Abstract
Cancer cells undergoing starvation- and treatment-induced autophagy were found to exhibit reduced intracellular lactate, reduced rates of steady-state lactate excretion and real-time pyruvate-lactate exchange, indicating that glycolytic metabolism was altered in autophagic cells. In this chapter we describe the technical details of the use of 1H-magnetic resonance spectroscopy (MRS) to measure endogenous cellular concentrations of lactate and glucose in autophagic cells and tissues; how to measure the rate of steady-state lactate excretion and glucose uptake by 1H-MRS in autophagic cells and details of the real-time measurement of [1-13C] pyruvate to lactate exchange in autophagic cells by 13C-MRS-DNP (dynamic nuclear polarization).
Keywords: Magnetic resonance spectroscopy, MRS, dynamic nuclear polarization, DNP, autophagy, lactate, glucose, pyruvate-lactate exchange, glycolysis
1.1. Introduction
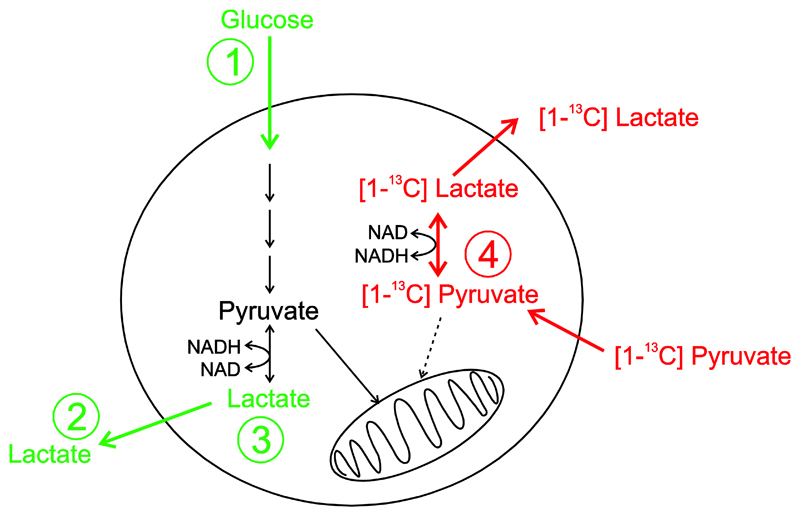
Glucose is metabolised by cells via glycolysis to produce pyruvate. The efficiency of this process is dependent on the rate of glucose uptake which is controlled by the glucose transporters (GLUTS) and the activity of the various enzymes along the glycolytic pathway. Pyruvate is then converted to acetyl-CoA by pyruvate dehydrogenase (PDH) or converted to oxaloacetate by pyruvate carboxylase (PC) and enters the TCA cycle to produce substrates for oxidative phosphorylation in the mitochondria. Alternative metabolic fates for pyruvate include transamination to form alanine or reduction in the cytosol to form lactate. The rate of reduction of pyruvate to lactate is dependent on the expression level and activity of lactate dehydrogenase (LDH) and co-factors such as oxidized (NAD+) and reduced nicotinamide adenine dinucleotide (NADH) (Figure 1), as well as the monocarboxylate (MCT) mediated transport of substrates into and out of the cell. If the availability of oxygen is restricted, then preferential conversion of pyruvate to lactate and concommittent transport of lactate out of the cell regenerates NAD+ in the cytosol which allows glycolysis to continue even in the absence of oxygen such as in ischemia or acute exercise.
Figure 1.
Schematic pathway diagram showing uptake of glucose mediated via the GLUT transporters and subsequent conversion via glycolysis to produce pyruvate and lactate. Pyruvate and lactate are both subject to monocarboxylate transport into and out of the cell. Measurements in green correspond to steady-state 1H MRS analysis of 1) glucose uptake, 2) lactate excretion and 3) intracellular lactate. Measurements in red correspond to real-time 13C MRS analysis of 4) pyruvate-lactate exchange kinetics.
A shift from oxidative phosphorylation to glycolysis and the conversion of pyruvate to lactate for energy production is favoured by cancer cells, even in the presence of freely available oxygen. In cancer there is increased transcriptional regulation of a number of glycolytic and mitochondrial enzymes including lactate dehydrogenase-A (LDH-A), pyruvate dehydrogenase kinase (PDK), among many others. This reprogramming of energy metabolism is called the ‘Warburg effect’ and has become a widely accepted hallmark of cancer. (Hanahan & Weinberg, 2000; Hanahan & Weinberg, 2011; Warburg, 1956)
Glycolytic metabolism was found to be altered in starvation- (amino acid and serum deprived) and treatment- (PI103 and dichloroacetate) induced autophagic cancer cells (Lin, Hill, Andrejeva, Boult, Troy, Fong, et al., 2014a; Lin, Andrejeva, Te Fong, Hill, Orton, Parkes, et al., 2014b). In these experiments induction of autophagy was confirmed in colorectal HT29 and HCT116 bax-ko cancer cells by overexpression of LC3II in western blots and the presence of double membrane autophagic vesicles by electron microscopy, whilst the absence of apoptosis was confirmed by the lack of cleaved PARP or change in caspase 3 expression. Reductions in the rate of lactate excretion and intracellular lactate were observed as well as reductions in pyruvate-lactate exchange kinetics measured in real-time. It was also shown that by replacing the amino acid and serum deprived media with full media or stopping the treatment there was a reversal of this phenomenon, which reported on cellular recovery from autophagy (Lin, et al., 2014a; Lin, et al., 2014b). In these studies, magnetic resonance spectroscopy (MRS) methods were used to examine the rates of lactate production or exchange in steady-state and in real-time, respectively. 1H-MRS analysis was used to measure cellular lactate levels in cell extracts and lactate secretion and glucose uptake in cell culture media. Dynamic nuclear polarization (DNP) and 13C-MRS of 13C-labelled pyruvate was used to measure the rate of pyruvate exchange with lactate in vitro and in vivo in real-time. The mechanisms behind these changes were found to be associated with decreased LDH activity in starvation-induced autophagy and an increased NAD+/NADH ratio in treatment-induced autophagy, suggesting a possible shift in glycolytic metabolism during autophagy (Lin, et al., 2014a; Lin, et al., 2014b).
