Abstract
Lysosomes are highly dynamic organelles that can move rapidly throughout the cell. They distribute in a rather immobile pool located around the microtubule‐organizing center in a “cloud,” and a highly dynamic pool in the cell periphery. Their spatiotemporal characteristics allow them to carry out multiple biological functions, such as cargo degradation, antigen presentation and plasma membrane repair. Therefore, it is not surprising that lysosomal dysfunction underlies various diseases, including cancer, neurodegenerative and autoimmune diseases. In most of these biological events, the involvement of lysosomes is dependent on their ability to move throughout the cytoplasm, to find and fuse to the correct compartments to receive and deliver substrates for further handling. These dynamics are orchestrated by motor proteins moving along cytoskeletal components. The complexity of the mechanisms responsible for controlling lysosomal transport has recently been appreciated and has yielded novel insights into interorganellar communication, as well as lipid‐protein interplay. In this review, we discuss the current understanding of the mechanisms of lysosomal transport and the molecular machineries that control this mobility.
Keywords: Arl GTPases, cholesterol, dendritic cells, dynein, endoplasmic reticulum, kinesins, late endosome, lysosome, membrane contact sites, mTOR, myosins, ORP1L, phosphatidylinositol phosphate, Rab GTPases, RNF26, TOLLIP
1. INTRODUCTION
Upon entry into the endosomal system, endocytic cargo first enters early endosomes, where it can be sorted for recycling back to the plasma membrane or targeted downstream to late endosomes.1 Late endosomes mature to or fuse with lysosomes, degradative organelles characterized by an acidic lumen and many hydrolytic enzymes. Lysosomes have long been considered as trash bins of cells and final state organelles. However, their role in various cellular processes from signal transduction to antigen presentation highlights the merits of this remarkable organelle.2 Lysosomes are broadly distributed throughout the cell, but overall they demonstrate two spatially distinct subsets: a relatively immobile perinuclear pool near the microtubule‐organizing center (MTOC) and a highly dynamic subset at the periphery of cells, some reaching as far as the plasma membrane.3 As their positioning reflects their characteristics, such as luminal pH, it is no surprise that these different spatial pools of lysosomes contribute to different biological means.4 For instance, peripheral lysosomes are shown to participate in plasma membrane repair and indicate nutrient availability by activating the mechanistic target of rapamycin complex 1 (mTORC1); meanwhile, perinuclear ones fuse with autophagosomes to generate auto‐lysosomes.5, 6, 7 Although these different pools of lysosomes have distinct characteristics and functions, cell‐wide homeostasis is maintained by regular mixing of these pools by trafficking and fusion processes. In general, lysosomal movement is not continuous but happens in a “stop‐and‐go” fashion, suggesting that it is constantly subjected to regulation.8 Recent findings reveal that various factors, such as interorganellar contact sites, play a direct role in regulating the positioning of late endosomes and lysosomes. In this review, we discuss the recent insights into the transport mechanisms of late endosomes and lysosomes (hereafter collectively referred to as lysosomes, unless stated otherwise) and machineries that regulate their positioning in cells.
2. WHERE IT ALL STARTS AND ENDS: THE PERINUCLEAR CLOUD OF ENDOSOMAL VESICLES
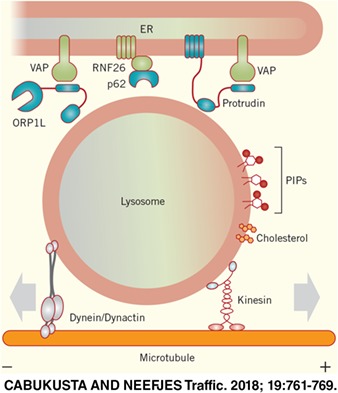
The steady‐state distribution of the endosomal system is not random but organized with dynamic endosomes at the cell periphery and relatively immobile ones in a “perinuclear cloud” around the MTOC (Figure 1). While endocytosed cargo enters the cell at the periphery, early endosomes move into the cloud and mature into late endosomes and lysosomes. From here, not only early and late endosomes but also trans‐Golgi network vesicles leave to move toward the cell periphery for their following destination. This perinuclear cloud is controlled by an endoplasmic reticulum (ER)‐located E3 ligase called RNF26 (Figure 3).3 RNF26 binds and ubiquitinates p62/SQSTM1 that then interacts with ubiquitin adaptors such as TOLLIP, EPS15 and TAX1BP1 on the various endosomal sets to arrest them in the perinuclear region. This organization increases the efficiency of endocytosed fluid phase material to enter lysosomes and activated epidermal growth factor to be degraded to terminate signaling.3 Therefore, the perinuclear cloud appears to be a site for efficient maturation of endosomes. At one point, endosomes leave the cloud, which involves the deubiquitination of SQSTM1/p62 by the deubiquitinating enzyme USP15 (Figure 3), and start their fast bidirectional microtubule‐based journey in the periphery along microtubules and finally the cortical actin cytoskeleton before reaching the cell surface (Figure 1).
Figure 1.
