on a different population. However, a network trained on a diverse population, such as Eastern Europeans or on multiple combined datasets demonstrated higher accuracy. Notably, by using the NHANES and Canadian datasets we could test a key requirement for aging clocks: the ability to predict mortality. Importantly, patients that were found to have an older blood-age than their chronological age had increased risk of dying and vice versa. A younger blood age could, therefore, be a useful outcome measure in interventions for healthy aging.
DL-based hematological aging clocks, even when trained on a limited feature space, demonstrate reasonably high accuracy in predicting chronological age. The application of DNNs to the prediction of chronological age and the quantification of biological age allows us to characterize nonlinear dependencies between blood parameters and age. Further, the population-adjusted aging clocks display high levels of generalization, resulting in increased performance when applied to chronological age prediction and biological age quantification of both ethnically-homogenous and heterogenous patient populations. Indeed, going forward we will include additional population specific blood biochemistry datasets in order to further increase the predictive power and general utility of DL-based hematologic aging clocks. Importantly, the continuously updated Aging.AI system is freely available on the www.aging.ai website.
Supplementary Material
Supplementary data is available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Conflict of interest
Polina Mamoshina, Kirill Kochetov, Evgeny Putin, Alexander Aliper, Alex Zhavoronkov are associated with the company, Insilico Medicine, Inc, engaged in drug discovery and aging research.
Acknowledgments
We would like to thank Prof. Yansung Park for establishing the international collaboration and help in data collection. We would like to acknowledge the tremendous support of Mr. Justin Riemer, Assistant Deputy Minister, and Dr. Larry Svenson, Executive Director, Analytics and Performance Reporting Branch, Alberta Health. We are also grateful to Dr. David Onyschuk and Elizabeth Dufraine for help in the generation of the administrative dataset. We are grateful to the editor and reviewers for their constructive input in this manuscript.
References
- 1. WHO | Life expectancy. 2016. http://www.who.int/gho/mortality_burden_disease/life_tables/situation_trends/en/. Accessed November 9, 2017.
- 2. Dwyer-Lindgren L, Bertozzi-Villa A, Stubbs RW, et al. . Inequalities in life expectancy among US counties, 1980 to 2014: Temporal trends and key drivers. JAMA Intern Med. 2017;177:1003–1011. doi:10.1001/jamainternmed.2017.0918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Health expenditure, total (% of GDP) | Data. https://data.worldbank.org/indicator/SH.XPD.TOTL.ZS. Accessed November 9, 2017.
- 4. Heuveline P. Demographic pressure, economic development, and social engineering: An assessment of fertility declines in the second half of the twentieth century. Popul Res Policy Rev. 2001;20:365–396. doi:10.1023/A:1013339124837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhavoronkov A, Litovchenko M. Biomedical progress rates as new parameters for models of economic growth in developed countries. Int J Environ Res Public Health. 2013;10:5936–5952. doi:10.3390/ijerph10115936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moskalev A, Chernyagina E, de Magalhães JP, et al. . Geroprotectors.org: a new, structured and curated database of current therapeutic interventions in aging and age-related disease. Aging (Albany NY). 20157:616–28. doi:10.18632/aging.100799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mak IW, Evaniew N, Ghert M. Lost in translation: Animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6:114–118. [PMC free article] [PubMed] [Google Scholar]
- 8. Aliper A, Belikov AV, Garazha A, et al. . In search for geroprotectors: In silico screening and in vitro validation of signalome-level mimetics of young healthy state. Aging (Albany NY). 2016;8:2127–2152. doi:10.18632/aging.101047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thomas I, Gregg B. Metformin; a review of its history and future: From lilac to longevity. Pediatr Diabetes. 2017;18:10–16. doi:10.1111/pedi.12473 [DOI] [PubMed] [Google Scholar]
- 10. Belsky DW, Huffman KM, Pieper CF, Shalev I, Kraus WE. Change in the rate of biological aging in response to caloric restriction: CALERIE Biobank analysis. J Gerontol A Biol Sci Med Sci. 2017;73:4–10. doi:10.1093/gerona/glx096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi:10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hannum G, Guinney J, Zhao L, et al. . Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi:10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peters MJ, Joehanes R, Pilling LC, et al. ; NABEC/UKBEC Consortium The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi:10.1038/ncomms9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Putin E, Mamoshina P, Aliper A, et al. . Deep biomarkers of human aging: Application of deep neural networks to biomarker development. Aging (Albany NY). 2016;8:1021–1033. doi:10.18632/aging.100968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shipony Z, Mukamel Z, Cohen NM, et al. . Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature. 2014;513:115–119. doi:10.1038/nature13458 [DOI] [PubMed] [Google Scholar]