In this chapter we will describe the technical details of how to measure endogenous cellular lactate and glucose in autophagic cells and tissues by 1H-MRS, how to measure the rate of lactate secretion and glucose uptake in autophagic cells by 1H-MRS as well as the measurement of the real-time exchange of 13C-labelled pyruvate to lactate in cells by 13C-MRS-DNP and the derivation of simple kinetic parameters from these measurements. The methods described here are generally applicable in cells and tissues to investigate cellular processes, cellular metabolism alternations following genetic perturbations or in disease state, as well as monitoring treatment response (Chung, Basetti & Griffiths, 2015; Chung & Griffiths, 2011).
1.2. Magnetic Resonance Spectroscopy
Magnetic Resonance Spectroscopy (MRS) is limited by low signal strength at thermal equilibrium due to low spin polarization. Nevertheless MRS and MR spectroscopic imaging offer chemically specific analysis of metabolite concentrations in body fluids, cell or tissue extracts, intact tissues or in vivo. In combination with metabolomics and statistical methods such as Principle Component Analysis (PCA), MRS has become a widely used tool to measure a wide range of metabolites in cell extracts, whole cells or tissue biopsy samples (Beckonert, Keun, Ebbels, Bundy, Holmes, Lindon, et al., 2007; Chung, Basetti & Griffiths, 2015; Chung & Griffiths, 2011)). Furthermore the ability to perform metabolic imaging in vivo using MRS (de Graaf, 2007) and the translation of these techniques clinically in humans allows for the development of molecular biomarkers of response to therapeutics that act directly on metabolic enzymes and transporters or through the inhibition of cell signalling pathways that transcriptionally control and regulate metabolic enzymes.
MRS exploits an intrinsic property of the nucleus known as spin. A wide range of biologically important isotopes possess a non-zero spin including 1H, 13C, 31P, 23Na, 15N among others. When placed in an external magnetic field of the spectrometer the spin states (angular momentum) of these nuclei become quantized. When excited by a radiofrequency pulse, a characteristic spectrum is generated dependent on the gyromagnetic ratio of the nucleus studied and the electronic environment of the molecule. Thus unique spectral peaks with distinct resonance frequencies known as chemical shift correspond with unique molecular environments. In addition, MRS is quantitative at thermal equilibrium in the sense that the peak integral is proportional to molecular concentration and can be measured directly via the addition of a reference compound of known concentration. Despite limited sensitivity 1H MRS is able to measure metabolite concentrations within the μmol range. In addition, incorporation of stable isotopes such as 13C can report on the steady-state activities and fluxes through different metabolic pathways. 13C isotopomer incorporation can be measured by MRS or mass spectrometry and is dependent on the respective labelling positions in starting substrates and their relative rates of incorporation through different pathways into reaction products and intermediates (Buescher, Antoniewicz, Boros, Burgess, Brunengraber, Clish, et al., 2015; DeBerardinis, Mancuso, Daikhin, Nissim, Yudkoff, Wehrli, et al., 2007).
1.3. Dynamic Nuclear Polarization
The sensitivity limitations of MRS can be overcome by using hyperpolarization techniques to transiently increase the spin polarization and thereby increase sensitivity. Techniques such as Dynamic Nuclear Polarization (DNP) enable significant MRS signal enhancements by a factor greater than 10,000 for low gyromagnetic ratio nuclei such as 13C and 15N in a range of endogenous biological metabolites (Ardenkjaer-Larsen, Fridlund, Gram, Hansson, Hansson, Lerche, et al., 2003). The ability to distinguish parent and downstream metabolites by virtue of a difference in chemical shift allows the measurement of the inter-conversion of metabolites in real-time in cellular systems, whole organ preparations, as well as in vivo, and thereby reports on the activity of endogenous enzymes and membrane transporters that catalyse their kinetic inter-conversion (Day, Kettunen, Gallagher, Hu, Lerche, Wolber, et al., 2007). For a more extensive discussion of DNP, including some of the challenges and limitations of the technique. a number of excellent reviews in this area have recently been published. (Chaumeil, Najac & Ronen, 2015; Comment & Merritt, 2014; Kurhanewicz, Vigneron, Brindle, Chekmenev, Comment, Cunningham, et al., 2011).
To date the most widely employed metabolic substrate for hyperpolarized 13C-MR imaging has been [1-13C] pyruvate (Golman, in't Zandt, Lerche, Pehrson & Ardenkjaer-Larsen, 2006). This is largely due to the very favourable spin lattice relaxation time (T1) of the C1 and C2 carbonyl carbons which is reported to be in the range 40-70s depending on the magnetic field strength (Keshari & Wilson, 2014), as well as the fast rates of MCT mediated entry of pyruvate into the cell. Pyruvate is located at the end point of glycolysis being subject to a number of metabolic fates including LDH mediated exchange with lactate, alanine transaminase (ALT) mediated exchange with alanine, one-way decarboxylation mediated by PDH to form CO2 or one-way carboxylation mediated by PC to form oxaloacetate. The ability to probe the metabolic fates of hyperpolarized pyruvate as well as its associated kinetic rates and metabolic fluxes is therefore of great current interest for imaging metabolic processes in vivo and is complementary to steady-state metabolomics type techniques. The apparent exchange rate constant of hyperpolarized [1-13C] pyruvate to lactate (kPL) has previously been shown to provide a potential metabolic biomarker for diagnosis and for assessing treatment response (Golman, in't Zandt & Thaning, 2006; Golman & Petersson, 2006; Park, Larson, Zierhut, Hu, Bok, Ozawa, et al., 2010; Ward, Venkatesh, Chaumeil, Brandes, VanCriekinge, Dafni, et al., 2010). This apparent rate has been shown to decrease following drug induced cell death, attributed to apoptosis with the activation of poly(ADP-ribose) polymerase (PARP) and depletion of the cofactors NAD(H) (Day, et al., 2007).
Apparent rate constants have also been shown to be dependent on NAD+/NADH ratios (Christensen, Karlsson, Winther, Jensen & Lerche, 2014), the expression and activity of LDH (Ward, et al., 2010) as well as on the activity of the MCT transporter family (Harris, Eliyahu, Frydman & Degani, 2009), which mediate pyruvate and lactate transport into and out of the cell. The rates of exchange mediated pyruvate-lactate conversion have further been shown to correlate with FDG uptake following etoposide treatment in an EL4 tumour model with concomitant decreases in NADH levels and GLUT3 expression (Witney, Kettunen, Day, Hu, Neves, Gallagher, et al., 2009). Kinetic assays using hyperpolarized pyruvate are therefore sensitive to a range of physicochemical properties of the cell that report on glycolytic fluxes, as well as being complementary to more conventional imaging techniques for probing glycolysis such as FDG PET imaging.
