Abstract
Purpose
The open structure of euchromatin renders it susceptible to DNA damage by ionizing radiation (IR) compared with compact heterochromatin. The effect of chromatin configuration on the efficacy of Auger electron radiotherapy was investigated.
Methods and Materials
Chromatin structure was altered in MDA-MB-468 and 231-H2N human breast cancer cells by suberoylanilide hydroxamic acid (SAHA), 5-aza-2-deoxycytidine, or hypertonic treatment. The extent and duration of chromatin structural changes were evaluated using the micrococcal nuclease assay. DNA damage (γH2AX assay) and clonogenic survival were evaluated after exposure to 111In-DTPA-hEGF, an Auger electron-emitting radiopharmaceutical, or IR. The intracellular distribution of 111In-DTPA-hEGF after chromatin modification was investigated in cell fractionation experiments.
Results
Chromatin remained condensed for up to 20 minutes after NaCl and in a relaxed state 24 hours after SAHA treatment. The number of γH2AX foci per cell was greater in MDA-MB-468 and 231-H2N cells after IR (0.5 Gy) plus SAHA (1 μM) compared with IR alone (16 ± 0.6 and 14 ± 0.3 vs. 12 ± 0.4 and 11 ± 0.2, respectively). More γH2AX foci were observed in MDA-MB-468 and 231-H2N cells exposed to 111In-DTPA-hEGF (6 MBq/μg) plus SAHA vs. 111In-DTPA-hEGF alone (11 ± 0.3 and 12 ± 0.7 vs. 9 ± 0.4 and 7 ± 0.3, respectively). 5-aza-2-deoxycytidine enhanced the DNA damage caused by IR and 111In-DTPA-hEGF. Clonogenic survival was reduced in MDA-MB-468 and 231-H2N cells after IR (6 Gy) plus SAHA (1 μM) vs. IR alone (0.6% ± 0.01 and 0.3% ± 0.2 vs. 5.8% ± 0.2 and 2% ± 0.1, respectively) and after 111In-DTPA-hEGF plus SAHA compared to 111In-DTPA-hEGF alone (21% ± 0.4% and 19% ± 4.6 vs. 33% ± 2.3 and 32% ± 3.7). SAHA did not affect 111In-DTPA-hEGF nuclear localization. Hypertonic treatment resulted in fewer γH2AX foci per cell after IR and 111In-DTPA-hEGF compared to controls but did not significantly alter clonogenic survival.
Conclusions
Chromatin structure affects DNA damage and cell survival after exposure to Auger electron radiation.
Keywords: Radiosensitivity, Auger electrons, Chromatin structure, Suberoylanilide hydroxamic acid
Introduction
Chromatin structure influences radiosensitivity when the extent of DNA damage in compact, transcriptionally inactive heterochromatin is compared with that in loose, transcriptionally active euchromatin after ionizing radiation (IR) (1–3). In euchromatin, which can be increased through histone depletion, the amount of DNA damage increases after IR compared with nonhistone depleted controls (4). By contrast, reduction in euchromatin caused by three-dimensional cell growth results in radioprotection compared with cells grown as a monolayer (5).
Interest in epigenetics has led to the development of drugs that modify the acetylation of histones or the methylation of CpG islands in gene promoters, leading to disrupted gene transcription and cell cycle progression (6, 7). Vorinostat (suberoylanilide hydroxamic acid, SAHA) inhibits histone deacetylases (HDAC), resulting in excessive acetylation of histones. 5-aza-2-deoxycytidine, which inhibits DNA methyltransferase, causes hypomethylation of CpG islands, resulting in chromatin relaxation and reexpression of silenced genes. Both drugs are used clinically: SAHA for cutaneous lymphoma and 5-aza-2-deoxycytidine for myelodysplastic syndrome (8). Several HDAC inhibitors sensitize cells to IR (9).
Although the impact of chromatin structure on cell sensitivity to IR has been studied extensively, little is known about its effect on DNA damage and cell killing caused by anticancer radio-pharmaceuticals. 111Indium-labeled human epidermal growth factor (111In-DTPA-hEGF) is an Auger electron–emitting agent that targets epidermal growth factor (EGF) receptor (EGFR)-overexpressing cells (10). 111In-DTPA-hEGF binds the EGFR, internalizes, translocates to the nucleus, and associates with chromatin. Nuclear localization of 111In-DTPA-hEGF is required for cytotoxicity because 111In releases short-range Auger electrons as it decays (11). We hypothesized that chromatin configuration could affect the accessibility of DNA to intranuclear radionuclides and so may be an important determinant of the cytotoxicity of Auger electron radiotherapy.
Methods and Materials
Cell culture
Human breast cancer cell lines MDA-MB-468 and 231-H2N were obtained from American Type Culture Collection and Robert Kerbel, Sunnybrook Health Sciences Centre, Toronto, Canada, respectively. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS; Invitrogen, Paisley, UK), 100 units/mL penicillin, and 100 μg/mL streptomycin at 37°C in 5% CO2. Hypertonic medium was prepared by adding NaCl to FBS-containing medium (0.15 M NaCl) to a final concentration of 0.5 M. Reagents were from Sigma-Aldrich, Dorset, UK, unless stated otherwise.
Radiopharmaceutical
111In chloride (PerkinElmer, Waltham, MA) was used to synthesize 111In-diethylene triamine pentaacetic acid (DTPA)-hEGF using a kit provided by Raymond Reilly, University of Toronto, Canada. Specific activity was 6 to 67 MBq/μg hEGF. Radiochemical purity was 50–90%.