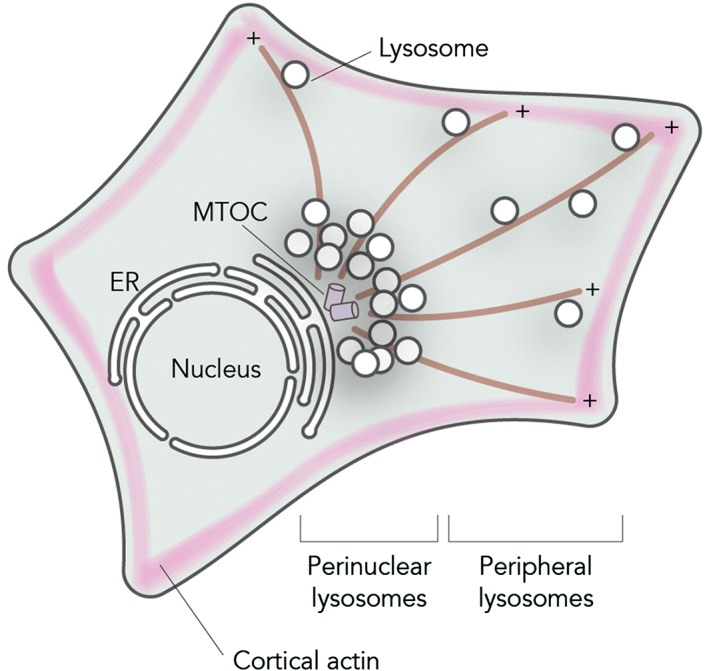
Intracellular distribution of lysosomes. Lysosomes can be found in two intracellular locations: a relatively immobile pool of perinuclear lysosomes clustered around the MTOC and a set of peripheral lysosomes moving fast along microtubules in a stop‐and‐go manner or are found attached to peripheral actin networks
3. MECHANISMS OF LYSOSOMAL TRANSPORT
Intracellular movement of proteins, RNA and organelles is as fundamental to multicellular organisms as membrane compartmentalization of cellular processes. Large particles do not move by free diffusion but require active directional support.9 Serving this purpose, three principle families of motor proteins are known—kinesins, dyneins and myosins. While kinesin and dynein motor proteins move on microtubules, myosin motor proteins move on actin filaments. Fast transport is typically microtubule‐based, myosin motors move more slowly along actin.10 There is also a spatial difference as microtubule networks span the entire cytosol while actin fibers are more (but not exclusively) concentrated under the plasma membrane to determine cell shape and motility. Directional intracellular transport is most prominent in polarized cells, such as neurons, where the delivery of organelles and proteins from cell body to axon occurs over long distances and is essential for neuronal function.11 In general, microtubules are radially distributed: with their minus‐end positioned at the perinuclear region, starting at the MTOC, and their plus‐end pointing toward the cell periphery.12
Long‐distance intracellular transport of organelles occurs along the microtubule fibers by the activity of kinesin and cytoplasmic dynein‐1 (here after referred to as dynein) motor proteins, and the balance between retrograde dynein and anterograde kinesin motor transport determines the net directionality of movement and maturation of endosomes.13 Since lysosomes move in a bidirectional manner along microtubules, this implies that they move by alternating activities of the dynein motor and one or more of the kinesin motors. The kinesin, dynein and myosin motors usually do not bind directly to selected organelles but their interaction is generally mediated by small GTPases, their effector proteins and lipids (reviewed in Reference 14). When in their active GTP‐bound state, small GTPases can mark defined organelles and then function as scaffolds for the assembly of transport and fusion machineries on target membranes.15
While the human genome encodes over 40 kinesin proteins, there is only one dynein protein that has been implicated in transport of cytoplasmic cargoes.16 Dynein associates with the multiprotein complex dynactin to facilitate minus‐end transport of organelles and other substrates. Organelles have their own solution for recruiting motor proteins, but the general mechanism that is employed is usually shared. Rab7 is a small GTPase that—when activated in its GTP‐bound state—marks late endosomes, lysosomes and lysosome‐related organelles. Rab7 then recruits effector proteins such as the Rab‐interacting lysosomal protein (RILP), which binds to the p150glued subunit of dynactin and dynein light intermediate chain, then acquiring the dynein‐dynactin complex for minus‐end transport (Figure 2).17, 18, 19 Under more specific conditions such as starvation, the dynein/dynactin motor can interact with ALG2 or JIP4 on lysosomal membranes to drive minus‐end transport of lysosomes (Figure 2, discussed further below).20
Figure 2.
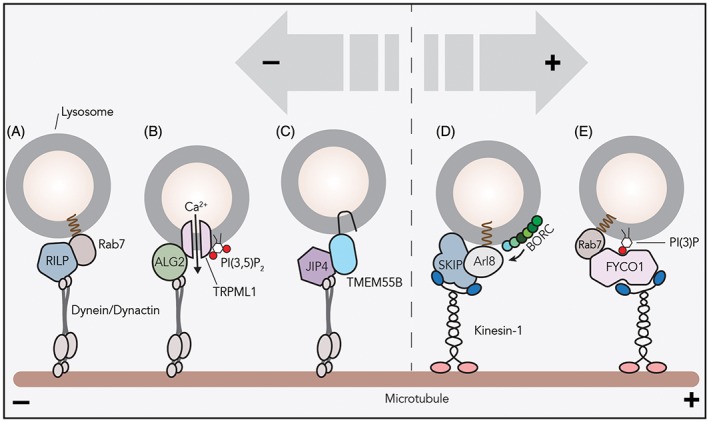
Many mechanisms of lysosomal transport. Various mechanisms of lysosomal recruitment of motor proteins are reported. A, The small GTPase Rab7 recruits effector RILP and the dynein/dynactin motor to lysosomes for minus‐end transport. B, Calcium plays a crucial role in lysosomal positioning. PI(3,5)P2 on lysosomal‐delimiting membranes activates the TRPML1 channel to stimulate calcium efflux. Then, the cytosolic calcium sensor ALG2 recruits the dynein/dynactin complex to TRPML1‐containing lysosomes. C, Transmembrane protein TMEM55B interacts with the dynein adaptor JIP4 and facilitates minus‐end transport. TMEM55B levels are controlled transcriptionally: depletion of nutrients or cholesterol upregulates TMEM55B transcription via autophagy‐associated transcription factors. D, BORC, a multisubunit complex on the lysosomal membrane, recruits Arl8b to lysosomes. The small GTPase Arl8b interacts with effector SKIP for lysosomal localization of kinesin‐1. E, The kinesin adaptor FYCO1 interacts with active Rab7 and PI(3)P on lysosomal membranes to recruit kinesin‐1 to lysosomes
Anterograde transport requires one or more kinesin motors to be employed and multiple mechanisms are identified. One of these systems also uses the Rab7 GTPase. Activated Rab7 can also bind the effector FYCO1 that in combination with phosphatidylinositol 3‐phosphate (PI(3)P) recruits the kinesin‐1 motor (Figure 2, discussed further below).21, 22 Alternatively, the lysosomal multiprotein complex BORC can activate the small GTPase Arl8b that then recruits the effector SKIP to engage in kinesin‐driven plus‐end transport (Figure 2).23, 24 Arl8b/SKIP can recruit two distinct kinesin types for anterograde transport of lysosomes: kinesin‐1 (KIF5B) prefers to move on perinuclear microtubules enriched in acetylated tubulin, whereas kinesin‐3 (KIF1Bβ and KIF1A) ensure movement on peripheral microtubules where tyrosinated α‐tubulin is abundant, further underscoring the complexity and dynamicity of lysosomal movement.25 The effector SKIP is also involved in anterograde transport of lysosome‐related organelles, such as lytic granules, Salmonella containing phagosomes and melanosomes.26, 27, 28 However, in transport of more mature melanosomes, SKIP and kinesin‐1 are recruited by GTPase Rab1A instead of Arl8b.27
Why do lysosomes contain two, maybe even more, mechanisms to drive plus‐end transport? It is currently unclear whether Rab7 and Arl8 occupy the identical lysosomes, but one possibility is that different GTPase/adaptor complexes function on different sets of lysosomes destined for distinct biological functions. For instance, FYCO1‐driven plus‐end trafficking results in cell growth and protrusion formation, suggesting that Rab7/FYCO1 works on lysosomes destined for plasma membrane fusion.22 Also, BORC‐regulated transport of lysosomes toward the periphery is required for efficient autophagosome‐lysosome fusion at the perinuclear region.29 Possibly, BORC/Arl8/SKIP drives retrograde trafficking and maturation of lysosomes that are transported back to perinuclear area. Although both models are plausible, the biological implications of Arl8/SKIP‐ and FYCO1/Rab7‐mediated outward trafficking are unclear.
