Abstract
Adrenocorticotrophic hormone (ACTH)‐secreting pituitary adenomas give rise to a severe endocrinological disorder, comprising Cushing's disease, with multifaceted clinical presentation and treatment outcomes. Experimental studies suggest that the disease variability is inherent to the pituitary tumour, thus indicating the need for further studies into tumour biology. The present study evaluated transcriptome expression pattern in a large series of ACTH‐secreting pituitary adenoma specimens in order to identify molecular signatures of these tumours. Gene expression profiling of formalin‐fixed, paraffin‐embedded specimens from 40 human ACTH‐secreting pituitary adenomas revealed the significant expression of genes involved in protein biosynthesis and ribosomal function, in keeping with the neuroendocrine cell profile. Unsupervised cluster analysis identified 3 distinct gene profile clusters and several genes were uniquely overexpressed in a given cluster, accounting for different molecular signatures. Of note, gene expression profiles were associated with clinical features, such as the age and size of the tumour. Altogether, the findings of the present study show that corticotroph tumours are characterised by a neuroendocrine gene expression profile and present subgroup‐specific molecular features.
Keywords: ACTH‐secreting adenomas, Cushing's disease, gene expression profiling, neuroendocrine tumours, POMC
1. INTRODUCTION
Cushing's disease, that is adrenocorticotrophic hormone (ACTH)‐secreting pituitary adenoma, is a rare and complex endocrine disease. Hypersecretion of ACTH by the tumour gives rise to excess cortisol secretion by the adrenal glands, which, in turn, promotes a variety of clinical signs encompassing osteoporosis, muscle atrophy, hypertension and diabetes. These patients carry a considerable disease burden, and mortality, if untreated, is high.1 Studies into the pathophysiology of corticotroph tumours have shed light on the involvement of individual factors, such as epidermal growth factor (EGF),2 ubiquitin‐specific peptidase 8 (USP8),3, 4 Brahma‐related gene 1 (Brg‐1),5 although little is known about other, possible alterations in these tumours.
Of note, an increasing body of evidence indicates that ACTH‐secreting adenomas differ markedly among themselves regarding their responses to the main modulators of corticotroph secretion, such as corticotrophin‐releasing factor and steroids,6, 7 as well as ancillary modulators, such as vasopressin analogues, gonadotrophin‐releasing hormone.8, 9 Differences in in vitro ACTH secretory patterns have also been observed in response to dopamine or somatostatin receptor agonists,10, 11 which obviously translates into different responsiveness to these drugs. Furthermore, somatic mutations and individual polymorphisms have been linked to differences in tumour size and secretory pattern.12, 13
It is therefore apparent that wide‐range evaluation of gene expression patterns in corticotroph adenomas is needed to understand which features characterise tumoral corticotrophs and underlie the phenotypical diversity and thus, possibly, pave the way for a personalised clinical strategy. The present study evaluated gene expression profiles in a large series of human ACTH‐secreting adenomas. We used RNA extracted from formalin‐fixed, paraffin‐embedded archival samples collected during transsphenoidal surgery, thus overcoming limitations as a result of the rarity of the disorder.
2. MATERIALS AND METHODS
2.1. Sample collection
Forty formalin‐fixed, paraffin‐embedded adenomatous samples were retrieved and sections were microdissected under a surgical microscope. Specimens were collected from the centre of the biopsy to minimise RNA degradation as a result of nucleic acid oxidation. Immunohistochemistry and neuropathological examination confirmed the diagnosis and the absence of normal pituitary tissue on sections. MIB‐1 staining was <3%, attesting to typical corticotroph adenoma (grade I). Overall, specimens had been stored 2‐15 years. Clinical data from patient presentation, surgical outcome and follow‐up were collected. The entire series comprised 9 men and 31 women, aged 17–69 years, 29 microadenomas (diameter ≤10 mm) and 11 macroadenomas (diameter >10 mm), 6 surgical failures and the remainder surgical remissions. Mean ± SD follow‐after surgery up was 140.3 ± 13.6 months (range 2‐15 years). The study was approved by the Ethical Committee of the Istituto Auxologico Italiano.
2.2. RNA formalin‐fixed, paraffin‐embedded sample extraction
Four 20‐μm thick sections from each specimen were extracted using Recover All Total Nucleic Acid Isolation Kit (Life Technologies, Carlsbad, CA, USA). Tissue blocks were deparaffinised in xylene, RNA‐bound proteins removed by protease, and RNA captured on glass‐filters and eluted. Recovery of RNA averaged 1.5‐2 μg per sample. To maximise recovery of adequate transcripts and reduce RNA damage through oxydation, slices were cut from the center of the paraffin block.