Drugs
SAHA (Cayman Chemical, Ann Arbor, MI) and 5-aza-2-deoxycytidine were dissolved in dimethyl sulfoxide (DMSO) at 1.5 mM and in phosphate-buffered saline (PBS) at 4.38 μM, respectively; sterilized through a Millex GV 0.22-μM filter (Millipore, MA); and stored at −80°C. The maximum concentration of DMSO in cells was 0.066%.
Western blot analysis
Cells were cultured with or without SAHA (1 μM, 24 hours) and resuspended in lysis buffer (2% SDS, 100 mM dithiothreitol, 10% glycerol, and 60 mM Tris pH 6.8). The bicinchoninic acid protein assay (Pierce, Thermo Scientific, IL) was used to enable equal protein loading. Gel electrophoresis was performed, followed by Western blotting. Membranes were blocked in 5% milk/PBS/Tween (0.1%) and probed with anti-H3Ac (Active Motif, Rixensart, Belgium) or EGFR (Santa Cruz Biotechnology, CA) antibodies. β-actin and H3 antibodies (both Abcam, Cambridge, MA) were used as loading controls. Secondary horseradish peroxidase-conjugated antibodies (Promega, Southampton, UK, and Santa Cruz Biotechnology, CA) were used to detect proteins by enhanced chemiluminescence (Pierce, Thermo Scientific). Results were quantified by densitometry (Image J, National Institutes of Health, USA), and fold induction was normalized to control samples.
Micrococcal nuclease assay
231-H2N cells were cultured with or without SAHA (1 μM, 24 hours) or DMSO or 0.5 M NaCl (30 minutes) and washed in fresh isotonic medium, and nuclei were isolated from 1 x 107 cells either immediately or at selected times after washoff. Micrococcal nuclease (MNase) digestion of chromatin was performed as described (12). Briefly, nuclei were digested at 25°C with 0.5U MNase in 1 mM CaCl2-containing digestion buffer. Digestion was terminated with ethylenediaminetetraacetic acid (pH 8.0) every minute for 5 minutes, and proteinase K and SDS were then added. DNA was purified, separated alongside a 100-bp marker (New England Biolabs, Hitchin, UK) by electrophoresis in 1.2% agarose gel containing ethidium bromide, and visualized by ultraviolet transillumination. The intensity of bands was analyzed by Image J and normalized to total lane intensity.
γH2AX assay
Cells were incubated in serum-free medium with or without SAHA (0.5 or 1 μM), 5-aza-2-deoxycytidine (50 or 100 nM) for 24 hours, or in serum-free medium overnight followed by hypertonic or isotonic medium for 30 minutes, when fresh isotonic medium was added. Serum-free medium was used to distinguish radiation-induced γH2AX foci from replication-associated foci. Cells were irradiated (0.5 or 1 Gy; 1 Gy/min; IBL 637 137Cs irradiator, CIS Bio-International, France) and allowed to recover for 30 minutes at 37°C, or incubated with 111In-DTPA-hEGF (12 nM; 6 or 18 MBq 111In/μg hEGF) or DTPA-hEGF (12 nM) for 1 hour at 37°C. Cells were fixed with 4% paraformaldehyde (Fisher Scientific, Leicestershire, UK), permeabilized with 1% Triton X-100, and blocked in 2% bovine serum albumin in 0.1% Triton X-100/PBS. Cells were probed with anti-γH2AX (Millipore) and AF488-tagged secondary antibodies (Invitrogen) and mounted in 4′,6-diamidino-2-phenylindole/Vectashield (Vector Laboratories, Peterborough, UK). Images of 100 cells were acquired using oil immersion (×63) optics (Zeiss Axioplan 2, Hertfordshire, UK). γH2AX foci per cell were counted using Image J.
Clonogenic survival
Cells incubated with or without 1 μM SAHA (24 hours) or 0.5 M NaCl (30 minutes) were exposed to IR (0–6 Gy), 111In-DTPA-hEGF (100 pM, 6 MBq/μg or 12 nM, up to 67 MBq/μg) or DTPA-hEGF (100 pM or 12 nM). After 24 hours in 111In-DTPA-hEGF, cells were washed and fresh medium was added. After 7 to 14 days, cultures were stained with 1% methylene blue plus ethanol (Fischer Scientific), and colonies (≥50 cells) were counted. The surviving fraction (SF) was calculated by dividing the number of colonies by the number of cells seeded times plating efficiency (PE). Data were normalized to SF in controls.
Internalization and intracellular distribution of 111In
Cells were incubated with or without SAHA (24 hours), and 3 × 105 cells were exposed to 111In-DTPA-hEGF (12 nM, 1 hour). Unbound, cytoplasmic, and nuclear radioactive fractions were collected as described previously and measured by gamma counter (PerkinElmer Wizard 1470, Waltham, MA) (13). Results were expressed as a percentage of total 111In-DTPA-hEGF added to the medium.
Statistical analysis
Two-way analysis of variance was used for multiple comparisons, and unpaired two-tailed t tests were used for analysis of clonogenic and 111In localization data using GraphPad Prism (GraphPad software Inc., LaJolla, CA) with a 5% significance level. Results were expressed as means ± SEM of three experiments unless stated otherwise.