2. Measurement of intracellular lactate and glucose level in autophagic cells or tumour extracts
Intracellular lactate and glucose metabolites can be extracted from cultured cells or tissues using a dual-phase extraction method detailed by Tyagi et al (Tyagi, Azrad, Degani & Salomon, 1996).
2.1. Dual phase extraction of cultured cells
-
1)
Seed and treat the cells according to your desired experimental schedule. At least 10 million cells are ideally needed but using smaller number of cells is also possible by acquiring the NMR spectra for longer.
-
2)
Set up 2 extra parallel flasks of cells per condition for cell counting.
-
3)
Remove the culture medium from the cell culture flask at a given time-point using a pipette and store 1ml of the medium in a -20 °C freezer. Media samples are later analysed by 1H-MRS to obtain information on the level of lactate secretion and glucose uptake (see Section 3).
-
4)
Trypsinize the parallel flasks of cells and count the number of cells per flask using standard cell counting protocol.
-
5)
For the cell extraction experiment, wash the cells three times with 10ml ice-cold saline and remove each wash carefully with a pipette.
-
6)
Put 3ml of ice-cold methanol into each flask (for T75 flasks) to cover the cells and keep the flasks on ice for 5-10 mins. The cells are then scraped using a cell scraper and the cell/methanol suspensions are placed into a clean centrifuge tube.
-
7)
Add 3ml of ice-cold chloroform into the tube and vortex the tube vigorously for 30 sec.
-
8)
Pipette 3ml of ice-cold de-ionised water into the tube and again vortex the tube vigorously for 30 sec.
-
9)
Centrifuge the samples for ~5000g at 4°C for 20 mins for phase separation. The final chloroform:methanol:water ratio should be 1:1:1 (v/v/v). The upper methanol-water phase contains the water-soluble cellular metabolites, the middle phase contains the protein pellet and the bottom chloroform phase contains the cellular lipids.
-
10)
Keep and store the cell pellets (the middle phase) at -80°C (for protein concentration determination if required).
-
11)
Pipette the upper methanol-water phase into a clean centrifuge tube and add ~5mg Chelex 100 to remove divalent ions.
-
12)
Centrifuge the sample for ~5000g at 4°C for 5 mins to separate the beads from the solution and transfer the clear supernatant to a clean centrifuge tube.
-
13)
Add 10μL of universal pH indicator solution.
-
14)
Store the supernatant in a -80°C freezer until freeze-drying.
2.2. Dual-phase extraction of tissues
-
1)
Weigh the freeze-clamped or snap frozen tissue sample and record the wet tissue weight.
-
2)
Grind the tissue sample into fine powder in liquid nitrogen using a pestle and mortar and place the ground tissue sample in a clean centrifuge tube.
-
3)
Put 3ml of ice-cold methanol into a clean centrifuge tube with the ground tissue sample and vortex the tube vigorously for 30 sec.
-
4)
Continue with the extraction procedures as described in Section 2.1 from Step 5 onwards.
2.3. Preparation of extracted cell or tissue samples for 1H-MRS analysis
-
1)
Freeze-dry the methanol-water phase of the cell or tumour extract samples (directly from the -80°C freezer) in a freeze-drier until the samples are in a powder form.
-
2)
Reconstitute the freeze-dried sample in 650 μl D2O and 50 μl of 0.75% sodium 3-trimethylsilyl-2,2,3,3-tetradeuteropropionate (TSP) in D2O and centrifuge the sample for ~5000g at 4°C for 5 mins.
-
3)
Put 600 μl of the sample into a 5 mm NMR tube and adjust the sample to pH7 using 0.1 M potassium hydroxide and 0.6% perchloric acid.
2.4. 1H-MRS of extracted cell and tissue samples
-
1)
1H-MR spectra of the extracted samples are acquired at 25°C using a pulse acquire MR sequence with water suppression (1D NOESY presat sequence) in a Broad-Band-Inverse (BBI) NMR probe on a 500 or 600 MHz NMR system. For cell extract and tissue extract samples typical NMR acquisition parameters are: 7500Hz spectral width, 32768 time domain points, 2.7s repetition time and 256 scans.
-
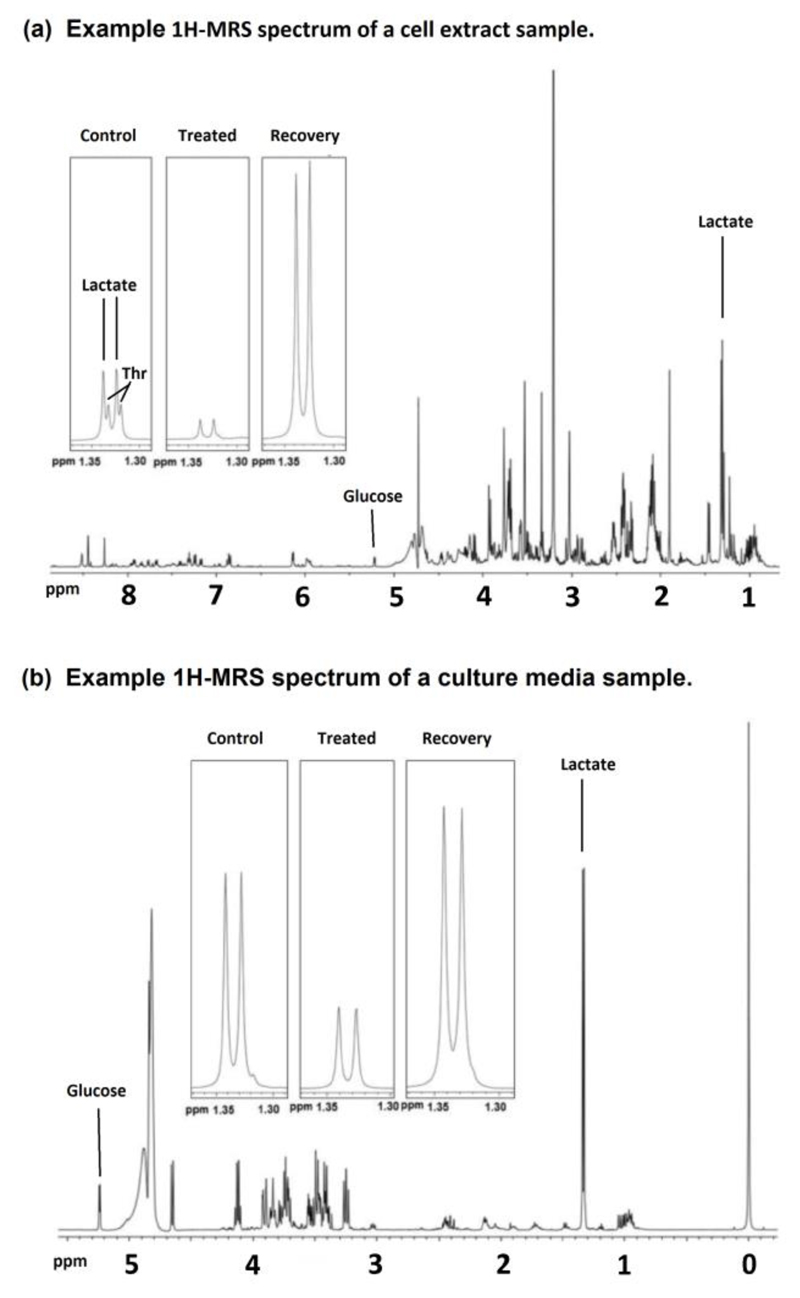
2)
A 1H-MR spectrum from a cell extract sample is shown in Figure 2(a).