Lysosomes prefer to move in a bidirectional stop‐and‐go manner by the alternating activities of kinesin and dynein motor proteins. They move along microtubules rather inefficiently, considering that their goal is to move from the perinuclear cloud to the plasma membrane. It is unclear why organelles move this way. One possibility is that lysosomes on microtubule tracks encounter various roadblocks such as other organelles like the ER or that the motor proteins are confused when getting at a crossing of two microtubule fibers. Alternatively, exhaustion of motor proteins might require their replacement or this movement is the result of a tug of war among multiple opposite‐directed motors. In fact, this type of stop‐and‐go transport is observed for all intracellular vesicles and appears to be intrinsic to vesicular transport of all kinds. Their complicated and apparently inefficient movement is however unexplained.
4. LYSOSOMAL POSITIONING REGULATED BY OTHER ORGANELLES
Membrane contact sites are cellular domains where two organelles interact to facilitate molecular function and promote functional integration. As a network, the ER reaches to the farthest corners of the cell and interacts with many organelles including the plasma membrane.30, 31 In fact, the ER is the largest intracellular organelle and contains the majority of cellular lipids. There is growing evidence that the ER forms membrane contact sites with almost all organelles and the plasma membrane.32 As the center stage of membrane contact sites, the ER is also tightly associated with endosomal biology. ER‐endosome contact sites take part in various molecular functions, including lipid trafficking, growth factor signaling, organelle fission, kinesin and dynein motor lysosomal transport and endosomal positioning.1, 2, 18, 22, 33
One of the ways by which ER‐lysosome contact sites control directionality of transport involves the lipid cholesterol. Cholesterol is an essential structural component of cellular membranes and has unique biophysical properties. It can alter membrane fluidity and form lipid microdomains in membranes.34 While some proteins are excluded from these microdomains, others are enriched.35 Rab7 and dynein may cluster into such cholesterol‐rich microdomains, at least on phagosomal membranes.36 Cholesterol may be involved in the formation of intraluminal vesicles in late endosomal multivesicular bodies.37, 38, 39 While mutations in proteins involved in cholesterol homeostasis are associated with various diseases, including lysosomal storage disorders, some lysosomal storage diseases show accumulating cholesterol in lysosomes, and it is now becoming clear that lysosome‐ER contact sites function in cholesterol homeostasis.40, 41, 42, 43 In one of the best‐understood mechanisms, cholesterol levels in lysosomes regulate the positioning of this organelle. The cytosolic oxysterol‐binding protein‐related protein 1 (ORP1L) localizes to lysosomes by interacting with the small GTPase Rab7 and phosphoinositides via its pleckstrin homology (PH) domain.44 When cholesterol is plenty in lysosomes, the cholesterol‐interacting domain of ORP1L, ORD, clamps down on cholesterol in the lysosomal‐limiting membrane and Rab7 effector RILP can recruit the dynein‐dynactin motor complex to facilitate minus‐end transport (Figure 3).17 This machinery is responsible for the vesicle clustering phenotype observed in many lysosomal storage disorders such as Niemann‐Pick disease type C, which are characterized by accumulation of cholesterol.18 In cholesterol‐low conditions, ORP1L undergoes a conformational change that exposes its FFAT motif to be seized by the ER‐bound VAP proteins.18 As a result of this newly formed membrane contact sites, the lysosome is tethered to the ER and the dynein‐dynactin motor is displaced from RILP. Within the ER contact sites formed by ORP1L/VAP interaction between the two compartments, cholesterol is transferred from lysosomes to the ER.42, 45 Overall, ORP1L‐regulated endosomal motility is responsible for interacting with the ER protein VAP that then determines clustering of lysosomes in cholesterol‐rich conditions and their immobilization in a scattered state when cholesterol is depleted.
Figure 3.
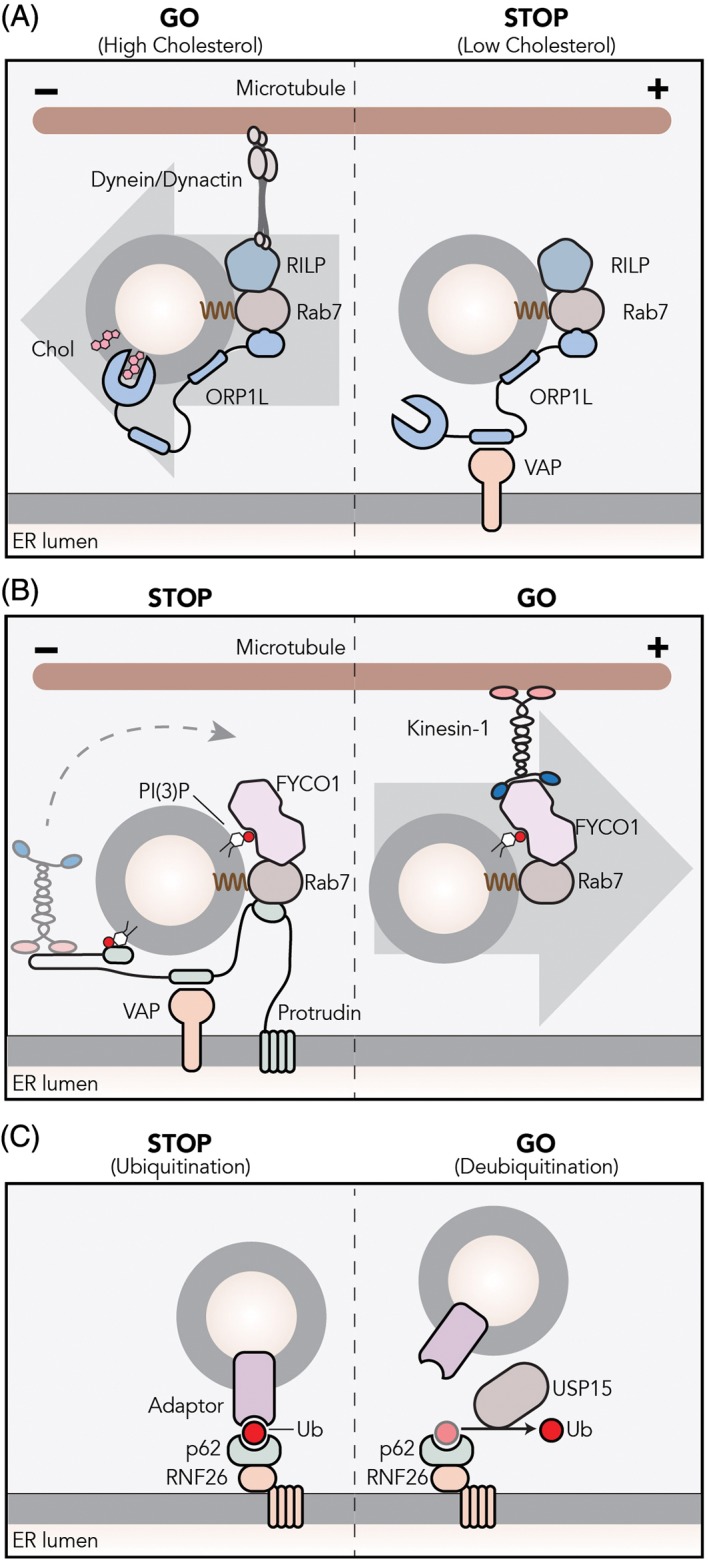
ER‐lysosome membrane contact sites control lysosomal transport and positioning. A, The Rab7 effector ORP1L localizes to lysosomes by interacting with Rab7 and phosphoinositides on the lysosomal membrane. When cholesterol is sufficient on the lysosomal‐ or autophagosomal‐delimiting membrane, ORP1L found in a closed conformation that mediates dynactin/dynein complex assembly and minus‐end transport via RILP. Under low‐cholesterol conditions, ORP1L adopts an open conformation and exposes an FFAT motif that allows interaction with the ER protein VAP. The resulting membrane contact site forces the release of the dynein/dynactin complex. B, ER‐bound protrudin interacts with the late Rab7 and PI(3)P and loads kinesin‐1 to Rab7‐effector FYCO1 on PI(3)P‐containing lysosomes. C, The ubiquitination of p62/SQSTM1 adaptor by the ER‐localized ubiquitin ligase RNF26 is recognized by TOLLIP that has a ubiquitin‐binding domain. This complex dictates perinuclear positioning of lysosomes. The cytosolic deubiquitinating enzyme USP15 mediates the release of lysosomes