Results
Western blot analysis and chromatin digestion
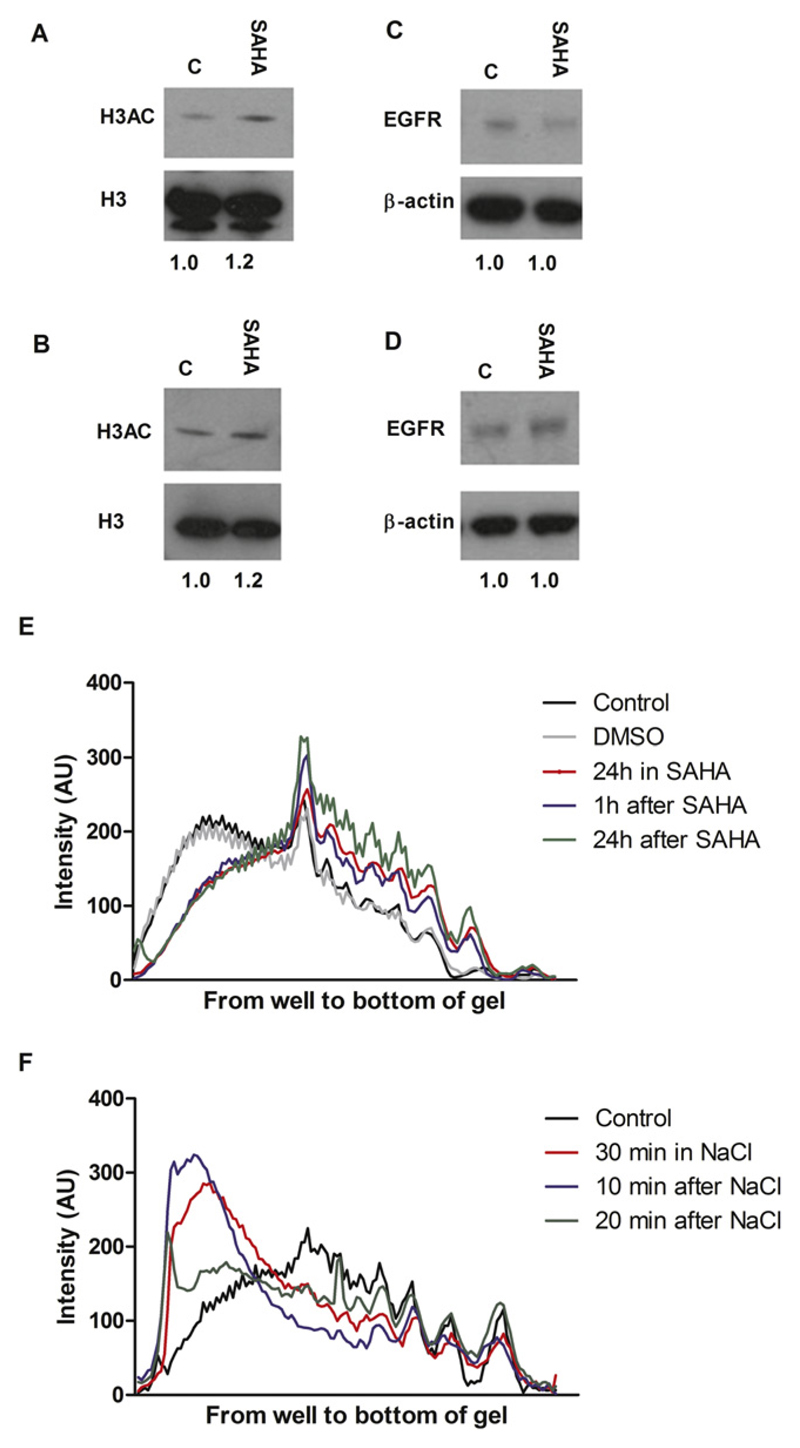
Western blot analysis showed that SAHA caused increased histone 3 acetylation, a marker of chromatin relaxation, without altering EGFR expression (Fig. 1A–D). MNase digestion confirmed that chromatin was relaxed up to 24 hours after SAHA washoff, compared with nontreated or DMSO-treated controls (Fig. 1E). Hypertonic treatment (30 minutes) resulted in condensation of chromatin and protection against digestion for up to 20 minutes after washoff (Fig. 1F and Fig. E1).
Fig. 1.
Western blots of (A, B) acetylated histone 3 (H3Ac) and (C, D) epidermal growth factor receptor (EGFR) protein expression in (A, C) MDA-MB-468 and (B, D) 231-H2N cells incubated with or without 1 μM suberoylanilide hydroxamic acid (SAHA). β-actin and H3 were used as loading controls. H3Ac:H3 and EGFR:β-actin signal ratios are shown below. (E) Analysis of micrococcal nuclease digestion pattern in 231-H2N cells at 1 minute after treatment with SAHA for 1 or 24 hours and (F) 10 and 20 minutes after NaCl treatment (See also Fig. E1).
