Abstract
PARP inhibitors (PARPi), a cancer therapy targeting poly(ADP-ribose) polymerase, are the first clinically approved drugs designed to exploit synthetic lethality, a genetic concept proposed nearly a century ago. Tumors arising in patients who carry germline mutations in either BRCA1 or BRCA2 are sensitive to PARPi because they have a specific type of DNA repair defect. PARPi also show promising activity in more common cancers that share this repair defect. However, as with other targeted therapies, resistance to PARPi arises in advanced disease. In addition, determining the optimal use of PARPi within drug combination approaches has been challenging. Nevertheless, the preclinical discovery of PARPi synthetic lethality and the route to clinical approval provide interesting lessons for the development of other therapies. Here, we discuss current knowledge of PARP inhibitors and potential ways to maximize their clinical effectiveness.
DNA damage and its repair or lack thereof is central to the induction of mutations, which drive the development of nearly all cancers. Healthy cells defend themselves against the deleterious effects of DNA damage through an inter-related series of molecular pathways, the DNA damage response (DDR), that recognize DNA damage, stall the cell cycle and mediate DNA repair, thus maintaining the integrity of the genome. Key to the DDR are the Poly(ADP-ribose) Polymerase 1 and 2 (PARP1 and PARP2) enzymes, DNA damage sensors and signal transducers that operate by synthesizing negatively charged, branched poly(ADP-ribose) (PAR) chains (PARylation) on target proteins as a form of post-translational modification (1). PARP1 binds damaged DNA at single strand DNA breaks (SSBs) and other DNA lesions, an event that causes a series of allosteric changes in the structure of PARP1 that activate its catalytic function (1–5) (Fig. 1). This leads to the PARylation and recruitment of DNA repair effectors such as XRCC1 as well as the remodelling of chromatin structure around damaged DNA as part of the DNA repair process. PARP1 eventually PARylates itself (autoPARylation) - the negative charge that PAR chains impart upon PARP1 likely causes its release from repaired DNA (1–5) (Fig. 1B).
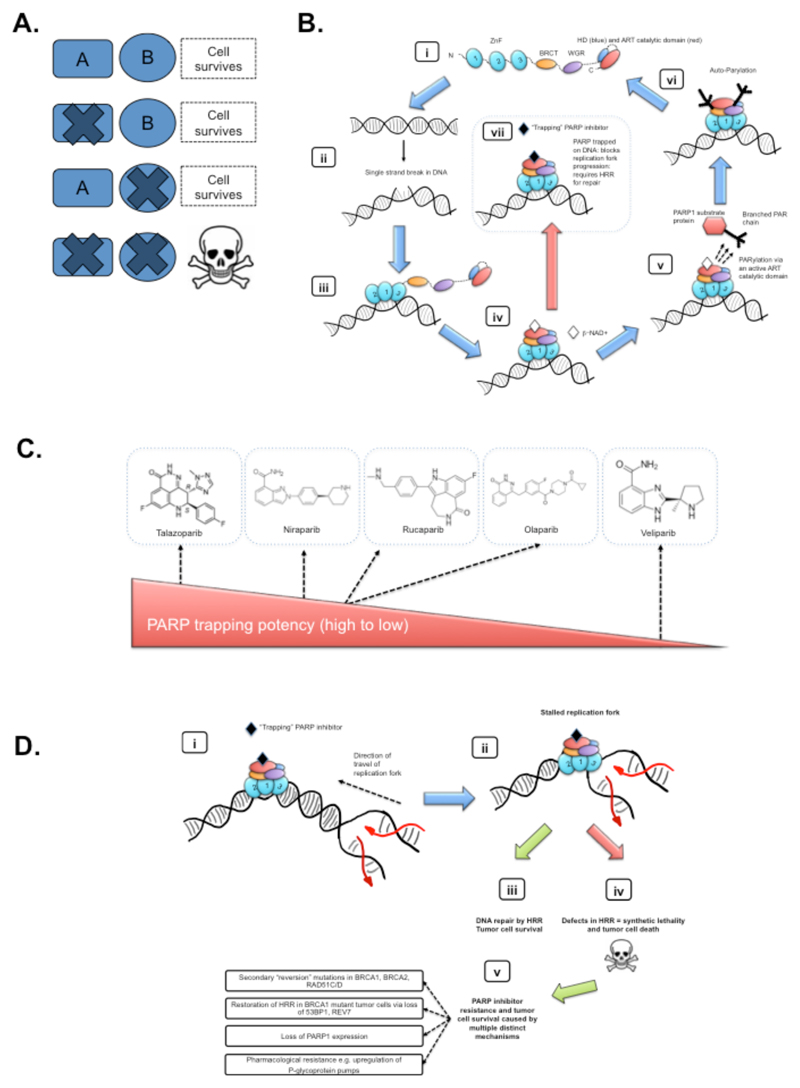
Fig. 1. Mechanism of action of PARPi.