- 16. Murabito JM, Zhao Q, Larson MG, et al. . Measures of biologic age in a community sample predict mortality and age-related disease: The framingham offspring study. J Gerontol Series A. 2017;glx144. doi:10.1093/gerona/glx144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aliper AM, Csoka AB, Buzdin A, et al. . Signaling pathway activation drift during aging: Hutchinson-Gilford Progeria Syndrome fibroblasts are comparable to normal middle-age and old-age cells. Aging (Albany NY). 2015;7:26–37. doi:10.18632/aging.100717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Belsky DW, Moffitt TE, Cohen AA, et al. . Eleven telomere, epigenetic clock, and biomarker-composite quantifications of biological aging: Do they measure the same thing?Am J Epidemiol. 2017;kwx346. doi:10.1093/aje/kwx346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cohen AA, Morissette-Thomas V, Ferrucci L, Fried LP. Deep biomarkers of aging are population-dependent. Aging (Albany NY). 2016;8:2253–2255. doi:10.18632/aging.101034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rogers RG, Everett BG, Saint Onge JM, et al. . Social, behavioral, and biological factors, and sex differences in mortality. Demography. 2010;47:555–578. doi:10.1353/dem.0.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pilling LC, Atkins JL, Bowman K, et al. . Human longevity is influenced by many genetic variants: evidence from 75,000 UK Biobank participants. Aging (Albany NY). 2016;8:547–560. doi:10.18632/aging.100930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeng Y, Nie C, Min J, et al. . Novel loci and pathways significantly associated with longevity. Sci Rep. 2016;6:21243. doi:10.1038/srep21243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of deep learning in biomedicine. Mol Pharm. 2016;13:1445–1454. doi:10.1021/acs.molpharmaceut.5b00982 [DOI] [PubMed] [Google Scholar]
- 24. Maas AL, Hannun AY, Ng AY. Rectifier nonlinearities improve neural network acoustic models. Proc. ICML. 30;2013. https://pdfs.semanticscholar.org/367f/2c63a6f6a10b3b64b8729d601e69337ee3cc.pdf. [Google Scholar]
- 25. Koushik J, Hayashi H. Improving stochastic gradient descent with feedback November 2016. http://arxiv.org/abs/1611.01505. Accessed April 16, 2017.
- 26. Srivastava N, Hinton G, Krizhevsky A, et al. . Dropout: A simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- 27. Therneau TM, Lumley T.. Survival: survival analysis; 2017. https://CRAN.R-project.org/package=survival. [Google Scholar]
- 28. Doweiko JP, Nompleggi DJ. Role of albumin in human physiology and pathophysiology. JPEN J Parenter Enteral Nutr. 1991;15:207–211. doi:10.1177/0148607191015002207 [DOI] [PubMed] [Google Scholar]
- 29. Don BR, Kaysen G. Serum albumin: Relationship to inflammation and nutrition. Semin Dial. 2004;17:432–437. doi:10.1111/j.0894-0959.2004.17603.x [DOI] [PubMed] [Google Scholar]
- 30. Gom I, Fukushima H, Shiraki M, et al. . Relationship between serum albumin level and aging in community-dwelling self-supported elderly population. J Nutr Sci Vitaminol (Tokyo). 2007;53:37–42. doi:10.3177/jnsv.53.37 [DOI] [PubMed] [Google Scholar]
- 31. Luevano-Contreras C, Chapman-Novakofski K. Dietary advanced glycation end products and aging. Nutrients. 2010;2:1247–1265. doi: 10.3390/nu2121247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Musch W, Verfaillie L, Decaux G. Age-related increase in plasma urea level and decrease in fractional urea excretion: Clinical application in the syndrome of inappropriate secretion of antidiuretic hormone. Clin J Am Soc Nephrol. 2006;1:909–914. doi:10.2215/CJN.00320106 [DOI] [PubMed] [Google Scholar]
- 33. Salive ME, Cornoni-Huntley J, Guralnik JM, et al. . Anemia and hemoglobin levels in older persons: Relationship with age, gender, and health status. J Am Geriatr Soc. 1992;40:489–496. doi:10.1111/j.1532–5415.1992.tb02017.x [DOI] [PubMed] [Google Scholar]
- 34. Stauder R, Thein SL. Anemia in the elderly: Clinical implications and new therapeutic concepts. Haematologica. 2014;99:1127–1130. doi:10.3324/haematol.2014.109967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Horvath S, Gurven M, Levine ME, et al. . An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171. doi:10.1186/s13059-016-1030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Waisman NY, Waisman N, Golubovsky MD, et al. . Differences in the parameters of longevity and its sex-specificity in human populations and modeling them in drosophila. Adv Gerontol. 2013;3:268–276. doi:10.1134/S2079057013040097 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.