HDAC inhibition and DNA demethylation increased IR- and 111In-DTPA-hEGF-induced γH2AX foci
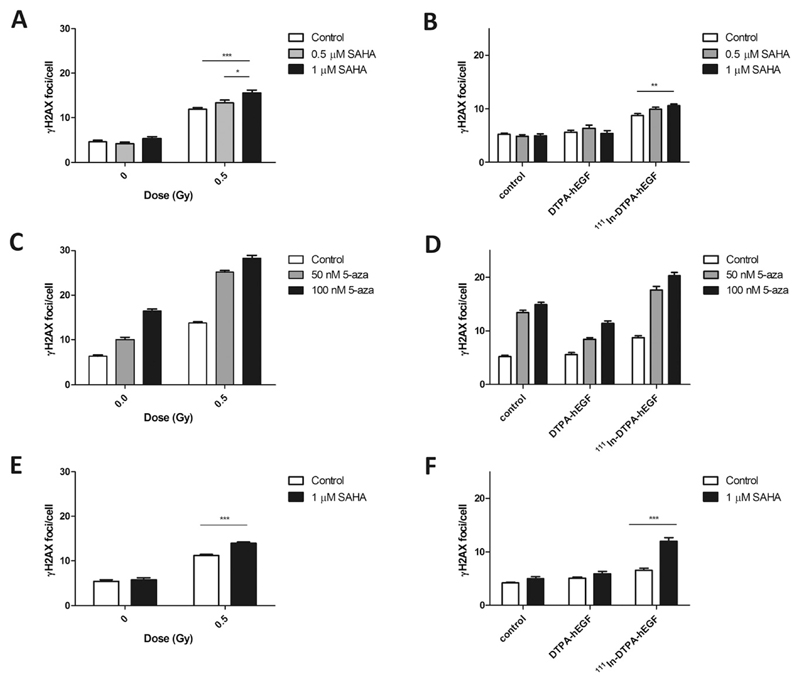
The number of γH2AX foci per cell in MDA-MB-468 cells increased significantly after γ-irradiation compared with controls (12 ± 0.4 vs. 5 ± 0.4, respectively; p < 0.0001; Fig. 2A). Pretreatment of cells with 0.5 or 1 μM SAHA further increased the number of γH2AX foci per cell (13 ± 0.6 and 16 ± 0.6, respectively). However, SAHA did not affect the number of γH2AX foci per cell in nonirradiated cells (Fig. 2A, B). Exposure of MDA-MB-468 cells to 111In-DTPA-hEGF caused a significant increase in γH2AX foci per cell compared with controls (9 ± 0.4 vs. 5 ± 0.3; p < 0.0001; Fig. 2B). Pretreatment with 0.5 and 1 μM SAHA plus 111In-DTPA-hEGF further increased γH2AX foci (10 ± 0.4 and 11 ± 0.3, respectively). DTPA-hEGF alone had no effect on γH2AX foci (Fig. 2B). In 231-H2N, the number of γH2AX foci per cell increased after SAHA before IR compared with IR alone (14 ± 0.3 vs. 11 ± 0.2; Fig. 2E). The number of γH2AX foci per cell was greater in 231-H2N cells exposed to 111In-DTPA-hEGF plus SAHA than in cells treated with 111In-DTPA-hEGF alone (12 ± 0.7 vs. 7 ± 0.3; Fig. 2F).
Fig. 2.
(A–D) MDA-MB-468 and (E–F) 231-H2N cells were exposed to suberoylanilide hydroxamic acid (SAHA) or 5-aza-2-deoxycytidine, irradiated, and stained for γH2AX after 30 minutes (A, C, E). Alternatively, after drug treatment, cells were incubated with 111In-DTPA-hEGF (6 MBq/μg; 12 nM) or DTPA-hEGF (12 nM) for 1 hour and stained for γH2AX (B, D, F). All comparisons in (C and D) were significantly different (p < 0.001).
5-aza-2-deoxycytidine (50 or 100 nM) treatment of MDA-MB-468 cells increased DNA damage after IR (25 ± 0.3 and 28 ± 0.6 γH2AX foci per cell, respectively) or exposure to 111In-DTPA-hEGF (18 ± 0.7 and 20 ± 0.6, respectively) (Fig. 2C, D; p < 0.0001). Interestingly, 5-aza-2-deoxycytidine (50 or 100 nM) alone induced γH2AX foci (13 ± 0.5 and 15 ± 0.5 vs. 5 ± 0.3; p < 0.0001; Fig. 2D). Inasmuch as it would be difficult to distinguish the contribution of 5-aza-2-deoxycytidine from that of IR or 111In-DTPA-hEGF to γH2AX focus induction, the effect of 5-aza-2-deoxycytidine in 231-H2N cells was not tested. Less DNA damage was observed in MDA-MB-468 cells treated with 5-aza-2-deoxycytidine (50 and 100 nM) plus DTPA-hEGF compared with cells incubated with 5-aza-2-deoxycytidine alone (8 ± 0.3 and 11 ± 0.4; p < 0.0001; Fig. 2D). Neither 111In-citrate nor DMSO induced γH2AX foci (not shown).