(A) Schematic of synthetic lethality. In its simplest form, the simultaneous alteration of two genes or proteins (shown here as A and B) causes cell death, whilst alteration of either gene/protein alone does not. When the concept is applied to cancer treatment, where gene A represents an oncogene, tumor suppressor gene or oncogenic process/pathway, gene B, once identified, becomes a candidate therapeutic target that can be used to target tumor cells with dysfunction in A. (B) A model describing the PARP1 catalytic cycle. (i) In its non-DNA bound state, PARP1 exists in a relatively disordered conformation, commonly referred to as “beads on a string” (4). The domain structure of PARP1 is shown, including three Zinc-finger related domains (ZnF 1, 2 and 3), BRCT, WGR and catalytic domain encompassing two subdomains; a helical domain (HD) and an ADP-ribosyltransferase (ART) catalytic domain. In this non-DNA bound state, HD acts as an auto-inhibitory domain preventing binding of the PARP-superfamily co-factor, β-NAD+, to its ART binding site (5). (ii) Damage of the DNA double helix often causes the formation of single strand DNA breaks (SSBs, pre-damaged and damaged DNA structures are shown); SSBs cause a change in the normal orientation of the double helix, which in-turn, (iii) provides a binding site for DNA binding PARP1 ZnF domains. The interaction of ZnF 1,2 and 3 with DNA initiates a step-wise assembly of the remaining PARP1 protein domains onto the PARP1/DNA nucleoprotein structure, shown in (iv); this process leads to a change in HD conformation, and resultant loss auto-inhibitory function, thus allosterically activating PARP1 catalytic activity (5). (v) ART catalytic activity drives the PARylation of PARP1 substrate proteins (branched PAR chains are shown on a target protein), mediating the recruitment of DNA repair effectors, chromatin remodelling and eventually DNA repair. (vi) PARP1 autoParylation (likely in cis at SSBs but possibly in trans at other DNA lesions (4)) finally causes the release of PARP1 from DNA and the restoration of a catalytically inactive state (shown in (i)). (viii) Several clinical PARPi, each of which binds the catalytic site, prevent the release of PARP1 from DNA, “trapping” PARP1 at the site of damage, potentially removing PARP1 from its normal catalytic cycle. These images are schematic; detailed structures and models of PARP1/DNA nucleoprotein complexes are described elsewhere [(4, 5) and references therein]. (C) Clinical PARP inhibitors. Chemical structures of five clinical PARPi are shown. The ability of each PARPi to trap PARP1 on DNA differs (talazoparib being the most potent PARP1 trapping inhibitor, veliparib being the least potent) and somewhat correlates with cytotoxic potency (22–24). (D) A model of PARP inhibitor synthetic lethality. Trapped PARP1/DNA nucleoprotein complexes impair the progression of replication forks. (i) schematic of trapped PARP1 on DNA in front of a replication fork; newly synthesized DNA is shown in red. (ii) The replication fork is impeded by trapped PARP1. This normally induces a DNA damage response. (iii) Homologous recombination repair (HRR), involving BRCA1 and BRCA2 tumor suppressor proteins, is the optimal DNA repair process for repairing and restarting replication forks stalled by PARPi and also involves the use of additional “BRCAness” proteins (see main text). In the absence of effective HRR, cells use DNA repair processes that can potentially generate large-scale genomic rearrangements, which often leads to tumor cell death and synthetic lethality. (v) Even where HRR is defective, PARPi resistance occurs. Multiple mechanisms cause PARPi resistance, but can be broadly classified into the examples shown (see also main text).
An understanding of the functions of PARP1 and PARP2 in the DDR drove long-standing efforts to develop small molecule PARP1/2 inhibitors (PARPi) (Fig. 1C) (6). The original rationale was that PARPi could sensitize tumor cells to conventional treatments that cause DNA damage, including multiple chemotherapy or radiotherapy approaches, which remain the backbone of treatment for most cancer patients. By inhibiting PARP-mediated repair of DNA lesions created by chemo- or radiotherapy, greater potency might be achieved. About 30 years ago, small molecule nicotinamide analogs were shown to inhibit PARylation and to enhance the cytotoxicity of dimethyl sulphate, a DNA damaging agent (7–9). Subsequent drug discovery efforts led to the development of clinical PARPi including veliparib (Abbvie), rucaparib (Pfizer/Clovis), olaparib (KuDOS/AstraZeneca) and niraparib (Merck/Tesaro). More recently, a second generation, more potent PARPi, talazoparib (Lead/Biomarin/Medivation/Pfizer) has also been developed (10). These PARPi all interact with the binding site of the PARP enzyme cofactor, β-NAD+, in the catalytic domain of PARP1 and PARP2, but, as discussed later, have differing effects in terms of their cytotoxic potency and ability to “trap” PARP1 on DNA.
Carriers of deleterious heterozygous germline mutations in the BRCA1 and BRCA2 genes have significantly elevated risks of developing breast, ovarian and other cancers (11–13). Because the wild-type BRCA allele is lost during tumorigenesis, these genes are considered classical tumor suppressors. Both BRCA1 and BRCA2 proteins are critical to the repair of double strand DNA breaks by a process called homologous recombination repair (HRR), a form of DNA repair that uses a homologous DNA sequence to guide repair at the DSB. HRR is generally a “conservative” mechanism, in that it restores the original DNA sequence at the site of DNA damage (14). When cells become HRR deficient, whether driven by defects in BRCA1, BRCA2 or other pathway components, non-conservative forms of DNA repair predominate, such as Non-Homologous End Joining (NHEJ). These processes either fuse broken DNA ends at the DSBs without using a homologous DNA sequence to guide repair or fuse regions of DNA close the site of the DSB that exhibit short regions of DNA sequence homology, deleting the intervening DNA sequence. The preferential use of these non-conservative repair mechanisms in the absence of HRR therefore often leads to DNA alterations including deletions of genetic material (15–17). Some of the mutations that arise in this way may foster cancer initiation or progression potentially explaining at least in part why mutations in BRCA1 and BRCA2 increase cancer risk; additional roles of BRCA1 and BRCA2 in processes such as chromatin remodeling and transcriptional regulation may also be relevant to pathogenesis (18).