The ER is also involved in targeting lysosomes toward the periphery. The ER‐resident protein protrudin forms membrane contact sites with late endosomes and loads kinesin‐1 on another Rab7 effector FYCO1, allowing plus‐end transport of late endosomes (Figure 3).21, 22 Later, these endosomal compartments fuse with the plasma membrane to induce protrusion formation and neurite outgrowth, two processes that require local deposition of membranes.
What advantage does ER‐based regulation of motor protein loading on lysosomes provide? One option is that the ER‐lysosome membrane contact sites provide a protected area for the transfer of nutrients and other means of information between the outside world (the content of lysosomes) and the cell interior (ER). Especially hydrophobic nutrients such as cholesterol and lipids, but also ions and other molecules can be exchanged efficiently within the space of the membrane contact sites between the two organelles. This may provide additional information for movement, maturation and fusion of lysosomes. Our current understanding of the nature of the communication between lysosomes and the ER is limited. However, these findings modify the old concept that cells consist of autonomous organelles. Rather, cells contain highly interactive organelles that in combination define many processes including motor protein‐controlled lysosomal transport.
5. LYSOSOMAL POSITIONING IN RESPONSE TO NUTRIENTS
Adaptation to changes in the environment is vital for all organisms. At the cellular level, the plasma membrane and the endosomal system are the platforms where cells have the opportunity to evaluate the contents of their surrounding medium, such as nutrients. mTORC1 is a master growth regulator that becomes activated at the cytosolic side of lysosomes in response to nutrients. In sufficient nutrient conditions, mTORC1 is recruited to lysosomal membranes. Here, it is activated by various amino acids, as well as other molecules such as cholesterol.46, 47, 48, 49, 50 Then, mTORC1 phosphorylates downstream proteins to induce cell growth and protein synthesis while inhibiting autophagy.46, 51, 52 In contrast, starvation inactivates and releases mTORC1 from lysosomes and activates autophagy. For autophagy, cytosolic proteins and organelles are engulfed by double‐membrane vesicles called autophagosomes that ultimately fuse with lysosomes to deliver material for degradation in order to restore the cellular nutrient pool. Therefore, lysosomes are intimately involved in nutrient responses, both in plenty and in starvation.
Lysosomal positioning is intimately associated with mTORC1 activity and nutrient levels. For example, mTORC1 activation under nutrient‐rich conditions causes scattering of lysosomes to the periphery.6 Upon starvation, peripheral lysosomes move toward the perinuclear region, where autophagosome‐lysosome fusion occurs. Proper maturation of autophagosomes relies on their minus‐end dynein motor‐mediated transport toward the perinuclear area.53 Following starvation, the lysosomal transmembrane protein TMEM55B recruits the dynein adapter JIP4 and then the dynein‐dynactin complex to lysosomes inducing minus‐end transport (Figure 2). TMEM55B protein levels are transcriptionally upregulated via mTORC1 upon starvation or cholesterol accumulation to induce perinuclear clustering.54 Once clustered, membrane contact sites formed between lysosomes and Golgi drive their immobilization at the perinuclear region.55
Removal of inactive mTORC1 from the lysosomal membrane upon starvation follows inactivation of the Ragulator complex. Ragulator can interact with the BORC complex to inactivate Arl8b‐dependent late endosomal scattering by SKIP/kinesin 1. Thus, amino acid depletion strengthens the BORC‐Ragulator interaction to ameliorate perinuclear clustering of lysosomes. However, as the BORC‐Ragulator‐controlled lysosomal positioning in response to amino acids appears to be independent of mTORC1, the exact mechanism is not fully understood.56, 57
6. LYSOSOMAL POSITIONING REGULATED BY PHOSPHOINOSITIDES
Lipids form an effective barrier against the extracellular milieu. Yet some lipids act as intracellular signals for several homeostatic processes. Since their discovery as primary messengers in the 1980s, phosphatidylinositol phosphates (PIPs) are at the center of attention in lipid signaling. As one of the major phospholipid species in mammalian cells, phosphatidylinositol can be phosphorylated at multiple positions and as a result, several different PIP species can be formed. These different PIPs are recognized by defined PH, Phox homology (PX) or FYVE domains in various proteins, and this interaction is often required for activation and/or localization of a series of proteins. Overall, distinctive subcellular localization and their comparably rapid and reversible interconversion harness PIPs as primary messengers in intracellular signaling and membrane trafficking.