HDAC inhibition decreased clonogenic survival after IR or 111In-DTPA-hEGF
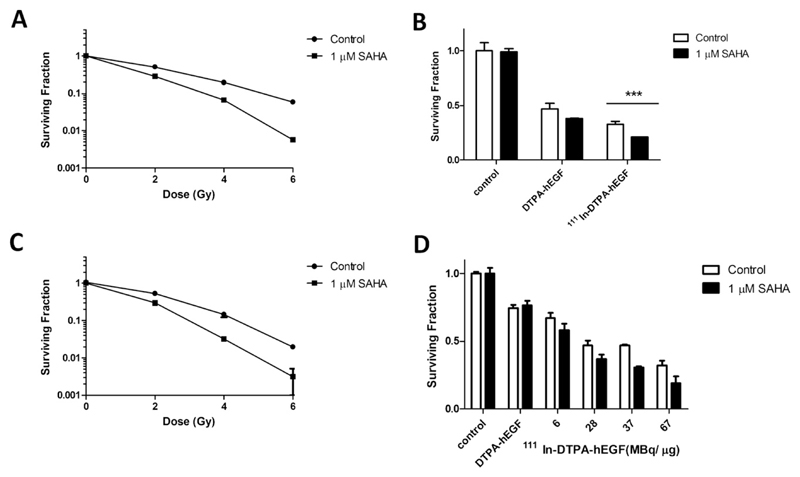
Radiation (6 Gy) reduced SF in MDA-MB-468 and 231-H2N cells significantly (5.8% ± 0.2 and 2% ± 0.1, respectively; p < 0.001; Fig. 3A, C). Incubation with SAHA before IR further decreased the SF (0.6% ± 0.01 and 0.3% ± 0.2, respectively). In MDA-MB-468 cells, 12 nM DTPA-hEGF caused greater reduction in SF than in 231-H2N cells (not shown); therefore, 100 pM DTPA-hEGF was used in clonogenic assays for MDA-MB-468 cells. In MDA-MB-468 and 231-H2N cells, exposure to DTPA-hEGF (100 pM and 12 nM, respectively) resulted in reduced SF (50% ± 5 and 75% ± 2.4, respectively; p < 0.0001), (Fig. 3B, D). In MDA-MB-468 cells, 111In-DTPA-hEGF (30 MBq/μg) reduced SF to 33% ± 2.3, whereas higher specific activities (up to 67 MBq/μg) were needed to produce an effect of similar magnitude (SF 32% ± 3.7) in 231-H2N cells. Incubation of MDA-MB-468 and 231-H2N cells with SAHA before exposure to 111In-DTPA-hEGF (at 30 or 67 MBq/μg) lowered SF further (21% ± 0.4% and 19% ± 4.6, respectively). In 231-H2N cells, the effect of SAHA on SF at different specific activities of 111In-DTPA-hEGF was significant compared with cells treated with 111In-DTPA-hEGF only (p = 0.023). In both cell lines, SAHA decreased PE by 10%; DMSO did not affect PE (not shown).
Fig. 3.
Clonogenic survival for (A, B) MDA-MB-468 and (C, D) 231-H2N cells. Cells were incubated with suberoylanilide hydroxamic acid (SAHA) followed by (A, C) IR, (B, D) 111In-DTPA-hEGF (30 MBq/μg; 100 pM or 0–67 MBq/μg; 12 nM, respectively) or DTPA-hEGF (100 pM or 12 nM). Results are means ± SEM of three experiments or means ± SEM of triplicates in one experiment. Error bars are too small to be seen in (A and D).
SAHA did not affect intracellular localization of 111In
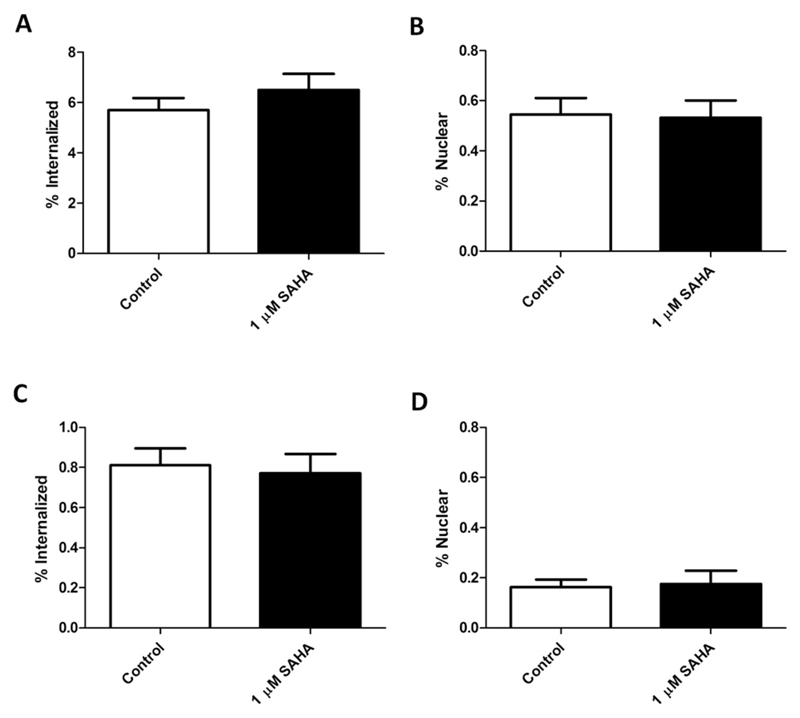
In SAHA-treated MDA-MB-468 cells, the amount of total administered 111In-DTPA-hEGF that internalized into the cell and nucleus was 6.6% ± 0.5 and 0.5% ± 0.01, respectively, compared with 5.9% ± 0.3 and 0.6% ± 0.02 in controls (Fig. 4A, B). In 231-H2N cells incubated with SAHA, 0.8% ± 0.07 and 0.2% ± 0.03 of 111In-DTPA-hEGF internalized into the cell and nucleus, respectively, compared with 0.8% ± 0.07 and 0.2% ± 0.02 in controls (Fig. 4C, D).
Fig. 4.
Internalization and nuclear localization of 111In in (A, B) MDA-MB-468 and (C, D) 231-H2N cells treated with suberoylanilide hydroxamic acid (SAHA) and 111In-DTPA-hEGF (12 nM).