In 2005, two groups described the Synthetic Lethal (SL) interaction (Fig. 1A) between PARP inhibition and BRCA1 or BRCA2 mutation, suggesting a novel strategy for treating patients with BRCA-mutant tumors (19, 20). SL is a concept introduced nearly a century ago by geneticists to describe the situation whereby a defect in either one of two genes has little effect on the cell or organism but a combination of defects in both genes results in death (21). The demonstration that BRCA-mutant tumor cells were as much as 1,000 times more sensitive to PARPi than BRCA-wild type cells (depending on the PARPi used and the experimental format) (19) provided the impetus for PARPi to be tested in clinical trials as single agents. Originally it was proposed that the mechanism underlying the SL interaction was that PARP inhibition caused persistent SSBs which, when encountered by a replication fork sometimes resulted in collapse of the fork, potentially creating a DSB (19). However, this initial model has recently been modified as a result of data suggesting that some PARPi (especially rucaparib, olaparib, niraparib and talazoparib) “trap” PARP1 on DNA, preventing autoPARylation and PARP1 release from the site of damage and therefore interfering with the catalytic cycle of PARP1 (22–24) (Fig. 1B,D). This trapped PARP1 protein has been suggested to be the relevant cytotoxic lesion at least for some PARPi, a situation analogous to the mechanism of action of cancer drugs that inhibit Topoisomerase II, which also “trap” a DNA repair protein on the double helix. Supporting this contention, PARP1 defective cells appear to be resistant to PARPi (22, 25). Clinically used PARPi differ in their ability to trap PARP1; talazoparib is approximately 100-fold more potent than niraparib in this respect, which in turn traps PARP1 more potently than olaparib and rucaparib (23) (Fig. 1C). In contrast, veliparib appears to have a limited ability to trap PARP1, despite its ability to inhibit PARylation (Fig. 1C). These differences in PARP1 trapping, rather than simply the ability to inhibit PARylation, may be a better predictor of in vitro cytotoxicity in BRCA mutant cells, with talazoparib having the most profound cytotoxic effects (10, 22, 23). Differences in PARP1 trapping activity may also have to be a consideration when designing combination therapies involving PARPi (see below); it is possible that the ability to trap PARP1 may produce unacceptable toxicity when some PARPi are combined with conventional doses of cytotoxic chemotherapies. It is important to note that PARP1 and PARP2 have multiple important roles beyond the DDR such as transcription, apoptosis and immune function; the anti-tumor efficacy of PARPi might also reflect alterations in these functions (3).
As with other targeted therapies, acquired resistance to PARPi therapy has been observed in most patients with advanced cancer (Fig. 1D). Multiple potential mechanisms of resistance to PARPi have been identified through in vitro experimentation. These include inactivation of the DNA repair proteins 53BP1 (26) or REV7 (27), which result in the restoration of HRR and loss of a number of proteins, including PARP1 itself (25), which are involved in maintaining replication fork stability (28). Secondary “revertant” mutations in BRCA1 or BRCA2 that restore the open reading frame of the genes and restore sufficient HRR function have also been shown to lead to PARPi resistance (29, 30); such mutations also cause clinical resistance to platinum-based chemotherapy (29, 31–34). The latter mechanism is the only one to be fully clinically validated as a mechanism of resistance to PARPi. As with other targeted therapies, the selective pressure that PARPi provide in BRCA-defective tumor cells drives the emergence of resistant clones, at least in advanced cancers. Alternative treatment strategies that suppress or at least delay the emergence of resistant clones are therefore now required. Moreover, as some of these mechanisms cause resistance to both PARPi and platinum-based drugs, careful consideration needs to be given to the therapies given to patients both prior to and after PARPi treatment.
The term “BRCAness” has been used to describe tumors that have not arisen from a germline BRCA1 or BRCA2 mutation (gBRCAm) but nonetheless share certain phenotypes, in particular an HRR defect, with these hereditary cancers (34, 35). For example, somatically occurring mutations in either BRCA1 or BRCA2 at least partially phenocopy gBRCAm in terms of causing an HRR defect, as does somatic promotor hypermethylation of BRCA1 or mutations in other genes that are involved in DSB repair and the stability of replication forks (34, 35). Consistent with the HRR defect, tumors with BRCAness might also share therapeutic vulnerabilities with gBRCAm tumors, such as platinum salt sensitivity (36). Shortly after the demonstration that BRCA1 or BRCA2 mutant cells were highly susceptible to PARPi, deficiencies in a number of tumour suppressor genes involved in HRR such as ATM, ATR, PALB2, and the FANC gene family, were also shown to confer sensitivity to PARPi (34, 37). Large-scale cancer genome sequencing projects also revealed that somatic mutations in genes involved in HRR occur in a wide spectrum of tumors (34), in particular in significant fractions of high-grade serous ovarian cancer (HGS-OVCa) (38), advanced prostate cancer (39) and pancreatic cancer (40, 41). These and other cancers with HRR mutations are therefore candidates for testing PARPi efficacy. Considerable effort has also been invested in other molecular profiling approaches, such as transcriptional and mutational signature profiling, that might identify BRCAness and PARPi sensitive tumors (34) as well as considering prior platinum sensitivity as a predictor of BRCAness and subsequent PARPi responses (36). Using mutational signatures (i.e. the number and type of mutations found in a tumor) to identify BRCAness is based on the observation that the genomes of tumors in cancer patients with germ-line BRCA gene mutations (gBRCAm) tend to display a characteristic pattern of mutations, a mutational “scar” that includes large-scale genomic rearrangements, that likely reflects the preferential use of non-conservative DNA repair mechanisms in the absence of HRR over the lifetime of the disease; similar mutational scars are also apparent in tumors that do not have BRCA gene mutations, raising the possibility that the presence of such a “BRCAness scar” could be used to predict clinical responses to agents such as PARPi or platinum salts in patients without gBRCAm (34). Which of these molecular profiling-based biomarkers will be most effective in predicting clinical responses to drugs such as PARPi, however, is not yet clear. For example, whilst mutational scars of BRCAness, as they are currently measured, probably reflect the mutational processes that alter the genome in the absence of HRR over the entire lifetime of a tumor, they might not provide an accurate estimation of whether HRR is still defective in tumor cells at the time that treatment is delivered, a likely pre-requisite for a profound and sustained anti-tumor response to a PARPi, or even, how plastic or reversible the extant HRR defect is. Although a further refinement of how such scars are quantified might allow more accurate predictions to be made (for example, by assessing the clonality of BRCAness mutational scars across multiple clones within a single tumor), one solution to this problem might be to directly measure HRR functionally in tumor specimens using approaches such as the localisation or activity of key DNA repair proteins that are involved in this DNA repair process, such as RAD51 (34, 42, 43) (Fig. 2A). Although this has not as yet been addressed, using functional HRR biomarkers alongside BRCAness scar assessments, might turn out to be the most effective way of identifying BRCAness.