-
3)
Commercially available software packages, such as Bruker Topsin-3.2 (Coventry, UK) or MestRe-C-4.9.9.6 (Santiago de Compostela, Spain) are used to analyse the MR spectra.
-
4)
Spectra are first processed by using exponential multiplication with a line broadening factor (lb=0.3Hz), followed by Fourier- transform, zero and first order phase correction, baseline correction and spectral peak integration of lactate and glucose.
-
5)Calculation of cellular lactate or glucose concentrations from a 1H-MR spectrum of cell or tumour extract:
Where: [M]sample is the cellular concentration of lactate or glucose. NTSP is the number of protons giving rise to the signal integral of TSP, NTSP = 9. NMet is the number of protons giving rise to the signal integral of lactate or glucose; NMET = 3 for lactate and NMET = 1 for glucose. ITSP and Isample are the signal integrals of TSP and lactate or glucose, respectively. [TSP] is the concentration of TSP. Wsample is the sample cell number, protein concentration or tissue wet weight. -
6)
Calculate the mean, standard deviation and standard error of metabolite concentrations for control and treated groups (minimum n=3 per group). Perform 2-tails student T-test on control and treatment groups to assess statistical significance. *p<0.05 is taken to be significant.
Figure 2.
Example 1H-MRS spectra of (a) a cell extract sample and (b) a culture media sample from HCT116 Bax-ko cells. Expanded lactate signals are shown for control, 24hr starved and 48hr recovered HCT116 Bax-ko cells. Key: Thr – Threonine. Figure is adapted from Lin et al. (Lin, et al., 2014b).
3. Measurements of the rate of lactate secretion and glucose uptake in cultured cells
3.1. Preparation of media samples for 1H-MRS analysis
-
1)
Put 500 μl of the culture medium (see Section 2.1, step 2) into a 5 mm NMR tube.
-
2)
Add 50 μl D2O and 50 μl 0.75% TSP in D2O in the sample.
3.2. 1H-MRS of media samples
-
1)
1H-MR spectra of the media samples are acquired at 25°C using a pulse acquire MR sequence with water suppression (1D NOESY presat sequence) in a Broad-Band-Inverse (BBI) NMR probe on a 500 or 600 MHz NMR system. For cell culture media samples typical NMR acquisition parameters are: 7500Hz spectral width, 32768 time domain points, 2.7s repetition time and 64 scans. NMR spectra from fresh media samples are also acquired under the same conditions.
-
2)
A 1H-MR spectrum from a culture media sample is shown in Figure 2(b).
-
3)
Commercially available software packages, such as Bruker Topsin-3.2 (Coventry, UK) or MestRe-C-4.9.9.6 (Santiago de Compostela, Spain) are used to analyse the MR spectra.
-
4)
Spectra are first processed by using exponential multiplication with a line broadening factor (Ib=0.3Hz), then followed by Fourier- transform, zero and first order phase correction, baseline correction and spectral peak integration of lactate and glucose.
-
5)The rates of lactate being secreted into the culture media or glucose being taken up from the media are measured from a 1H-MR spectrum of the culture medium that has been incubated with the cells for a given time with respect to the fresh culture medium:
Where: KUptake/Secretion is the rate of glucose uptake from or lactate secreted into the culture media by the cells. A positive value indicates the rate of glucose being taken up from the media by the cells and a negative value shows the rate of lactate being secreted into the media by the cells. ISM is the signal integral of lactate or glucose in the starting media. ICM is the signal integral of lactate or glucose in the media incubated with the cells. ITSP is the signal integral of TSP. NTSP is the number of proton giving rise to the signal integral of TSP, NTSP = 9. NMet is the number of proton giving rise to the signal integral of lactate or glucose; NMET = 3 for lactate and NMET = 1 for glucose. [TSP] is the concentration of TSP. Wsample is the sample cell number or protein concentration. VTot is the total volume of media incubated with the cells. Vtube is the volume of media placed into the NMR tube (500µl as described in Section 3.1). T is the length of time since the last media change. -
6)
Calculate the mean, standard deviation and standard error of the rates of excretion or uptake for control and treated groups (minimum n=3 per group). Perform 2-tails student T-test on control and treatment groups to assess statistical significance. *p<0.05 is taken to be significant.
As an addendum, we further note that the protocols outlined above in sections 2.1 – 3.2 offer a general methodology for extracting metabolites from cells or tissues as well as their measurement in culture media by MRS. The protocols may therefore readily be extended to steady-state 13C-labelling experiments which require similar extraction protocols to those described here. We do not discuss this methodology further in the current context but would also be of interest in the study of autophagic cells, and would be complementary to the real-time hyperpolarized 13C-MR labelling experiments, as discussed below.