2.3. RNA quality control
RNA (200 ng) was reverse‐transcribed (Superscript‐Vilo cDNA synthesis kit; Life Technologies) and amplified by real‐time polymerase chain reaction (PCR) for ribosomal protein L13A (RPL13A),14 using Taqman probe Hs03043885_g, (Applied Biosystems, Foster City, CA, USA). All 40 samples yielded the expected 81‐bp transcript at <30 cycles, attesting to adequate quality RNA.
2.4. Whole genome‐DASL HT
The ILLUMINA Whole Genome DASL High‐Throughput assay (WG‐DASL‐HT; Illumina, San Diego, CA, USA) for RNA extracted from formalin‐fixed, paraffin‐embedded tissues was used. The Human HT_12 v4 Bead Chip evaluates 29 285 probes (20 815 genes) and is highly accurate with respect to estimating differences in gene expression by head‐to‐head comparison with quantitative real‐time PCR.15 Each sample carried 300 ng of RNA. Fluorescence was imaged and data captured into HiScan, a high‐resolution laser imager (Illumina). Microarray data have been deposited with Gene Expression Omnibus (accession number GSE93825).
2.5. USP8 sequencing
Approximately 100 ng of RNA was reverse‐transcribed (Superscript VILO cDNA synthesis kit; Life Technologies) with the oligonucleotide primers: 5′‐CTTGACCCAATCACTGGAAC‐3′ (forward); 5′‐TTACTGTTGGCTTCCTCTTCTC‐3′ (reverse)13 for amplification of USP8 exon 14, the most frequent site of mutations reported so far.13 Touch‐down PCR was performed using GO TAQ DNA polymerase (Promega, Madison, WI, USA) at 64–57°C annealing. PCR products were purified by ExoProStar Illustra enzyme (Ge Healthcare, Chicago, IL, USA) and Sanger sequencing performed using the ABI PRISM Big DYE Terminator V3.1 cycle sequencing kit (Applied Biosystems) on ABI PRISM 3500 analyser.
2.6. Data analysis
Genome Studio software (Illumina) was used to detect genes significantly expressed in all specimens (ie, genes with detection P < 0.05 based on fluorescence intensity [average signal]), thus establishing a common expression profile. Subsequently, we attempted to identify different gene expression profiles in human corticotroph adenomas using unsupervised cluster analysis. This approach looks for similarities among gene expression profiles and identifies subgroups of samples on the basis of similarity between expression profiles.16, 17 Unsupervised methods for clustering among disease samples do not require prior knowledge on samples themselves or control tissues. Unsupervised cluster analysis was performed with hClust package (RStudio Inc., Boston, MA, USA) using the Ward minimum variance method18 and patients organised in accordance with their gene expression profile. Dissimilarity between clusters was measured by Euclidean distance.
Comparisons of clinical variables between clusters (ANOVA followed by Fisher's least significant difference or chi‐squared test, as appropriate) were performed with statview, version 4.5 (Abacus Concepts, Berkeley, CA, USA). P < 0.05 was considered statistically significant. Quantitative variables are given as the mean ± SEM.
2.7. Differential gene expression analysis
Once clusters had been established on the basis of similarity in gene expression profiles, differential gene expression analysis enabled the identification of genes that are expressed to a greater or lesser extent in each cluster. Clusters were analysed for differential expression analysis by rank invariant normalisation (Genome Studio; Illumina). Only genes with Benjamini and Hochberg P ≤ 0.05 were considered and Diff Scores were calculated based on P value transformation (ie, Diff Score ≥13 [up‐regulated gene] and ≤−13 [down‐regulated gene] P = 0.05) according to the difference between average signals in reference cluster vs the other 2 clusters. A Volcano plot was used to illustrate differential expression because it visualises the magnitude of over‐ and underexpression of a given gene plotted vs the significance of the comparison. This double‐filtering approach allows significantly differentially expressed genes to be easily identified.19
2.8. Real‐time gene expression analysis
RNA (100 ng) was reverse‐transcribed (Superscript‐Vilo cDNA synthesis kit; Life Technologies) and quantitative real‐time PCR was performed on a 7900 HT sequence Detection System (Applied Biosystems), using Platinum Quantitative PCR Supermix‐UDG with premixed ROX. Taqman assay (Applied Biosystems) was used for detection of the following genes: BCAS2 (probe Hs00903014_g1), SCG5 (probe Hs00161638_m1), IGFBP5 (probe Hs00181213_m1) and RPLP0 (probe Hs99999902_m1). Basal expression data () were calculated and normalised to RPLP0; differences between clusters were established by a Mann‐Whitney test.