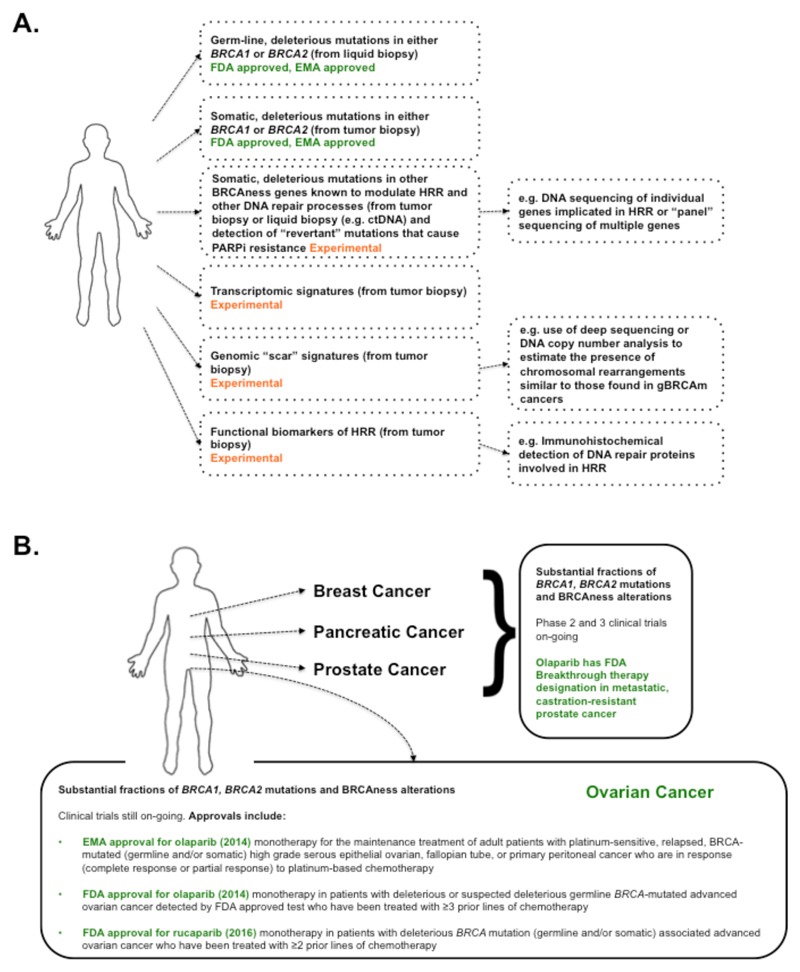
Fig. 2. Clinical PARPi synthetic lethality.
(A) Predictive biomarkers of PARP inhibitor sensitivity. Companion FDA and EMA approved tests, which detect the presence of either germ-line or somatic BRCA1 or BRCA2 mutations, are currently used to identify patients likely to respond to PARPi therapy. Experimental biomarkers (i.e. non-approved biomarkers where the sensitivity (true positive rate/proportion of positives that are correctly identified) and/or specificity (true negative rate) are not as yet clear) are currently in development. These include approaches that estimate the presence/absence of an HRR defect via the identification of DNA mutations in BRCAness genes that control tumor cell responses to PARPi, approaches that estimate an HRR defect by identifying the extent and type of chromosomal alterations often found in BRCA mutant and BRCAness tumors and also functional biomarkers that use the visualization of key proteins involved in HRR as a predictor of the ability to repair PARPi-induced DNA lesions. (B) Clinical assessment of PARPi synthetic lethality. Most clinical trials assessing PARPi synthetic lethality have focused on tumor types that exhibit significant fractions of either germ-line BRCA1 or BRCA2 mutations or other candidate BRCAness defects. Regulatory bodies including the FDA and EMA have recently approved PARPi to be used in ovarian cancer patients with either BRCA1 or BRCA2 mutations (as shown) with these being detected via companion diagnostic assays.
The Clinical Development of PARPi
PARPi first entered human clinical trials as a low-dose rucaparib/full-dose temozolomide combination (44). Based on the preclinical data showing an SL interaction between PARP inhibition and BRCA mutation status (19, 20), a phase 1 clinical trial of olaparib, including patients with germ-line BRCA1 or BRCA2 mutations (gBRCAm), was initiated. 63% of the gBRCAm mutant patients demonstrated a clinical benefit, thus confirming the SL hypothesis (45). Dose-limiting myelosuppression and central nervous system side effects were observed in some patients, but overall, side effects were less severe than those seen with conventional chemotherapy regimens (45). Expansion of this trial to 50 gBRCAm patients with gynecological malignancies confirmed efficacy and demonstrated a correlation between a favorable response to prior platinum chemotherapy and subsequent olaparib response; this observation was consistent with the hypothesis that platinum-based drugs and PARPi target similar molecular defects (36). Phase 2 trials involving patients with gBRCAm breast, ovarian, pancreatic or prostate cancers confirmed that olaparib offered significant clinical benefit (46–48) (Fig. 2B). The response rate in gBRCAm ovarian cancer patients (48) was sufficient for the United States Food and Drug Administration (FDA) to approve olaparib as a treatment for patients with advanced ovarian cancer who had already received three or more prior lines of therapy (49) (Fig. 2B).