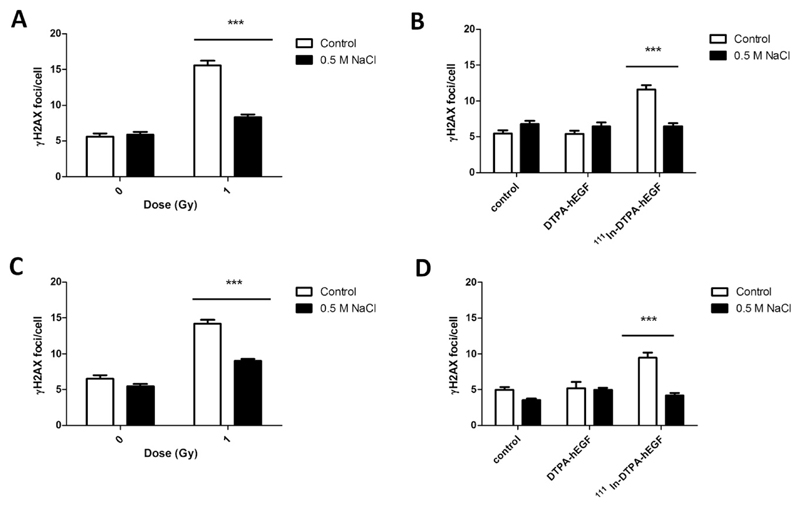
Chromatin condensation after NaCl reduced DNA damage but not clonogenic survival
Incubation of MDA-MB-468 cells with NaCl before IR resulted in a significant decrease in the number of γH2AX foci compared with IR alone (8 ± 0.4 vs. 16 ± 0.7). Exposure to 111In-DTPA-hEGF (18 MBq/μg; 12 nM) significantly increased γH2AX foci compared with untreated cells (12 ± 0.6 vs. 5 ± 0.5, p < 0.0001), and this was reversed by NaCl (6 ± 0.5). Similarly, fewer γH2AX foci were induced in 231-H2N cells when exposed to NaCl before IR or 111In-DTPA-hEGF compared with IR or 111In-DTPA-hEGF alone (9 ± 0.3 and 4.2 ± 0.3 vs. 14 ± 0.6 and 9 ± 0.7; Fig. 5C, D). NaCl alone did not affect the number of γH2AX foci per cell (Fig. 5).
Fig. 5.
(A, B) MDA-MB-468 and (C, D) 231-H2N cells were incubated with hypertonic medium, exposed to (A, C) IR, and stained for γH2AX after 30 minutes. (B, D) After hypertonic treatment, cells were incubated with 111In-DTPA-hEGF (18 MBq/μg; 12 nM) or DTPA-hEGF (12 nM) and stained for γH2AX immediately.
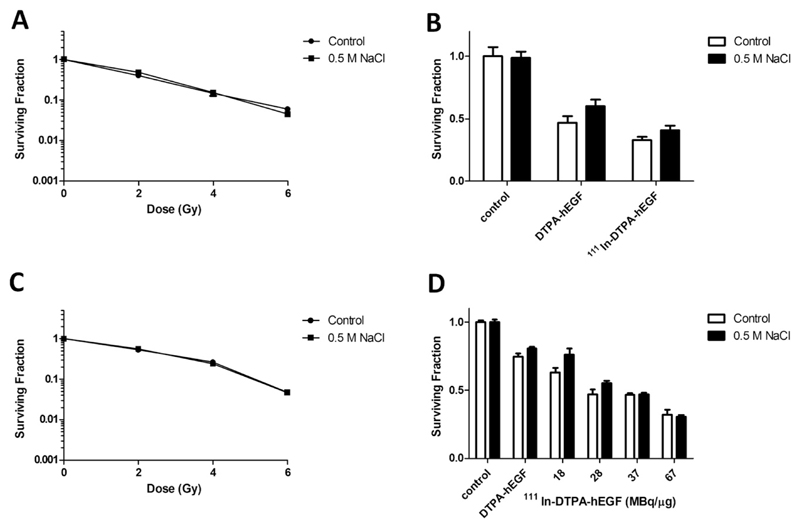
IR (6 Gy) reduced the SF in both MDA-MB-468 and 231-H2N significantly (5.9% ± 0.3 and 4.7% ± 0.2) compared with controls (p < 0.001; Fig. 6A, C). Incubation with NaCl before IR (6 Gy) did not affect the SF (4.5 ± 0.03% and 4.7% ± 0.2, respectively). In MDA-MB-468 and 231-H2N cells, exposure to DTPA-hEGF resulted in a significantly lower SF (50% ± 5 and 75% ± 2.4, respectively; p < 0.0001) compared with controls (Fig. 6B, D). SF decreased further after treatment of MDA-MB-468 and 231-H2N cells with 111In-DTPA-hEGF (33% ± 2.3 [at 30 MBq/μg] and 32% ± 3.7 [at 67 MBq/μg], respectively; p < 0.0001). Incubation of cells with NaCl before 111In-DTPA-hEGF did not statistically significantly affect the SF (41 ± 3.6 [at 30 MBq/μg] and 31% ± 1.2 [at 67 MBq/μg], p = 0.08 and p = 0.12, respectively). NaCl decreased PE by 40% (not shown).
Fig. 6.
Clonogenic assays were performed on (A, B) MDA-MB-468 and (C, D) 231-H2N cells. Cells were incubated with (A, C) NaCl plus IR, (B, D) 111In-DTPA-hEGF (30 MBq/μg; 100 pM or up to 67 MBq/μg; 12 nM) or DTPA-hEGF (100 pM or 12 nM). Results are means ± SEM of three experiments or as means ± SEM of triplicates within one experiment. Error bars are too small to be seen in (A and C).