PI(3)P is the hallmark of the endosomal system, including early and late endosomes.58 The localization of the early endosomal antigen EEA1 is facilitated by its interaction with Rab5 and PI(3)P on early endosomes.59, 60 This, in turn, causes further recruitment of Rab5 and its adaptor phosphatidylinositol kinase Vps34 increasing PI(3)P levels on early endosomal membranes (Figure 4).61, 62 This increased PI(3)P concentration on early endosomes is one of the factors critical for early endosomal maturation. Indeed, Rab5‐to‐Rab7 transition in maturing endosomes is induced by PI(3)P‐mediated recruitment of Rab7GEF to these membranes.63, 64 Rab5 thus controls the next step in endosome formation, the recruitment of active GTP‐loaded Rab7. PI(3)P levels have broader effects including the recruitment of ESCRT complex components to prepare for intraluminal vesicle formation.65
Figure 4.
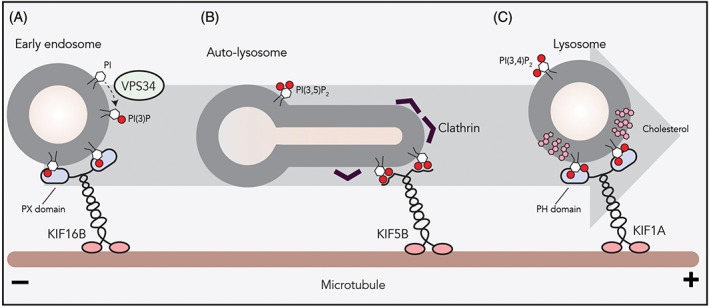
The role of phosphoinositides on lysosomal transport. A, PI(3)P, a phosphoinositide marking the endosomal system, is generated on the early endosomal membrane by the PI kinase VPS34. PI(3)P on early endosome serves as a docking station for KIF16B (kinesin‐3) to drive plus‐end transport. B, Clathrin on autophagosomal membranes causes clustering of PI(4,5)P2 that allows the binding of the KIF5B (kinesin‐1) onto the autophagosome membrane. KIF5B‐driven plus‐end transport results in autophagosome tubulation and proto‐lysosome formation. C, Formation of membrane microdomains by cholesterol and sphingomyelin or by glycosphingolipids on lysosomal membranes facilitates the interaction between the PH domain of KIF1A (kinesin‐3) and PI(4,5)P2
Similarly, late endosomal PI(3)P contributes to the formation of ER membrane contact sites by engaging ORP1L and protrudin.22, 44 The ER‐bound protein protrudin interacts with late endosomal Rab7 and PI(3)P to hand over kinesin‐1 to FYCO1. FYCO1 also interacts with Rab7 and PI(3)P on late endosomes to facilitate plus‐end transport.22 Nutrients stimulate the catalytic activity of the phosphatidylinositol kinase Vps34 to produce PI(3)P.66, 67 Conversely, the inhibition of Vps34 blocks the peripheral trafficking of late endosomes and results in perinuclear clustering even under nutrient‐rich conditions.68 This suggests that Vps34‐generated PI(3)P on endosomal membranes is required for plus‐end transport and mTORC1 activation in a protrudin/FYCO1/kinesin‐dependent manner. Interestingly, deprivation of nutrients causes late endosomal recruitment of a PI(3)P kinase that produces autophagosome‐related PI(3,4)P2 (phosphatidylinositol 3,4‐bisphoshate), mediating perinuclear clustering and mTORC1 suppression.69
Further maturation of late endosomes to lysosomes is accompanied by conversion of PI(3)P to PI(3,5)P2 (phosphatidylinositol 3,5,‐bisphosphate) and this conversion is usually associated with degradative sorting. PI(3,5)P2 acts upstream of the lysosomal calcium channel TRPML1 and activates it (Figure 2).70 TRPML1 increases cytosolic calcium levels to activate ALG2 and induces the formation of TRPML1/ALG2/dynein complexes on lysosomal membranes for minus‐end trafficking. TRPML1 activation by lysosomal PI(3,5)P2 can be inhibited by autophagosome‐associated PI(3,4)P2.71 In general, these processes should be controlled in time and space by lipid kinases and phosphatases.
Recruitment by PIPs often requires interaction of soluble protein domains with lipid head groups. Various PIP‐binding PH, PX and FYVE domains are found in Rab and Arf/Arl‐effector protein and even motor proteins.16, 24, 72 For instance, late endosomal localization of ORP1L is brought by its interaction with Rab7 as well as its PIP‐interacting PH domain.18 Additionally, both protrudin and the Rab7‐effector FYCO1 contain FYVE domains to interact with PI(3)P. Why would two components of the same machinery have the same function? Possibly this feature, together with the ability to bind Rab7, is critical to mark a defined subset of late endosomes/lysosomes. Interestingly, the Arl8b effector on lysosomes SKIP, also named PLEKHM2, also possesses a PH domain but the specificity and function of this domain remains unclear.
While Rab7 and Arl8b effector proteins can have phosphoinositide‐interacting domains that may modulate their interactions with motor proteins, there are endosome‐interacting motor proteins that integrated this option. The kinesin‐3 family member KIF16B interacts with PI(3)P via its PX domain. Moreover, plus‐end transport of early endosomes by KIF16B requires PI(3)P generated by Vps34 on early endosomes (Figure 4).73, 74 Another kinesin‐3 motor, KIF1A, interacts with PI(4,5)P2 on vesicles to facilitate plus‐end trafficking. in vitro experiments demonstrated that the formation of lipid‐ordered microdomains by cholesterol on vesicles results in a 40‐fold increase in PIP‐mediated KIF1A trafficking (Figure 4).75 At the peak of starvation, the majority of lysosomes fuse with autophagosomes to create auto‐lysosomes, resulting in a markedly decreased number of lysosomes. To restore this drop in lysosome numbers, the kinesin‐1 motor KIF5B interacts with PI(4,5)P2 on auto‐lysosome membranes and initiates autophagosome tubulation toward microtubule plus‐end to form proto‐lysosomes (Figure 4). During this process, clathrin causes lipid clustering on autophagosomal membranes to form PI(4,5)P2‐enriched microdomains.76 These observations suggest that not only the presence of adaptor proteins or lipids, but also their spatial organization within the same vesicle can dictate vesicular trafficking.