A number of studies sought to extend these promising results to patients with BRCAness tumors: high-grade serous ovarian cancers (HGSOvCa), triple negative breast cancers (TNBC) and advanced prostate cancers. In HGSOvCa, olaparib was found to reduce the rate of disease recurrence and extend progression-free survival when used as a maintenance therapy in patients previously treated with platinum-based chemotherapy (50–52). These observations led to the approval of olaparib by the European Medicines Agency (EMA) as a maintenance treatment for BRCA mutant patients with platinum-sensitive, gynaecological cancers (high-grade serous epithelial ovarian, fallopian tube, or primary peritoneal cancers). In this particular case, olaparib was approved for use in patients with either germ-line or somatically-occurring BRCA mutations (gBRCAm or sBRCAm, respectively) (Fig. 2B). BRCAness-related clinical benefit has also been seen in metastatic, castration-resistant, prostate cancer: in the TO-PARP phase 2 clinical trial, about 30% of patients showed a clinical response to olaparib, half of whom had BRCA2 or ATM defects in their tumors (39). In TNBC, clinical responses to olaparib were somewhat mixed; while patients with BRCA1/2 mutant tumors showed some disease stabilization when treated with olaparib, there were no sustained responses in either BRCA1/2-mutant or non-mutant cohorts, suggesting that BRCAness might be less frequent in TNBC than in diseases such as HGSOvCa (53) or that other clinical parameters or prior treatment effects might be important.
The recent approval of olaparib for ovarian cancer was followed by promising phase 3 clinical results with niraparib in ovarian cancer patients as a maintenance therapy (54). Patients with platinum-sensitive, recurrent ovarian cancer were categorized according to the presence or absence of gBRCAm. Patients without a gBRCA mutation (non-gBRCAm) were further characterized to identify those without a gBRCAm whose tumors nevertheless exhibited a mutational pattern often seen in the genome of gBRCAm tumors - a BRCAness DNA scar, described above (55)). Those patients who received niraparib showed longer progression-free survival (PFS) than those who received placebo, with the strongest effects being seen in the gBRCAm group. In the non-gBRCAm cohort, niraparib also extended PFS, especially in patients whose tumors had somatic BRCA mutations (sBRCAm), who displayed a similar reduction in the risk of disease progression as those with gBRCAm. Overall, BRCAness-scar positive patients displayed longer PFS compared to BRCAness-scar negative patients, although the ultimate predictive value of this scar biomarker still requires clarification in larger phase 3 trials (54, 56).
Recently, rucaparib has also been shown to extend PFS in a maintenance setting (57). In the ARIEL2 phase 2 trial (NCT01891344), patients with platinum-sensitive, high-grade ovarian carcinoma were classified into three groups according to the genomic features of their tumors, including an assessment of a proposed BRCAness-related DNA mutational scar assessed by calculating the extent of chromosomal loss of heterozygosity (LOH) across the genome: (i) BRCA mutant (gBRCAm or sBRCAm); (ii) BRCA wild-type but with high genomic LOH; and (iii) BRCA wild-type with low LOH. PFS was greatest for the BRCA mutant group, with the high LOH group then displaying slightly more benefit from rucaparib treatment than in the LOH low group (57). However, the fraction of patients exhibiting durable responses in these BRCA wild-type groups was still below that expected with standard platinum-salt based combination chemotherapies, suggesting that further work is still required to justify the use of PARPi in these BRCA wild-type patients, as opposed to platinum based treatments (56, 57). A subsequent phase 3 clinical study assessing rucaparib (ARIEL 3, NCT01968213) will no doubt clarify this issue and also confirm whether the distinction in clinical responses between LOH high and low groups can be replicated and whether a refined criteria for converting a genome-wide loss of heterozygosity score into a clinically applicable BRCAness biomarker can be achieved (56). Nevertheless, these clinical results, when taken together with additional phase 2 data in HGSOvCa (NCT01482715 (58)) have led to FDA approval for rucaparib for use in patients with gBRCAm or sBRCAm advanced ovarian cancer who have been treated with two or more chemotherapies (Fig. 2B).
Talazoparib is also starting to show clinical potential. In a recent study, thirteen early-stage breast cancer patients with germline mutations in either BRCA1 or BRCA2 were treated for two months with talazoparib in a “neoadjuvant” setting, i.e. prior to chemotherapy and surgery. All patients displayed a reduction in tumor volume after two months (59). This study is now being expanded to assess the effects of 4-6 months of neoadjuvant talazoparib therapy. Similar neoadjuvant studies assessing rucaparib in breast cancer are also underway.
It would be premature to draw conclusions about which PARPi are most effective in particular patient subsets; at present, the different clinical trial designs and distinct patient populations used to assess different PARPi preclude such direct comparisons. However, a burgeoning dissection of the biochemical and cellular effects of different PARPi is beginning to influence ideas on how different PARPi might be used clinically. For example, although talazoparib can kill BRCA mutant cells in vitro at a dose 200 times lower than the dose needed for olaparib and rucaparib [an effect that correlates with the superior PARP1 trapping activity of talazoparib (23)], the in vitro therapeutic ratio achieved with in BRCA1/2 defective cells vs. wild type cells is similar for all three PARPi (10). It is therefore unclear whether the enhanced trapping ability of talazoparib compared to rucaparib and olaparib will in itself lead to an improved therapeutic ratio (10). Furthermore, mass spectrometry based analysis of PARPi binding proteins suggests that different PARPi have distinct interaction profiles (60). However, it is not clear how or whether differences in the “off-target” profile might alter the anti-tumor effects of PARPi, or contribute to adverse side effects. An understanding of which PARPi is the preferred agent in a given clinical situation will likely be informed by a future comparison of mature phase 3 clinical outcome data.