4. Hyperpolarization methods for dissolution DNP using pyruvic acid
Dynamic Nuclear Polarization (DNP) exploits the high spin polarization of unpaired electrons in the form of stable free-radicals at very low temperature. When placed in a strong magnetic field and cooled in liquid helium to temperatures <4.2K, unpaired electrons are polarized to near unity owing to their large gyromagnetic ratio. By irradiating the sample close to the electron paramagnetic resonance (EPR) frequency (ωe ± ωC) (where ωe and ωC are the Larmor frequencies of the electron and carbon at a field strength of 3.35T, respectively) polarization is transferred from unpaired electrons to surrounding nuclei. This generates a large population difference or hyperpolarization of the associated nuclear spins, and a significant enhancement of the NMR signal. In 2003 Ardenkjaer-Larsen et al. showed that hyperpolarization generated by DNP at low temperature in the solid-state could be conserved if the sample was rapidly melted and dissolved using an aqueous physiological buffer (Ardenkjaer-Larsen, et al., 2003; Golman, et al., 2006; Wolber, Ellner, Fridlund, Gram, Johannesson, Hansson, et al., 2004). The concentration of the hyperpolarized molecule in solution is typically in the mM range following dissolution, depending on the amount of substrate polarized and the volume of buffer employed for the dissolution. The enhanced signal enables acquisition of 13C spectra with a single acquisition and therefore to derive kinetic rate constants for interconversion of metabolites in real-time.
Samples are polarized in a commercially available Hypersense system (Oxford Instruments). This system consists of a superconducting magnet operating at 3.35T. In the bore of the magnet there is a variable temperature insert (VTI) containing a microwave cavity fed via a microwave guide tube from a 94GHz source. The sample chamber within the VTI is filled with liquid helium supplied from the main cryostat of the magnet and fed along a capillary tube with feedback control from a needle valve operated from a stepper motor outside the magnet. The VTI is connected to a rotary vacuum pump to reduce the pressure in the VTI and achieve an operating temperature of 1.4K.
4.1. Sample preparation for DNP
[1-13C] pyruvic acid (99% isotopically enriched) is prepared as a stock solution containing 15 mM trityl free radical OX63 (tris[8-carboxyl-2,2,6,6-benzo(1,2-d:4,5-d)-bis(1,3)dithiole-4-yl] methyl sodium salt, Oxford Instruments, UK) and 1mM of gadolinium MRI contrast agent in the form of Dotarem (Gadoteric acid: gadolinium complex of 1, 4, 7, 10 tetraazacyclododedane-N,N’,N”,N’” tetraacetic acid).
-
1)
1g bottle of pyruvic acid is used to prepare a stock (Sigma-Aldrich, 99% [1-13C], MW = 89g/mol). Measure the total volume using a Gilson pipette (~850μl/g) and put into an Eppendorf.
-
2)
Weigh 18.4mg of trityl radical (OX063, Oxford Instruments, UK, MW = 1426.8 g/mol) into a separate Eppendorf using an analytical balance and dissolve by adding the pyruvic acid and vortexing for 2mins.
-
3)
Make a stock of Dotarem (gadoterate meglumine, Guerbet, France, 0.5 M concentration) and dilute 1:5 with deionised water to make a 100mM solution.
-
4)
Add 8.5μl of 100mM Dotarem stock to the pyruvic acid stock and vortex for 2mins to achieve a final concentration of 1mM [Gd].
-
5)
Wrap the stock polarization solution in foil and keep in a -20C freezer in the dark to prevent photolysis of the radical.
-
6)
Thaw the stock when required for an experiment and vortex to ensure a homogeneous solution.
-
7)
For in vitro cell experiments, pipette and weigh 14μl (18mg) of the polarization solution into a sample cup corresponding to 200μmol of 13C labelled pyruvic acid.
4.2. Operation of the DNP polarizer
-
1)
Initialise the Hypersense system by pressing “Cooldown” tab. The pump is turned on to evacuate the VTI and once a vacuum is achieved (10-20s) the needle valve gradually opens from 0% (closed) to 99% (fully open) to start filling the VTI with liquid helium.
-
2)
Within ~5mins the system achieves operating conditions with the VTI filled to 65% helium and a temperature of 1.4K. The system is ready to insert a sample.
-
3)
Attach the sample cup containing polarization solution to the end of the insertion stick.
-
4)
Press the “insert sample” button. An over pressure of helium gas is blown into the VTI to prevent contamination from condensation of air. The sample holder is pneumatically raised out of the helium bath and the access valve opens to allow insertion of the sample.
-
5)
Insert the sample stick through the access valve to the bottom of the sample holder and pull the sample release button from the stick to leave the sample cup in situ. Remove the sample stick and press “Finished”.
-
6)
Once inserted the access valve is closed to re-equilibrate to the preset helium level (65%) and temperature (1.4K).
-
7)
Run the Hypersense polarimeter and press “Solid state build-up” to enable build-up monitoring of the polarization using the integrated 13C NMR coil for detection.
-
8)
Select ”polarize sample” and enter a measurement interval (typically 300s) and check “finished”. The microwave source is turned on with the selected frequency of ωe – ωC = 94.1GHz. Polarization build-up is recorded every 5mins. For [1-13C] pyruvic acid polarization typically takes ~1hour (build-up time constant of the order 15mins).
4.3. Dissolution
-
1)
For in vitro cell experiments employing pyruvic acid make up a stock aqueous phosphate buffer containing 1 mM EDTA and 50mM Sodium lactate (Sigma Aldrich, MW = 112G). The in vitro cell experiment includes unlabelled lactate in the buffer (adapted from that presented by Day et al. (Day, et al., 2007) to ensure that the LDH dynamics are at or close to equilibrium so that both forwards and reverse reactions are taking place and therefore that both NAD and NADH are concomitantly interconverted.
-
2)
Inject 4ml of aqueous buffer into the solvent heater vessel at the top of the dissolution stick.
-
3)
For in vitro cell experiments pipette 20μl of 10M NaOH into the bottom of the collection vessel. Attach collection vessel to the exit tube of the polarizer.
-
4)
Position the dissolution stick at the entrance port to the VTI. Press “Run Dissolution” button. The buffer is heated to high temperature (~200°C) whilst gradually elevating the pressure to 10bar.