7. ACTIN‐BASED TRANSPORT FOR THE FINAL CARGO DELIVERY
Actin cytoskeleton and its associated motor protein myosins drive morphological changes of the plasma membrane for cell motility, differentiation and cell division. Actin fibers form a dense network under the plasma membrane and determine cell shape (Figure 1). There are about 31 active myosin motors identified with different functions and expression patterns. When arriving at the plus‐end of microtubules, lysosomes must pass through the actin mesh to be able to fuse with the plasma membrane. Analogous to some microtubule‐based motors, positioning and plasma membrane fusion of lysosomes are under the control of myosin motors coordinated by small GTPases.5, 77, 78 Rab27A, through its effector Slp2, recruits the myosin motors myosin Va and myosin VIIa to accumulate melanosomes at the cell periphery, under the melanocyte plasma membrane.79, 80, 81, 82 This machinery also controls the docking of other lysosome‐related organelles such as cytolytic granules to actin in cytotoxic T cells and insulin secretory granules in pancreatic β cells.83, 84 Interestingly, Rab27A mutations cause type 2 Griscelli syndrome that results in a defect in secretion of melanosomes.82, 85 It is likely that actin tethering is required for the fusion of membranes and secretion of cargo of lysosomes and lysosome‐related organelles. In addition to forming a well‐defined cortical network at the cell periphery, actin is also found in the cell's interior. In fact, non‐cortical actin and myosin play a critical role in the biology of lysosomes. Myosin Ia facilitates the delivery of endocytosed material to lysosomes and myosin Ic is involved in the fusion of autophagosomes with lysosomes.86, 87
In their process of maturation to generate an immune response, dendritic cells boost their major histocompatibility complex (MHC) class II surface expression by mass export of intracellular MHC class II pools.88, 89 A genome‐wide gene depletion screen identified the small GTPase Arl14 located on MHC class II‐containing intracellular lysosomes in control of this process. Arl14, through its effector ARF7EP, recruits myosin Ie and inactivation of this system results in immature dendritic cells with increased MHC class II surface expression at the cost of the intracellular pools.90 Myosin1e is a single‐headed motor protein that does not walk. This Arl14‐ARF7EP‐Myosin1e system arrests MHC class II‐containing lysosomes in dendritic cells. While recent reports reveal that the actin‐based control of lysosomal positioning and maturation is as complex as that of microtubule‐based control, the crosstalk between these two motor systems is likely even more complex and less understood.
8. CONCLUDING REMARKS AND PERSPECTIVES
Our current understanding is that lysosomal positioning and mobility relies on a complex interplay of interactions that involve microtubule‐ and actin‐based motors, protein complexes and membrane contact sites between organelles in response to nutrient levels and lipid distribution in membranes. Although several molecular mechanisms responsible for these interactions have been identified, their dynamic interplay is unclear. A mobile lysosome will interact with the ER, but also other organelles during its transit to or from the cell surface. Mechanisms responsible for recruitment of motor proteins and for perinuclear localization are partly identified, but our understanding of the control of lysosomal dynamics is still limited. Outstanding questions include the mechanisms of endosomal entry in and release from the perinuclear cloud, the bidirectional movement, the switch of motor proteins and the control of fusion in relation to motor protein transfer. It appears that dynein‐driven minus‐end transport of late endosomes and HOPS (homotypic fusion and protein sorting complex)‐mediated fusion are integrated processes.91 But is a coupling of transport and fusion of endosomes a more general process? Also the arrival at the site of actin cytoskeleton for fusion to the cell surface is unclear as well as the final fate of matured lysosomes. Although lysosomes are considered end‐stage compartments, their complex and dynamic biology remains a viable, relevant and exciting field of research.
Supporting information
Editorial Process
ACKNOWLEDGMENTS
The authors thank Ruud H. Wijdeven and Marlieke L. M. Jongsma for their critical input. This work was supported by a European Research Council (ERC) advanced grant to J.N.
Editorial Process File
The Editorial Process File is available in the online version of this article.
Cabukusta B, Neefjes J. Mechanisms of lysosomal positioning and movement. Traffic. 2018;19:761–769. 10.1111/tra.12587
Funding information European Research Council , Grant/Award Number: ERCOPE ‐ ERC‐ADV 694307
REFERENCES
- 1. Bakker J, Spits M, Neefjes J, Berlin I. The EGFR odyssey – from activation to destruction in space and time. J Cell Sci. 2017;130(24):4087‐4096. 10.1242/jcs.209197. [DOI] [PubMed] [Google Scholar]
- 2. Neefjes J, Jongsma MML, Berlin I. Stop or go? Endosome positioning in the establishment of compartment architecture, dynamics, and function. Trends Cell Biol. 2017;27(8):580‐594. 10.1016/j.tcb.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 3. Jongsma MLLM, Berlin I, Wijdeven RHHM, et al. An ER‐associated pathway defines endosomal architecture for controlled cargo transport. Cell. 2016;166(1):152‐166. 10.1016/j.cell.2016.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson DE, Ostrowski P, Jaumouillé V, Grinstein S. The position of lysosomes within the cell determines their luminal pH. J Cell Biol. 2016;212(6):677‐692. 10.1083/jcb.201507112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Encarnação M, Espada L, Escrevente C, et al. A Rab3a‐dependent complex essential for lysosome positioning and plasma membrane repair. J Cell Biol. 2016;213(6):631‐640. 10.1083/jcb.201511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korolchuk VI, Saiki S, Lichtenberg M, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13(4):453‐460. 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakamura S, Yoshimori T. New insights into autophagosome–lysosome fusion. J Cell Sci. 2017;130(7):1209‐1216. 10.1242/jcs.196352. [DOI] [PubMed] [Google Scholar]
- 8. Jordens I, Fernandez‐Borja M, Marsman M, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein‐dynactin motors. Curr Biol. 2001;11(21):1680‐1685. 10.1016/S0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 9. Reits EA, Neefjes JJ. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat Cell Biol. 2001;3(6):E145‐E147. 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- 10. Kapitein LC, van Bergeijk P, Lipka J, et al. Myosin‐V opposes microtubule‐based cargo transport and drives directional motility on cortical actin. Curr Biol. 2013;23(9):828‐834. 10.1016/J.CUB.2013.03.068. [DOI] [PubMed] [Google Scholar]