Potential PARPi Combination Therapies
As discussed above, the ability of PARPi to sensitize tumor cells to DNA damaging chemotherapies provided the initial rationale for developing clinically useful PARPi drugs. PARPi sensitize cells to the alkylating agent temozolomide as well as to topoisomerase poisons and inhibitors, effects that are now becoming mechanistically understood. Temozolomide sensitization is caused by PARP1 trapping whereas topoisomerase poison sensitivity is largely driven by the catalytic inhibition of PARP1 and less dependent upon trapping ability (22, 23), observations somewhat consistent with the failure of the relatively poor PARP1 trapping PARPi, veliparib, to enhance clinical temozolomide responses in BRCA gene mutant breast cancer patients, despite enhancing the effect of a carboplatin/paclitaxel combination (61). However, clinical experience with therapies that combine PARPi with chemotherapies has been, in general, mixed. Dose-limiting normal tissue toxicity is frequently observed, especially when the commonly-used dose of the chemotherapy regimen is combined with a PARPi (62). As some preclinical studies have confirmed that tumor cell growth inhibition can be achieved using high-dose PARPi combined with relatively low doses of chemotherapy, on-going clinical studies are assessing the tolerability and efficacy of similar “high PARPi/low chemo” approaches (e.g., NCT02049593; ClinicalTrials.gov). In addition, pre-clinical evidence suggests that a series of targeted agents, such as inhibitors of the phosphatidylinositol 3-kinase signaling pathway, can impart or enhance a BRCAness phenotype in tumor cells, thus causing sensitivity to PARPi (63); clinical studies testing the applicability of this approach are also now underway. In a distinct strategy, the concept of combining PARPi with targeted agents that impair the ability of tumor cells to stall the cell cycle to process and repair “trapped” PARP1 DNA lesions (e.g. WEE1, ATR and CHEK1/2 inhibitors) has gained some traction of late, being bolstered by the recent development of drug-like WEE1 and ATR inhibitors that now make this a testable hypothesis in clinical trials [e.g., NCT02264678, NCT02576444 (62)]. Finally, combinations of PARPi with immunotherapies such as anti-CTLA4 and anti-PD1/PDL-1 are now being tested clinically; these studies are based partly on the hypothesis that tumors with BRCA1, BRCA2 or BRCAness defects have a higher mutagenic burden and therefore potentially an elevated neo-antigen load, which is thought to produce a stronger anti-tumor immune response (64) (65). In addition there is evidence that BRCA deficiency may induce a STING-dependent innate immune response (66), which might also influence the anti-tumor effect of PARPi/immunotherapy drug combinations. In this regard, trials such as MEDIOLA (NCT02734004), a phase 1/2 trial of olaparib in combination with durvalumab (MEDI4736) an anti-PD-L1 immune checkpoint inhibitor, may be informative. The rationale for the identification of efficacious PARPi combinations has focused largely on enhancing the anti-tumor effect of PARPi by creating DNA damage or modulating DNA repair. However, PARPi could also be beneficially combined with drugs that act by targeting cancer-specific features that are superficially unrelated to the DDR or BRCA function; the promising combination of olaparib with the anti-angiogenic agent cediranib, is an example of this (67, 68). One potential advantage of such an approach is that therapy resistance could perhaps be mitigated with two orthogonal treatments.
In addition to identifying well-tolerated PARPi combination therapies, defining predictive biomarkers that facilitate patient stratification for such combinations is a key objective, as is identifying combination therapies that target PARPi resistance mechanisms and therefore provide more durable clinical responses. Finally, to deliver the best overall response, the scheduling of PARPi, and its sequencing with other drugs in the combination, should be given greater attention.
Conclusions and future prospects
The successful development of PARPi for BRCA mutant cancers provides proof-of-concept that SL interactions can be translated into cancer therapies. Although it remains to be seen how many other SL interactions can be clinically exploited, a number of lessons can be learned from the discovery and development of the PARPi/BRCA SL interaction. For example, SL interactions with therapeutic value are ideally: (i) associated with a therapeutic window defined by a biomarker that can be used to stratify patients for therapy; (ii) capable of highly penetrant effects, where the presence of the biomarker predicts profound sensitivity to inhibition of the SL target in the majority of cases; (iii) robust in the face of the molecular diversity and plasticity seen in human tumors; (iv) pharmacologically tractable; and (v) well understood in terms of molecular mechanism, as this can inform the development of biomarkers and an understanding of drug resistance mechanisms. In the case of PARPi, the scale of the therapeutic effect in in vitro pre-clinical models with BRCA mutations was compelling enough to drive early entry of the drugs into hypothesis-testing clinical trials. An early assessment of the robustness and penetrance of the BRCA/PARPi SL interactions indicated that these were highly penetrant and relatively resistant to genetic background effects and additional molecular alterations. The use of preclinical approaches to identify mechanisms of drug resistance alongside early stage clinical trials provided further insights into the mechanism of action of PARPi and revealed the basis of some of the incomplete clinical responses to PARPi in clinical trials (29, 30). Finally, a critical factor in the rapid translation to the clinic of the SL concept was the availability of potent drug-like PARPi at the same time as the identification of the BRCA SL (19, 20), highlighting the importance of medicinal chemistry and pharmacology to the successful application of PARP/BRCA SL and exemplifying how the convergence of otherwise distinct disciplines such as cancer genetics and drug discovery can be effective.
Ideally, the design and interpretation of clinical trials based on SL interactions should be based upon the biological hypothesis and robust pre-clinical data. Although there is a direct link between the recent approvals for PARPi in BRCA1/2 mutant cancers and the original pre-clinical data identifying the SL interactions over a decade ago, the clinical development of PARPi as a SL approach has not been straightforward. For example, a purported PARPi, iniparib, failed to elicit the expected clinical responses in a phase 3 trial, despite showing potential in early stage clinical assessment (69). As a result, questions were raised about the clinical potential of the entire drug class and the SL approach in general (69). In retrospect, the evidence supporting the mechanism of action of iniparib as a PARPi was not compelling (69). This reinforces the argument for the clinical development of drugs to be informed by robust pre-clinical biology. Of course, the pre-clinical and clinical investigation of PARPi SL effect is far from complete and we highlight in Box 1 a series of unanswered questions that – once addressed – could guide the optimal use of PARPi in the future. For example, a key observation has been that a fraction (approximately 15%) of ovarian cancer patients with BRCA1 or BRCA2 mutant tumors continue to be disease free more than five years after the initiation of PARPi treatment (52). Understanding the underlying reasons for these extended responses could help in the design of both predictive markers and therapeutic strategies.