-
5)
When the solvent pressure reaches 10bar the dissolution stick is pneumatically driven into the VTI (whilst applying an overpressure of helium gas to prevent contamination), engages with the sample cup and there is instantaneous mixing of the frozen hyperpolarized sample and hot buffer. The solution is ejected under pressure into the collection vessel followed by a chase gas to ensure complete retrieval of hyperpolarized solution.
-
6)
The hyperpolarized 13C signal decays with its characteristic solution state T1 and therefore must be used immediately. During transfer of the solution between the magnetic field of the polarizer and the field of the MR spectrometer the sample is subject to significant magnetic field gradients and zero field crossing points should be avoided or the sample maintained in a constant field during transit.
4.4. In vitro Cell assays
In the following protocol we describe an in vitro cancer cell suspension assay for assessing the rates of 13C- pyruvate to lactate exchange. Such approaches have been widely used in the literature. However there are some drawbacks that should be noted including the possibility of cell settling during the experiment which could lead to possible changes in field homogeneity over the course of an experiment. There is also a time delay between harvesting the cells and performing the assay which should be kept to a minimum to ensure good cellular viability whilst maintaining temperature constant at 37°C. In addition in the assay described here, cells are mixed with hyperpolarized pyruvate outside the bore of the magnet and the temporal data is therefore missing the initial 5-10s of the kinetic time-points which could potentially contain important information for the fitting. Some of these issues have been addressed with the introduction of bioreactors that allow in situ measurement of hyperpolarized 13C kinetics from cells grown on beads or encapsulated cells (Harris, et al., 2009; Keshari, Kurhanewicz, Jeffries, Wilson, Dewar, Van Criekinge, et al., 2010; Ward, et al., 2010).
-
1)
The NMR spectrometer is setup employing a Broad-Band-Observe (BBO) NMR probe and is tuned and matched on a non-hyperpolarized 13C-pyruvate solution in the same dissolution buffer with 10% D2O for the lock signal. After shimming on the deuterium lock signal a dummy thermal 13C-MR spectrum is acquired with the same acquisition parameters as the hyperpolarized experiment (below) and a receiver gain of 1024 to ensure functioning of the spectrometer. The test sample is then removed and the lock and sweep are turned off prior to the hyperpolarized experiment.
-
2)
Harvest cultured cells by trypsinization. Wash the cell pellet with culture media (without serum or pyruvate added) twice and spin down the cell pellet after each wash. Perform cell count measurement.
-
3)
Resuspend the cells in 500μl of culture media (serum and pyruvate free) in an Eppendorff tube and maintain at 37ºC. Use fresh cells within 10 minutes of harvesting to ensure good viability.
-
4)
Using a 1ml Gilson, pipette 500μl of cell suspension into a clean 5mm NMR tube. Maintain the cell temperature at 37°C using a heater block.
-
5)
Create a new dataset and run the NMR acquisition prior to inserting the sample. 13C spectra are acquired every 2 seconds in a pseudo two dimensional experiment using a single scan and a 10° flip angle and 128 spectra acquired in 4mins 16s total acquisition time. 13C spectra are acquired with WALTZ16 1H decoupling, a sweep width of 150 ppm (18797Hz), carrier frequency positioned at 150ppm, a time domain of 16384 data points in the FID yielding an acquisition time of 0.436s, a prescan delay of 1.564s to yield a repetition time of 2s and the receiver gain set to 32.
-
6)
Following dissolution immediately inject 100μl of hyperpolarized solution into the NMR tube. Mix vigorously by inverting the tube and shaking to yield a homogenous suspension of cells with a final concentration 8mM hyperpolarized [1-13C] pyruvate, 8mM unpolarized 12C lactate.
-
7)
Insert sample into the NMR spectrometer.
-
8)
At the end of the experiment spectra are Fourier transformed with an exponential line broadening factor (lb = 3Hz) and phase and baseline corrected. Integrate the peak areas of lactate and pyruvate over the time-course of the experiment using Bruker Topsin-3.2 (Coventry, UK) software.
-
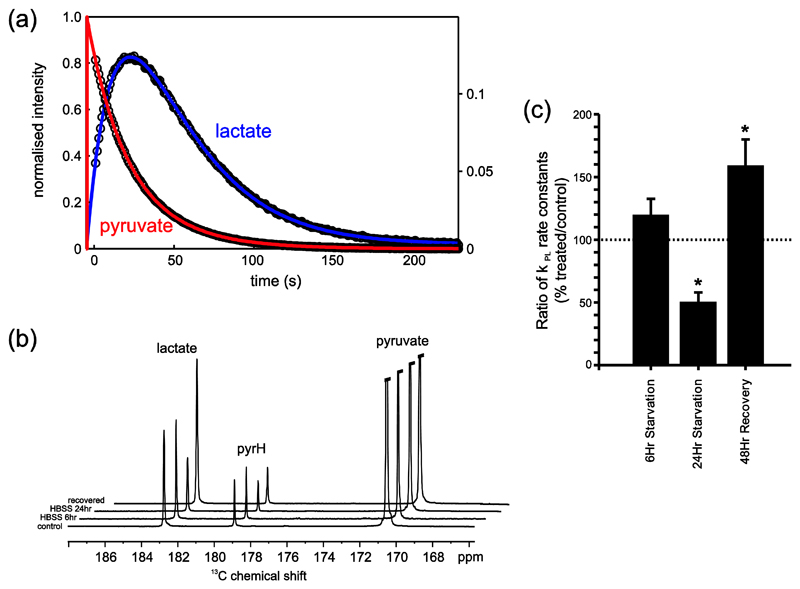
9)
Example fitting of the temporal data of pyruvate and lactate from a HCT116 bax-ko cell suspension is shown in Figure 3(a).
-
10)
Using Topspin software also calculate the sum projection of the time-series of spectra to yield a single 1D 13C-MR spectrum, save spectrum in a separate protocol.
-
11)
Example sum spectra showing the sum over the entire dynamic time series from a control HCT116 Bax-ko cell experiment, following 6hr starvation with HBSS media, following 24 hr starvation with HBSS media and after 24hr starvation followed by 48hr cell recovery in full media are illustrated in Figure 3(b).
Figure 3.