- 11. Farias GG, Guardia CM, De Pace R, Britt DJ, Bonifacino JS. BORC/kinesin‐1 ensemble drives polarized transport of lysosomes into the axon. Proc Natl Acad Sci USA. 2017;114(14):E2955‐E2964. 10.1073/pnas.1616363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanders AAWM, Kaverina I. Nucleation and dynamics of Golgi‐derived microtubules. Front Neurosci. 2015;9:431 10.3389/fnins.2015.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soppina V, Rai AK, Ramaiya AJ, Barak P, Mallik R. Tug‐of‐war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc Natl Acad Sci USA. 2009;106(46):19381‐19386. 10.1073/pnas.0906524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonifacino JS, Neefjes J. Moving and positioning the endolysosomal system. Curr Opin Cell Biol. 2017;47:1‐8. 10.1016/j.ceb.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513‐525. 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 16. Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10(10):682‐696. 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 17. Johansson M, Rocha N, Zwart W, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7‐RILP‐p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176(4):459‐471. 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rocha N, Kuijl C, van der Kant R, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7‐RILP‐p150Glued and late endosome positioning. J Cell Biol. 2009;185(7):1209‐1225. 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tan SC, Scherer J, Vallee RB. Recruitment of dynein to late endosomes and lysosomes through light intermediate chains. Mol Biol Cell. 2011;22(4):467‐477. 10.1091/mbc.E10-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X, Rydzewski N, Hider A, et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol. 2016;18(4):404‐417. 10.1038/ncb3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pankiv S, Alemu EA, Brech A, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end‐directed vesicle transport. J Cell Biol. 2010;188(2):253‐269. 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raiborg C, Wenzel EM, Pedersen NM, et al. Repeated ER‐endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520(7546):234‐238. 10.1038/nature14359. [DOI] [PubMed] [Google Scholar]
- 23. Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell. 2015;33(2):176‐188. 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosa‐Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin‐1. Dev Cell. 2011;21(6):1171‐1178. 10.1016/j.devcel.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guardia CM, Farias GG, Jia R, Pu J, Bonifacino JS. BORC functions upstream of kinesins 1 and 3 to coordinate regional movement of lysosomes along different microtubule tracks. Cell Rep. 2016;17(8):1950‐1961. 10.1016/j.celrep.2016.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tuli A, Thiery J, James AM, et al. Arf‐like GTPase Arl8b regulates lytic granule polarization and natural killer cell‐mediated cytotoxicity. Mol Biol Cell. 2013;24(23):3721‐3735. 10.1091/mbc.E13-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishida M, Ohbayashi N, Fukuda M. Rab1A regulates anterograde melanosome transport by recruiting kinesin‐1 to melanosomes through interaction with SKIP. Sci Rep. 2015;5(1):8238 10.1038/srep08238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boucrot E, Henry T, Borg J‐P, Gorvel J‐P, Méresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 2005;308(5725):1174‐1178. 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- 29. Jia R, Guardia CM, Pu J, Chen Y, Bonifacino JS. BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy. 2017;13(10):1648‐1663. 10.1080/15548627.2017.1343768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang C‐L, Chen Y‐J, Liou J. ER‐plasma membrane junctions: why and how do we study them? Biochim Biophys Acta Mol Cell Res. 2017;1864(9):1494‐1506. 10.1016/j.bbamcr.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang J, Zhao Z, Gu M, Feng X, Xu H. Release and uptake mechanisms of vesicular Ca2+ stores. Protein Cell. 2018. 10.1007/s13238-018-0523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17(2):69‐82. 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rowland AA, Chitwood PJ, Phillips MJ, Voeltz GK. ER contact sites define the position and timing of endosome fission. Cell. 2014;159(5):1027‐1041. 10.1016/j.cell.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387(6633):569‐572. 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 35. Rao M, Mayor S. Active organization of membrane constituents in living cells. Curr Opin Cell Biol. 2014;29(1):126‐132. 10.1016/j.ceb.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 36. Rai A, Pathak D, Thakur S, Singh S, Dubey AK, Mallik R. Dynein clusters into lipid microdomains on phagosomes to drive rapid transport toward lysosomes. Cell. 2016;164(4):722‐734. 10.1016/j.cell.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du X, Kazim AS, Brown AJ, Yang H. An essential role of Hrs/Vps27 in endosomal cholesterol trafficking. Cell Rep. 2012;1(1):29‐35. 10.1016/j.celrep.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 38. Edgar JR, Eden ER, Futter CE. Hrs‐ and CD63‐dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic. 2014;15(2):197‐211. 10.1111/tra.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trajkovic K, Hsu C, Chiantia S, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244‐1247. 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 40. Ikonen E. Mechanisms for cellular cholesterol transport: defects and human disease. Physiol Rev. 2006;86(4):1237‐1261. 10.1152/physrev.00022.2005. [DOI] [PubMed] [Google Scholar]
- 41. van der Kant R, Zondervan I, Janssen L, Neefjes J. Cholesterol‐binding molecules MLN64 and ORP1L mark distinct late endosomes with transporters ABCA3 and NPC1. J Lipid Res. 2013;54(8):2153‐2165. 10.1194/jlr.M037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eden ER, Sanchez‐Heras E, Tsapara A, Sobota A, Levine TP, Futter CE. Annexin A1 tethers membrane contact sites that mediate ER to endosome cholesterol transport. Dev Cell. 2016;37(5):473‐483. 10.1016/j.devcel.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilhelm LP, Wendling C, Védie B, et al. STARD3 mediates endoplasmic reticulum‐to‐endosome cholesterol transport at membrane contact sites. EMBO J. 2017;36(10):1412‐1433. https://doi.org/10.15252/embj.201695917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johansson M, Lehto M, Tanhuanpää K, Cover TL, Olkkonen VM. The oxysterol‐binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell. 2005;16(12):5480‐5492. 10.1091/mbc.E05-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao K, Ridgway ND. Oxysterol‐binding protein‐related protein 1L regulates cholesterol egress from the endo‐lysosomal system. Cell Rep. 2017;19(9):1807‐1818. 10.1016/j.celrep.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 46. Sancak Y, Bar‐Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator‐rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290‐303. 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chantranupong L, Scaria SM, Saxton RA, et al. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell. 2016;165(1):153‐164. 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Han JM, Jeong SJ, Park MC, et al. Leucyl‐tRNA synthetase is an intracellular leucine sensor for the mTORC1‐signaling pathway. Cell. 2012;149(2):410‐424. 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 49. Jewell JL, Kim YC, Russell RC, et al. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015;347(6218):194‐198. 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castellano BM, Thelen AM, Moldavski O, et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9‐Niemann‐Pick C1 signaling complex. Science. 2017;355(6331):1306‐1311. 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sancak Y, Peterson TR, Shaul YD, et al. The rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496‐1501. 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim E, Goraksha‐Hicks P, Li L, Neufeld TP, Guan K‐L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10(8):935‐945. 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wijdeven RH, Janssen H, Nahidiazar L, et al. Cholesterol and ORP1L‐mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat Commun. 2016;7:11808 10.1038/ncomms11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Willett R, Martina JA, Zewe JP, Wills R, Hammond GRV, Puertollano R. TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat Commun. 