Box 1. Some key unanswered questions about PARP inhibitors.
What proteins beyond BRCA1 and BRCA2 contribute to the processing of trapped PARP1, the drug’s key cytotoxic DNA lesion?
How does the inability to repair a trapped PARP1 lesion at the replication fork translate into cell death?
How do the roles of PARP1 in processes unrelated to DNA repair (e.g. inflammation, apoptosis, immune system) influence the anti-cancer activity of PARPi?
What is the relative predictive value of BRCA1, BRCA2 gene mutations or BRCAness biomarkers for platinum and/or PARPi responses - what is the best way to measure BRCAness?
What mechanisms distinguish “super-responders” who show profound and sustained responses to PARPi from those that do not?
What mechanisms operate clinically to drive development of resistance to PARPi in both BRCA mutant cancers and BRCAness cancers?
How can PARPi be optimally used in combination therapies?
Due to advances in technology, the systematic genome-wide identification of new SL interactions has now been achieved in yeast (70) and mapping each of the SL vulnerabilities associated with cancer driver genes and oncogenic processes in human cells is an ongoing activity; this raises the possibility that additional cancer-related SL interactions might be available for therapeutic exploitation. Critical to these efforts will be a greater understanding of the underlying principles of what triggers an SL interaction, the factors determining the robustness of such interactions (i.e. how easily are SL interactions reversed by other molecular changes) and how robust SL interactions can be predicted, rather than only empirically identified through large-scale genetic screens. For example, it has been suggested that robust SL interactions are enriched for pairs of genes that are closely connected on protein-protein interaction networks; i.e., those that directly interact or interact via one or two additional proteins or nodes, rather than being distantly connected via a larger number of intervening nodes (71). Likewise, proteins involved in similar functions (often predicted from overlapping protein-protein interaction networks) are hypothesized to have some shared SL interactions, leading to the development of algorithms that predict SL relationships (72). Integrating functional genomics and proteomics with computational network analysis approaches might therefore be useful to dissect these principles with the aim of streamlining the process of identifying highly penetrant SL effects with potential therapeutic value.
It took more than 10 years from the discovery of the PARPi/BRCA SL interaction to regulatory approval. Now, based on pre-clinical studies recently published and currently underway, PARPi remain a very active area of investigation. With the number of on-going clinical trials, there is optimism that in the short term there will be additional regulatory approvals for PARPi in multiple cancers. We suggest three broad areas, the “holy trinity” of personalized cancer therapy research, that require further investigation:
-
(i)
Identifying who to treat. This can be achieved by dissecting the mechanisms by which PARPi kill or inhibit tumor cells and using this information to develop refined, mechanism-based, biomarkers that allow patient stratification.
-
(ii)
Combating drug resistance. This can be achieved by identifying the mechanisms that cause PARPi resistance and biomarkers that predict it, by understanding how cancer heterogeneity and plasticity influence these processes, and by identifying clinical approaches to delay or prevent the emergence of the drug resistant phenotype.
-
(iii)
Optimizing combination therapy. This can be achieved by understanding the mechanistic basis of why some drug combinations have synergistic anti-tumor effects, how drug combinations can be used to target mechanisms of drug resistance and identifying predictive biomarkers of not only single agent PARPi responses but also PARPi combination therapy responses.
If these issues can be addressed, we believe that PARPi could eventually deliver considerable benefit to a substantial subset of cancer patients.
Acknowledgements
CJL acknowledges funding from Breast Cancer Now, Cancer Research UK and NHS funding to the NIHR Royal Marsden Hospital Biomedical Research Centre. AA is funded by Susan G. Komen for the Cure, The Breast Cancer Research Foundation, The BRCA Foundation and UCSF. We thank Dr. Dragomir Krastev (ICR, London) for assistance with figure generation and Prof. Andrew Tutt (ICR, London and King’s College London) for helpful suggestions.
Footnotes
Conflict of Interest Statement
AA and CJL are named inventors on patents describing the use of PARP inhibitors and as such stand to gain financially as part of the ICR “Rewards to Inventors” Scheme.
References
- 1.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 2.De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eustermann S, et al. Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol Cell. 2015;60:742–754. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawicki-McKenna JM, et al. PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain. Mol Cell. 2015;60:755–768. doi: 10.1016/j.molcel.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaremba T, Curtin NJ. PARP inhibitor development for systemic cancer targeting. Anticancer Agents Med Chem. 2007;7:515–523. doi: 10.2174/187152007781668715. [DOI] [PubMed] [Google Scholar]
- 7.Shall S. Proceedings: Experimental manipulation of the specific activity of poly(ADP-ribose) polymerase. J Biochem. 1975;77:2. [PubMed] [Google Scholar]
- 8.Purnell MR, Whish WJ. Novel inhibitors of poly(ADP-ribose) synthetase. Biochem J. 1980;185:775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terada M, Fujiki H, Marks PA, Sugimura T. Induction of erythroid differentiation of murine erythroleukemia cells by nicotinamide and related compounds. Proc Natl Acad Sci U S A. 1979;76:6411–6414. doi: 10.1073/pnas.76.12.6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Y, et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 2013;19:5003–5015. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King MC. "The race" to clone BRCA1. Science. 2014;343:1462–1465. doi: 10.1126/science.1251900. [DOI] [PubMed] [Google Scholar]
- 12.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 13.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 14.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- 16.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 17.Tutt A, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhillon KK, Bajrami I, Taniguchi T, Lord CJ. Synthetic lethality: the road to novel therapies for breast cancer. Endocr Relat Cancer. 2016;23:T39–55. doi: 10.1530/ERC-16-0228. [DOI] [PubMed] [Google Scholar]