Hyperpolarized 13C-data from an HCT116 Bax-ko cell assay. (a) Example fitting of the temporal data of pyruvate (red - left hand scale) and lactate (green - right hand scale) from which apparent exchange rate constants can be derived. (b) Example sum spectra showing the sum over the entire dynamic time series from a control cell experiment, following 6hr starvation with HBSS media, following 24 hr starvation with HBSS media and after 24hr starvation followed by 48hr cell recovery in full media. The spectrum displays peaks from pyruvate, lactate and pyruvate hydrate (Pyr H). (c) The mean ratios (treated/24hr vehicle-control) of the apparent forward reaction rate of pyruvate to lactate exchange kPL (derived from fitting of the experimental data and normalized to cell number) (± s.e.m.) for 6hr HBSS starvation, 24hr HBSS starvation and 24hr starvation followed by 48hr recovery in HCT116 Bax-ko cells. Minimum n=3 in each group. Statistically significant changes are indicated (*p≤0.05). Figure adapted from (Lin, et al., 2014b).
4.5. Kinetic modelling
Kinetic modeling is carried out by fitting the time-series of peak areas of pyruvate and lactate with the modified Bloch equations, as previously described (Hill, Orton, Mariotti, Boult, Panek, Jafar, et al., 2013). A number of alternative methods have been proposed in the literature for the kinetic analysis of hyperpolarized data, either in vitro such as the experiments presented here, ex vivo in perfused organs or in vivo. A recent paper has compared different quantitative and semi-quantitative methods for assessing 13C-pyruvate metabolism in vivo using a SpinLab system and imaging performed on a clinical scanner (Daniels, McLean, Schulte, Robb, Gill, McGlashan, et al., 2016). These include fitting directly to the differential equations describing a given reaction model (which may assume one-way or two way kinetics), fitting to the integrated solution to a given differential model, or semi quantitative indices such as the time-to-peak, ratio of the lactate to pyruvate peak heights or ratios of the areas under the curve of the lactate and pyruvate peaks. The different methods available for characterizing hyperpolarized kinetics are usually considered to be semi-quantitative, particularly for in vivo experiments, since it is difficult to measure absolute local concentrations of hyperpolarized substrate delivered nor the endogenous pool sizes of metabolites or co-factors involved in a given enzyme reaction. For in vitro experiments we are able to relate the kinetics to absolute concentration of substrate delivered which can be informative when comparing with steady-state measurements of endogenous pools of intracellular and excreted metabolites described previously.
-
1)
The pyruvate and lactate temporal data are fitted simultaneously using a program written in Matlab (Mathworks®, UK). Apparent rate constants of the reaction are derived by kinetic modelling to a first order 2-site exchange model (Day, et al., 2007; Spielman, Mayer, Yen, Tropp, Hurd & Pfefferbaum, 2009; Zierhut, Yen, Chen, Bok, Albers, Zhang, et al., 2010). The program yields estimates of the forward (kPL) and reverse (kLP) rate constants as well as estimates of the relaxation times of pyruvate (T1P) and lactate (T1L). Divide the fitted rates by the number of cells in the sample to yield an effective rate constant per million cells. The rate of pyruvate to lactate exchange in controls and autophagic cells can then be compared (see example in Figure 3(c)).
-
2)
Integrate the lactate and pyruvate peaks from the summed 13C-MR spectrum derived in Section 4.4, step 7. Calculate the ratio of the areas under the curve (AUC) corresponding to the ratio of the integrals of lactate/pyruvate. Divide the AUC by the number of cells in the sample to yield an effective AUC per million cells.
-
3)
Calculate the mean, standard deviation and standard error of kPL derived from the fitting and AUC for different treatment groups (minimum n=3 per group). Perform 2-tails student T-test applied to vehicle and treatment groups to assess statistical significance. *p<0.05 is taken to be significant.
5. Discussion
We have described here the use of in vitro steady-state 1H-MRS methods and real-time hyperpolarized 13C-MRS for probing glycolytic metabolism in cells undergoing autophagy. The methods report on the steady-state rates of glucose uptake and lactate excretion, endogenous concentration of lactate and the rate constants for the exchange of pyruvate to lactate. In turn these measurements are sensitive to a range of cellular parameters including GLUT transport, downstream rates of glycolysis, LDH expression and activity, the ratio of the cofactors NAD+ and NADH and MCT mediated influx and efflux of pyruvate and lactate. The link between autophagy and metabolism is poorly understood but it is likely that autophagy induces multiple cellular metabolic adaptations in cancer cells, one manifestation of which may be a reduced reliance on glycolysis, as reported in our studies (Lin, et al., 2014a; Lin, et al., 2014b). The experiments that we have proposed inform on glycolytic changes that may reflect not only the autophagic cell processes per se but also any direct effects that the treatments may have on metabolism. Thus observations using these techniques should be validated with standard biochemical assays. The methods that we describe here in the context of autophagy are of general applicability in other cell types and will be sensitive to a range of cellular processes and or treatment regimes.
Acknowledgements
This work is support by funding received from the CR-UK Cancer Imaging Centre in association with the MRC and Department of Health (England) grant C1060/A10334, C1060/A16464, NHS funding to the NIHR Biomedical Research Centre.