2017;8(1):1580 10.1038/s41467-017-01871-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Starling GP, Yip YY, Sanger A, Morton PE, Eden ER, Dodding MP. Folliculin directs the formation of a Rab34–RILP complex to control the nutrient‐dependent dynamic distribution of lysosomes. EMBO Rep. 2016;17(6):823‐841. https://doi.org/10.15252/embr.201541382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pu J, Keren‐Kaplan T, Bonifacino JS. A Ragulator‐BORC interaction controls lysosome positioning in response to amino acid availability. J Cell Biol. 2017;216(12):4183‐4197. 10.1083/jcb.201703094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Filipek PA, de Araujo MEG, Vogel GF, et al. LAMTOR/Ragulator is a negative regulator of Arl8band BORC‐dependent late endosomal positioning. J Cell Biol. 2017;216(12):4199‐4215. 10.1083/jcb.201703061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wallroth A, Haucke V. Phosphoinositide conversion in endocytosis and the endolysosomal system. J Biol Chem. 2018;293(5):1526‐1535. 10.1074/jbc.R117.000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107‐117. 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 60. Lawe DC, Patki V, Heller‐Harrison R, Lambright D, Corvera S. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3‐phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J Biol Chem. 2000;275(5):3699‐3705. 10.1074/jbc.275.5.3699. [DOI] [PubMed] [Google Scholar]
- 61. Schu P, Takegawa K, Fry M, Stack J, Waterfield M, Emr S. Phosphatidylinositol 3‐kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260(5104):88‐91. 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 62. Volinia S, Dhand R, Vanhaesebroeck B, et al. A human phosphatidylinositol 3‐kinase complex related to the yeast Vps34p‐Vps15p protein sorting system. EMBO J. 1995;14(14):3339‐3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cabrera M, Nordmann M, Perz A, et al. The Mon1‐Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin‐fold‐Rab interface and association with PI3P‐positive membranes. J Cell Sci. 2014;127(5):1043‐1051. 10.1242/jcs.140921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early‐to‐late endosome transition. Cell. 2010;141(3):497‐508. 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 65. Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458(7237):445‐452. 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 66. Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient‐regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280(38):33076‐33082. 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 67. Nobukuni T, Joaquin M, Roccio M, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH‐kinase. Proc Natl Acad Sci USA. 2005;102(40):14238‐14243. 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hong Z, Pedersen NM, Wang L, Torgersen ML, Stenmark H, Raiborg C. PtdIns3P controls mTORC1 signaling through lysosomal positioning. J Cell Biol. 2017;216(12):4217‐4233. 10.1083/jcb.201611073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Marat AL, Wallroth A, Lo WT, et al. mTORC1 activity repression by late endosomal phosphatidylinositol 3,4‐bisphosphate. Science. 2017;356(6341):968‐972. 10.1126/science.aaf8310. [DOI] [PubMed] [Google Scholar]
- 70. Dong X, Shen D, Wang X, et al. PI(3,5)P2 controls membrane trafficking by direct activation of mucolipin Ca2+ release channels in the endolysosome. Nat Commun. 2010;1(4):1‐11. 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang X, Li X, Xu H. Phosphoinositide isoforms determine compartment‐specific ion channel activity. Proc Natl Acad Sci USA. 2012;109(28):11384‐11389. 10.1073/pnas.1202194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kenniston JA, Lemmon MA. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29(18):3054‐3067. 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hoepfner S, Severin F, Cabezas A, et al. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121(3):437‐450. 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 74. Pyrpassopoulos S, Shuman H, Ostap EM. Adhesion force and attachment lifetime of the KIF16B‐PX domain interaction with lipid membranes. Mol Biol Cell. 2017;28(23):3315‐3322. 10.1091/mbc.E17-05-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109(3):347‐358. 10.1016/S0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Du W, Su QP, Chen Y, et al. Kinesin 1 drives autolysosome tubulation. Dev Cell. 2016;37(4):326‐336. 10.1016/j.devcel.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 77. Desnos C, Huet S, Fanget I, et al. Myosin va mediates docking of secretory granules at the plasma membrane. J Neurosci. 2007;27(39):10636‐10645. 10.1523/JNEUROSCI.1228-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):13‐19. 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 79. Hume AN, Seabra MC. Melanosomes on the move: a model to understand organelle dynamics. Biochem Soc Trans. 2011;39(5):1191‐1196. 10.1042/BST0391191. [DOI] [PubMed] [Google Scholar]
- 80. Jordens I, Westbroek W, Marsman M, et al. Rab7 and Rab27a control two motor protein activities involved in melanosomal transport. Pigment Cell Res. 2006;19(5):412‐423. 10.1111/j.1600-0749.2006.00329.x. [DOI] [PubMed] [Google Scholar]
- 81. Fukuda M. Synaptotagmin‐like protein (Slp) homology domain 1 of Slac2‐a/Melanophilin is a critical determinant of GTP‐dependent specific binding to Rab27A. J Biol Chem. 2002;277(42):40118‐40124. 10.1074/jbc.M205765200. [DOI] [PubMed] [Google Scholar]
- 82. Fukuda M. Rab27 effectors pleiotropic regulators in secretory pathways. Traffic. 2013;14(9):949‐963. 10.1111/tra.12083. [DOI] [PubMed] [Google Scholar]
- 83. Stinchcombe JC, Barral DC, Mules EH, et al. Rab27a is required for regulated secretion in cytotoxic T lymphocytes. J Cell Biol. 2001;152(4):825‐834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Waselle L, Coppola T, Fukuda M, et al. Involvement of the Rab27 binding protein Slac2c/MyRIP in insulin exocytosis. Mol Biol Cell. 2003;14(10):4103‐4113. 10.1091/mbc.E03-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ménasché G, Pastural E, Feldmann J, et al. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat Genet. 2000;25(2):173‐176. 10.1038/76024. [DOI] [PubMed] [Google Scholar]
- 86. Raposo G, Cordonnier MN, Tenza D, et al. Association of myosin I alpha with endosomes and lysosomes in mammalian cells. Mol Biol Cell. 1999;10(5):1477‐1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Brandstaetter H, Kishi‐Itakura C, Tumbarello DA, Manstein DJ, Buss F. Loss of functional MYO1C/myosin 1c, a motor protein involved in lipid raft trafficking, disrupts autophagosome‐lysosome fusion. Autophagy. 2014;10(12):2310‐2323. 10.4161/15548627.2014.984272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cella M, Engering A, Pinet V, Pieters J, Lanzavecchia A. Inflammatory stimuli induce accumulation of MHC class II complexes on dendritic cells. Nature. 1997;388(6644):782‐787. 10.1038/42030. [DOI] [PubMed] [Google Scholar]
- 89. Chow A, Toomre D, Garrett W, Mellman I. Dendritic cell maturation triggers retrograde MHC class II transport from lysosomes to the plasma membrane. Nature. 2002;418(6901):988‐994. 10.1038/nature01006. [DOI] [PubMed] [Google Scholar]
- 90. Paul P, van den Hoorn T, Jongsma MLM, et al. A genome‐wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell. 2011;145(2):268‐283. 10.1016/j.cell.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 91. van der Kant R, Fish A, Janssen L, et al. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci. 2013;126(15):3462‐3474. 10.1242/jcs.129270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Editorial Process