- 19.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 20.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 21.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Murai J, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murai J, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pommier Y, O'Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:362ps317. doi: 10.1126/scitranslmed.aaf9246. [DOI] [PubMed] [Google Scholar]
- 25.Pettitt SJ, et al. A genetic screen using the PiggyBac transposon in haploid cells identifies Parp1 as a mediator of olaparib toxicity. PLoS One. 2013;8:e61520. doi: 10.1371/journal.pone.0061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaspers JE, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 2013;3:68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu G, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray Chaudhuri A, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards SL, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 30.Barber LJ, et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J Pathol. 2013;229:422–429. doi: 10.1002/path.4140. [DOI] [PubMed] [Google Scholar]
- 31.Sakai W, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norquist B, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29:3008–3015. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med. 2013;19:1381–1388. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- 34.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 35.Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 36.Fong PC, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 37.McCabe N, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 38.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mateo J, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carnevale J, Ashworth A. Assessing the Significance of BRCA1 and BRCA2 Mutations in Pancreatic Cancer. J Clin Oncol. 2015;33:3080–3081. doi: 10.1200/JCO.2015.61.6961. [DOI] [PubMed] [Google Scholar]
- 42.Graeser M, et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res. 2010;16:6159–6168. doi: 10.1158/1078-0432.CCR-10-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukhopadhyay A, et al. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin Cancer Res. 2010;16:2344–2351. doi: 10.1158/1078-0432.CCR-09-2758. [DOI] [PubMed] [Google Scholar]
- 44.Plummer R, et al. A phase II study of the potent PARP inhibitor, Rucaparib (PF-01367338, AG014699), with temozolomide in patients with metastatic melanoma demonstrating evidence of chemopotentiation. Cancer Chemother Pharmacol. 2013;71:1191–1199. doi: 10.1007/s00280-013-2113-1. [DOI] [PubMed] [Google Scholar]
- 45.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 46.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 47.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 48.Kaufman B, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim G, et al. FDA Approval Summary: Olaparib Monotherapy in Patients with Deleterious Germline BRCA-Mutated Advanced Ovarian Cancer Treated with Three or More Lines of Chemotherapy. Clin Cancer Res. 2015;21:4257–4261. doi: 10.1158/1078-0432.CCR-15-0887. [DOI] [PubMed] [Google Scholar]
- 50.Ledermann J, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 51.Ledermann J, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 52.Ledermann JA, et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. 2016;17:1579–1589. doi: 10.1016/S1470-2045(16)30376-X. [DOI] [PubMed] [Google Scholar]
- 53.Gelmon KA, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 54.Mirza MR, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 55.Telli ML, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalez Martin A. Progress in PARP inhibitors beyond BRCA mutant recurrent ovarian cancer? Lancet Oncol. 2017;18:8–9. doi: 10.1016/S1470-2045(16)30621-0. [DOI] [PubMed] [Google Scholar]
- 57.Swisher EM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 58.Shapira-Frommer R, et al. A phase 2 open-label, multicenter study of single-agent rucaparib in the treatment of patients with relapsed ovarian cancer and a deleterious BRCA mutation. Ann Oncol. 2015;33(suppl):2476. [Google Scholar]
- 59.Litton JK, et al. A pilot study of neoadjuvant talazoparib for early-stage breast cancer patients with a BRCA mutation. Ann Oncol. 2016;27(Supplement 6):vi43–vi67. [Google Scholar]
- 60.Knezevic CE, et al. Proteome-wide Profiling of Clinical PARP Inhibitors Reveals Compound-Specific Secondary Targets. Cell Chem Biol. 2016;23:1490–1503. doi: 10.1016/j.chembiol.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isakoff SJ, et al. A randomized Phase II study of veliparib with temozolomide or carboplatin/paclitaxel versus placebo with carboplatin/paclitaxel in BRCA1/2 metastatic breast cancer: design and rationale. Future Oncol. 2017;13:307–320. doi: 10.2217/fon-2016-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drean A, Lord CJ, Ashworth A. PARP inhibitor combination therapy. Crit Rev Oncol Hematol. 2016;108:73–85. doi: 10.1016/j.critrevonc.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 63.Ibrahim YH, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2:1036–1047. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higuchi T, et al. CTLA-4 Blockade Synergizes Therapeutically with PARP Inhibition in BRCA1-Deficient Ovarian Cancer. Cancer Immunol Res. 2015;3:1257–1268. doi: 10.1158/2326-6066.CIR-15-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang J, et al. The PARP1 inhibitor BMN 673 exhibits immunoregulatory effects in a Brca1(-/-) murine model of ovarian cancer. Biochem Biophys Res Commun. 2015;463:551–556. doi: 10.1016/j.bbrc.2015.05.083. [DOI] [PubMed] [Google Scholar]
- 66.Parkes EE, et al. Activation of STING-Dependent Innate Immune Signaling By S-Phase-Specific DNA Damage in Breast Cancer. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ivy SP, Liu JF, Lee JM, Matulonis UA, Kohn EC. Cediranib, a pan-VEGFR inhibitor, and olaparib, a PARP inhibitor, in combination therapy for high grade serous ovarian cancer. Expert Opin Investig Drugs. 2016;25:597–611. doi: 10.1517/13543784.2016.1156857. [DOI] [PubMed] [Google Scholar]
- 68.Liu JF, et al. Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: a randomised phase 2 study. Lancet Oncol. 2014;15:1207–1214. doi: 10.1016/S1470-2045(14)70391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mateo J, Ong M, Tan DS, Gonzalez MA, de Bono JS. Appraising iniparib, the PARP inhibitor that never was--what must we learn? Nat Rev Clin Oncol. 2013;10:688–696. doi: 10.1038/nrclinonc.2013.177. [DOI] [PubMed] [Google Scholar]
- 70.Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- 71.Campbell J, et al. Large-Scale Profiling of Kinase Dependencies in Cancer Cell Lines. Cell Rep. 2016;14:2490–2501. doi: 10.1016/j.celrep.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Young JH, Marcotte EM. Predictability of Genetic Interactions from Functional Gene Modules. G3 (Bethesda) 2016 doi: 10.1534/g3.116.035915. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]