References
- Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckonert O, Keun HC, Ebbels TMD, Bundy JG, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nature Protocols. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- Buescher JM, Antoniewicz MR, Boros LG, Burgess SC, Brunengraber H, Clish CB, DeBerardinis RJ, Feron O, Frezza C, Ghesquiere B, Gottlieb E, et al. A roadmap for interpreting C-13 metabolite labeling patterns from cells. Current Opinion in Biotechnology. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil MM, Najac C, Ronen SM. Studies of Metabolism Using (13)C MRS of Hyperpolarized Probes. Methods in Enzymology. 2015;561:1–71. doi: 10.1016/bs.mie.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Christensen CE, Karlsson M, Winther JR, Jensen PR, Lerche MH. Non-invasive in-cell determination of free cytosolic [NAD+]/[NADH] ratios using hyperpolarized glucose show large variations in metabolic phenotypes. Journal of Biological Chemistry. 2014;289:2344–2352. doi: 10.1074/jbc.M113.498626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y-L, Basetti M, Griffiths JR. Metabolism and metabolomics by MRS. eMagRes. 2015;4:689–698. [Google Scholar]
- Chung YL, Griffiths JR. Metabolomic Studies on Cancer and on Anticancer Drugs by NMR Ex Vivo. In: Harris RK, Wasylishen RE, editors. Encyclopedia of Magnetic Resonance. Chichester: John Wiley; 2011. [Google Scholar]
- Comment A, Merritt ME. Hyperpolarized Magnetic Resonance as a Sensitive Detector of Metabolic Function. Biochemistry. 2014;53:7333–7357. doi: 10.1021/bi501225t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CJ, McLean MA, Schulte RF, Robb FJ, Gill AB, McGlashan N, Graves MJ, Schwaiger M, Lomas DJ, Brindle KM, Gallagher FA. A comparison of quantitative methods for clinical imaging with hyperpolarized C-13-pyruvate. NMR in Biomedicine. 2016;29:387–399. doi: 10.1002/nbm.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day SE, Kettunen MI, Gallagher FA, Hu DE, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM. Detecting tumor response to treatment using hyperpolarized C-13 magnetic resonance imaging and spectroscopy. Nature Medicine. 2007;13:1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- de Graaf RA. In Vivo NMR Spectroscopy. 2nd Edition. Chichester: John Wiley & Sons, Ltd; 2007. [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golman K, in't Zandt R, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized C-13 magnetic resonance imaging for in vivo tumor diagnosis. Cancer Research. 2006;66:10855–10860. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- Golman K, in't Zandt R, Thaning M. Real-time metabolic imaging. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11270–11275. doi: 10.1073/pnas.0601319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golman K, Petersson JS. Metabolic imaging and other applications of hyperpolarized C-13. Academic Radiology. 2006;13:932–942. doi: 10.1016/j.acra.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harris T, Eliyahu G, Frydman L, Degani H. Kinetics of hyperpolarized C-13(1)-pyruvate transport and metabolism in living human breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18131–18136. doi: 10.1073/pnas.0909049106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DK, Orton MR, Mariotti E, Boult JKR, Panek R, Jafar M, Parkes HG, Jamin Y, Miniotis MF, Al-Saffar NMS, Beloueche-Babari M, et al. Model Free Approach to Kinetic Analysis of Real-Time Hyperpolarized C-13 Magnetic Resonance Spectroscopy Data. Plos One. 2013;8 doi: 10.1371/journal.pone.0071996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshari KR, Kurhanewicz J, Jeffries RE, Wilson DM, Dewar BJ, Van Criekinge M, Zierhut M, Vigneron DB, Macdonald JM. Hyperpolarized C-13 Spectroscopy and an NMR-Compatible Bioreactor System for the Investigation of Real-Time Cellular Metabolism. Magnetic Resonance in Medicine. 2010;63:322–329. doi: 10.1002/mrm.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshari KR, Wilson DM. Chemistry and biochemistry of C-13 hyperpolarized magnetic resonance using dynamic nuclear polarization. Chemical Society Reviews. 2014;43:1627–1659. doi: 10.1039/c3cs60124b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, DeBerardinis RJ, Green GG, Leach MO, Rajan SS, Rizi RR, et al. Analysis of Cancer Metabolism by Imaging Hyperpolarized Nuclei: Prospects for Translation to Clinical Research. Neoplasia. 2011;13:81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Hill DK, Andrejeva G, Boult JKR, Troy H, Fong ACLFWT, Orton MR, Panek R, Parkes HG, Jafar M, Koh DM, et al. Dichloroacetate induces autophagy in colorectal cancer cells and tumours. British Journal of Cancer. 2014a;111:375–385. doi: 10.1038/bjc.2014.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Andrejeva G, Te Fong ACW, Hill DK, Orton MR, Parkes HG, Koh DM, Robinson SP, Leach MO, Eykyn TR, Chung YL. Reduced Warburg Effect in Cancer Cells Undergoing Autophagy: Steady-State H-1-MRS and Real-Time Hyperpolarized C-13-MRS Studies. Plos One. 2014b;9 doi: 10.1371/journal.pone.0092645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I, Larson PEZ, Zierhut ML, Hu S, Bok R, Ozawa T, Kurhanewicz J, Vigneron DB, VandenBerg SR, James CD, Nelson SJ. Hyperpolarized C-13 magnetic resonance metabolic imaging: application to brain tumors. Neuro-Oncology. 2010;12:133–144. doi: 10.1093/neuonc/nop043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman DM, Mayer D, Yen YF, Tropp J, Hurd RE, Pfefferbaum A. In Vivo Measurement of Ethanol Metabolism in the Rat Liver Using Magnetic Resonance Spectroscopy of Hyperpolarized [1-C-13]Pyruvate. Magnetic Resonance in Medicine. 2009;62:307–313. doi: 10.1002/mrm.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi RK, Azrad A, Degani H, Salomon Y. Simultaneous extraction of cellular lipids and water-soluble metabolites: evaluation by NMR spectroscopy. Magnetic Resonance in Medicine. 1996;35:194–200. doi: 10.1002/mrm.1910350210. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Ward CS, Venkatesh HS, Chaumeil MM, Brandes AH, VanCriekinge M, Dafni H, Sukumar S, Nelson SJ, Vigneron DB, Kurhanewicz J, James CD, et al. Noninvasive Detection of Target Modulation following Phosphatidylinositol 3-Kinase Inhibition Using Hyperpolarized (13)C Magnetic Resonance Spectroscopy. Cancer Research. 2010;70:1296–1305. doi: 10.1158/0008-5472.CAN-09-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witney TH, Kettunen MI, Day SE, Hu DE, Neves AA, Gallagher FA, Fulton SM, Brindle KM. A Comparison between Radiolabeled Fluorodeoxyglucose Uptake and Hyperpolarized C-13-Labeled Pyruvate Utilization as Methods for Detecting Tumor Response to Treatment. Neoplasia. 2009;11:574–U588. doi: 10.1593/neo.09254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolber J, Ellner F, Fridlund B, Gram A, Johannesson H, Hansson G, Hansson LH, Lerche MH, Mansson S, Servin R, Thaning M, et al. Generating highly polarized nuclear spins in solution using dynamic nuclear polarization. Nuclear Instruments & Methods in Physics Research Section a-Accelerators Spectrometers Detectors and Associated Equipment. 2004;526:173–181. [Google Scholar]
- Zierhut ML, Yen YF, Chen AP, Bok R, Albers MJ, Zhang V, Tropp J, Park I, Vigneron DB, Kurhanewicz J, Hurd RE, et al. Kinetic modeling of hyperpolarized 13C1-pyruvate metabolism in normal rats and TRAMP mice. Journal of Magnetic Resonance. 2010;202:85–92. doi: 10.1016/j.jmr.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]