Discussion
Little is known about whether chromatin architecture influences the efficacy of molecularly targeted radiotherapeutic agents. It has been shown that fewer double-strand breaks are induced in supercoiled than in linear or nicked plasmid DNA when exposed to the Auger electron–emitting radionuclide, 125I (14). Also, cells are more sensitive to DNA-incorporated Auger electron–emitting agents than to those localized in the cytoplasm or at the cell membrane (14). These lines of evidence suggest that the efficacy of Auger electron–emitting radiopharmaceuticals is determined both by their intracellular distribution and by the accessibility of DNA.
In this study, chromatin structure was shown to affect the sensitivity of cells to the Auger electron–emitting radiopharmaceutical 111In-DTPA-hEGF. SAHA at a clinically relevant concentration resulted in increased histone 3 acetylation and digestion by micrococcal nuclease, indicating chromatin decondensation (Fig. 1) (7). Incubation of cells in SAHA before IR or 111In-DTPA-hEGF caused greater induction of γH2AX foci compared with IR or 111In-DTPA-hEGF alone (Fig. 2). Because pretreatment with SAHA did not lead to induction of doublestrand breaks by unlabeled DTPA-hEGF, the sensitization of cells to 111In-DTPA-hEGF by SAHA is attributed to greater chromatin accessibility, resulting in increased DNA damage induced by 111In as it decays. In these experiments, cells were incubated with 111In-DTPA-hEGF for 1 hour. This length of exposure was selected because induction of γH2AX foci by 111In-DTPA-hEGF increases linearly up to 4 hours (15).
The effect of altering chromatin structure through DNA demethylation was also explored. 5-aza-2-deoxycytidine led to increased γH2AX foci per cell after IR or 111In-DTPA-hEGF compared with either alone (Fig. 2). However, unlike SAHA, 5-aza-2-deoxycytidine itself was associated with an increase in γH2AX foci per cell (Fig. 2), as reported by others (16). Interestingly, the addition of DTPA-hEGF partially abrogated DNA damage by 5-aza-2-deoxycytidine. This observation is consistent with a role for EGF, acting via the EGFR, in DNA repair (17).
SAHA has been reported to downregulate EGFR expression (18). Given that 111In-DTPA-hEGF depends on EGFR for internalization, downregulation of EGFR would be expected to reduce its cytotoxicity. However, SAHA did not affect EGFR protein expression in the cell lines used (Fig. 1), nor did it affect 111In-DTPA-hEGF internalization or nuclear localization (Fig. 4). The amount of 111In internalized in 231-H2N cells was less than in MDA-MB-468 cells, corresponding well with lower expression of EGFR on the cell surface of 231-H2N cells, 1 × 105 EGFR per cell, compared with 5 × 106 EGFR per cell in MDA-MB-468 cells.
Addition of SAHA increased radiosensitivity to IR and 111In-DTPA-hEGF in clonogenic assays (Figs. 2, 3). However, it is unclear whether the effect of SAHA in either the γH2AX or the clonogenic assay resulted from a physical change in accessibility of the chromatin to either the radiopharmaceutical or the DNA repair proteins, from altered expression of DNA repair genes, or from a combination of these. We speculate that any effect of SAHA or NaCl on the induction of DNA damage after a 1-hour exposure to 111In-DTPA-hEGF is predominantly due to altered physical accessibility to the DNA. This is consistent with a report by Singh et al. showing that the repair capacity of SAHA-treated cells is similar to that of controls until 60 minutes after the first IR dose in split-dose experiments (19). In clonogenic assays, however, cells were exposed to 111In-DTPA-hEGF for 24 hours. It is likely, therefore, that in addition to increased physical accessibility of chromatin caused by SAHA, altered DNA repair capacity would influence the cellular response to 111In-DTPA-hEGF in clonogenic assays.
Of note, MDA-MB-468 cells were more sensitive to DTPA-hEGF than previously noted (15). This may have been due to the increase in EGFR expression that has been reported to occur in this cell line during cell culture (20). We performed 111In-DTPA-hEGF binding experiments and found that the number of EGFR/cell was high reaching 5x106 copies per cell (data not shown).
Pretreatment with NaCl, which causes chromatin compaction, resulted in fewer γH2AX foci per cell after IR or 111In-DTPA-hEGF compared with cells treated with either alone, although the number of foci was still greater than in controls (Fig. 5). Interestingly, hypertonic treatment increased cell survival after 111In-DTPA-hEGF, although this did not reach statistical significance (Fig. 6). One possible explanation for why the increase in cell survival was modest is that chromatin condensation by NaCl, which persists for up to 20 minutes (as shown in MNase assays), results in decreased accessibility of repair proteins to DNA. It is thus possible that radioprotection afforded by chromatin compaction was offset by a reduction in DNA repair, and the net effect was that clonogenic survival did not change significantly.
These results indicate that current interest in HDAC inhibitors as sensitizers to external radiation is also relevant to targeted radiotherapy. The effect of SAHA on DNA damage was greater when combined with 111In-DTPA-hEGF compared with IR, suggesting that alteration of chromatin configuration might be more effective in sensitizing cells to Auger electron–emitting radio-nuclides than to IR.
Supplementary Material
Supplementary material for this article can be found at www.redjournal.org.
Summary.
The effect of chromatin structure on the cytotoxicity of radiopharmaceuticals is largely unknown. The evidence presented here suggests that the relaxation of chromatin, through exposure to HDAC inhibitors for example, enhances the cytotoxic effect of 111In-DTPA-hEGF, an Auger electron-emitting anticancer radiopharmaceutical. In contrast NaCl-induced chromatin condensation resulted in a radioprotective outcome. These results indicate that the current interest in combining external radiation with HDAC inhibitors is also relevant to molecularly targeted radiation therapy.
Acknowledgment
The authors thank Raymond Reilly for the DTPA-hEGF kit and Bart Cornelissen and Aaron Goodarzi for helpful discussions.
Supported by Cancer Research-UK (C14521/A6245), by the Oxford Experimental Cancer Medicine Centre, and by the NIHR Oxford Biomedical Research Centre.
Footnotes
Conflict of interest: none.
References
- 1.Cowell IG, Sunter NJ, Singh PB, et al. gammaH2AX foci form preferentially in euchromatin after ionising radiation. PLoS One. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magnander K, Hultborn R, Claesson K, et al. Clustered DNA damage in irradiated human diploid fibroblasts: influence of chromatin organization. Radiat Res. 2010;173:272–282. doi: 10.1667/RR1891.1. [DOI] [PubMed] [Google Scholar]
- 3.Vasireddy RS, Karagiannis TC, El-Osta A. Gamma-radiation-induced gammaH2AX formation occurs preferentially in actively transcribing euchromatic loci. Cell Mol Life Sci. 2010;67:291–294. doi: 10.1007/s00018-009-0181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heussen C, Nackerdien Z, Smit BJ, et al. Irradiation damage in chromatin isolated from V-79 Chinese hamster lung fibroblasts. Radiat Res. 1987;110:84–94. [PubMed] [Google Scholar]
- 5.Storch K, Eke I, Borgmann K, et al. Three-dimensional cell growth confers radioresistance by chromatin density modification. Cancer Res. 2010;70:3925–3934. doi: 10.1158/0008-5472.CAN-09-3848. [DOI] [PubMed] [Google Scholar]
- 6.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 7.Karagiannis TC, El-Osta A. Modulation of cellular radiation responses by histone deacetylase inhibitors. Oncogene. 2006;25:3885–3893. doi: 10.1038/sj.onc.1209417. [DOI] [PubMed] [Google Scholar]
- 8.Batty N, Malouf GG, Issa JP. Histone deacetylase inhibitors as antineoplastic agents. Cancer Lett. 2009;280:192–200. doi: 10.1016/j.canlet.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 9.De Schutter H, Nuyts S. Radiosensitizing potential of epigenetic anticancer drugs. Anticancer Agents Med Chem. 2009;9:99–108. doi: 10.2174/187152009787047707. [DOI] [PubMed] [Google Scholar]
- 10.Reilly RM, Kiarash R, Cameron RG, et al. 111In-labeled EGF is selectively radiotoxic to human breast cancer cells overexpressing EGFR. J Nucl Med. 2000;41:429–438. [PubMed] [Google Scholar]
- 11.Howell RW. Radiation spectra for Auger-electron emitting radionuclides: Report No. 2 of AAPM Nuclear Medicine Task Group No. 6. Med Phys. 1992;19:1371–1383. doi: 10.1118/1.596927. [DOI] [PubMed] [Google Scholar]
- 12.Micrococcal nuclease: Southern blot assay. Nat Meth. 2005;2:719–720. [Google Scholar]
- 13.Cornelissen B, Hu M, McLarty K, et al. Cellular penetration and nuclear importation properties of 111In-labeled and 123I-labeled HIV-1 peptide immunoconjugates in BT-474 human breast cancer cells. Nucl Med Biol. 2007;34:37–46. doi: 10.1016/j.nucmedbio.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Kassis AI. Molecular and cellular radiobiological effects of Auger emitting radionuclides. Radiat Prot Dosimetry. 2010;143:241–247. doi: 10.1093/rpd/ncq385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Z, Chen Z, Bailey KE, et al. Relationship between induction of phosphorylated H2AX and survival in breast cancer cells exposed to 111In-DTPA-hEGF. J Nucl Med. 2008;49:1353–1361. doi: 10.2967/jnumed.108.051805. [DOI] [PubMed] [Google Scholar]
- 16.Palii SS, Van Emburgh BO, Sankpal UT, et al. DNA methylation inhibitor 5-aza-2 ′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen DJ, Nirodi CS. The epidermal growth factor receptor: A role in repair of radiation-induced DNA damage. Clin Cancer Res. 2007;13:6555–6560. doi: 10.1158/1078-0432.CCR-07-1610. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, Shaw PG, Davidson NE. Inhibition of histone deacetylase suppresses EGF signaling pathways by destabilizing EGFR mRNA in ER-negative human breast cancer cells. Breast Cancer Res Treat. 2009;117:443–451. doi: 10.1007/s10549-008-0148-5. [DOI] [PubMed] [Google Scholar]
- 19.Singh S, Le H, Shih SJ, et al. Suberoylanilide hydroxyamic acid modification of chromatin architecture affects DNA break formation and repair. Int J Radiat Oncol Biol Phys. 2010;76:566–573. doi: 10.1016/j.ijrobp.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agelopoulos K, Greve B, Schmidt H, et al. Selective regain of egfr gene copies in CD44+/CD24-/low breast cancer cellular model MDA-MB-468. BMC Cancer. 2010;10:78. doi: 10.1186/1471-2407-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.