2.9. Functional annotation and Gene Ontology
Two approaches were used to extract biological features from gene lists: david, version 6.720 was used to annotate and classify significant genes and perform functional annotation clustering. The minimum value of the enrichment score for significant clusters was ≤1.3. Clusters were annotated to Gene Ontology (GO) project, including Molecular Function (MF), Cellular Component (CC), Biological Process (BP), Kyoto Encyclopaedia of Genes and Genomes (KEGG) and Protein Information Resource (SP_PIR). Furthermore, cytoscape with Cluego plug‐in21 was used to identify molecular interaction networks. The biological pathways included were: GO project, KEGG and Inter Pro for protein domains.22 K score = 0.4 and Benjamini and Hochberg P value ≤0.001 were used for cytoscape analysis of significantly expressed genes. Of note, the group leading term is the most significant and genes can be included in several terms.22 The size of nodes reflects statistical significance of terms, whereas the degree of connectivity between terms (ie, edge) is calculated using Kappa statistics.23
3. RESULTS
3.1. Clustering analysis
Figure 1 shows unsupervised clustering and heat map profiling of significantly expressed genes in all 40 corticotroph adenoma samples. Of 20 815 genes, 1259 genes were expressed at P ≤ 0.05 and formed 5 distinct gene groups with decreasing expression. Of note, genes known to be associated with the corticotroph phenotype (eg, POMC, TBX19 [ie, Tpit]) were highly expressed (group 2). Conversely, the expression of genes associated with other anterior pituitary cells (for example, GH, PRL, PIT1, LHB, FSHB and CGA) was not significant across specimens, thus confirming absent contamination by normal anterior pituitary tissue.24, 25, 26 Highest enrichment scores were annotated to protein biosynthesis, ribosomes and RNA processing by GO, KEGG and SP‐PIR (see Supporting information, Table S1). In detail, the highest enrichment score was annotated to protein biosynthesis, followed by ribosomal function in group 1. The second‐ranked cluster revealed significant enrichment in ribosomal function, RNA processing, membrane transport and lumen, ATP metabolic processes and cytoskeleton organisation. Enrichment scores were lower in groups 3 and 4, mainly annotated to vesicle and lumen function, protein transport and localisation. Enrichment in group 5 was the lowest, with lumen and protease the only annotated functions (see Supporting information, Table S1).
Figure 1.
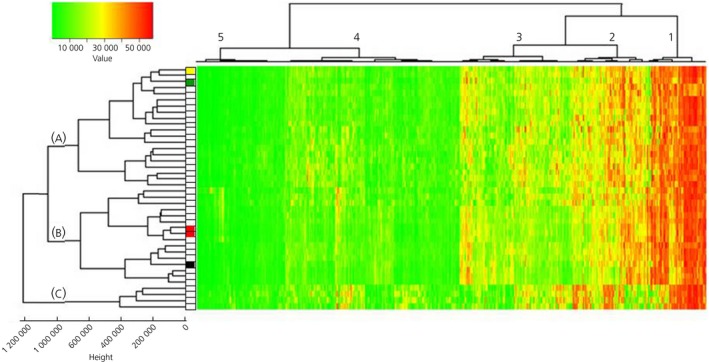
Unsupervised clustering of 1259 significantly expressed genes. Colour coding of gene expression values is shown in the upper left corner. USP8 mutation status is indicated to the left of the heatmap: white squares identify wild‐type sequence, whereas coloured squares indicate USP8 variants: red, variant c.2159C>G; green, variant c.2152T>C; black, variant c.2155_2157delTCC; yellow, variant c.2157_2171delCCCAGATATAACCCA. Height of nodes represents dissimilarity between clusters as measured by Euclidean distance
cytoscape analysis revealed that significantly expressed genes formed consistent networks related primarily to protein targeting to the endoplasmic reticulum (ER), cellular macromolecule metabolic processes, RNA and polyRNA binding (Figure 2). Other networks were related to cellular transport of the protein to the ER and Golgi system and to membrane activity, such as vesicle forming and coating. Networks related to post‐transcriptional regulation of gene expression and the energy derivation by oxidation of organic compounds were also identified. Altogether, functional annotation by both approaches identified major enrichment of pathways related to protein synthesis and intracellular transport. Of note, this analysis identified genes that characterise the expression profile of corticotroph adenomas per se, and not genes uniquely expressed in tumoral corticotrophs.
Figure 2.
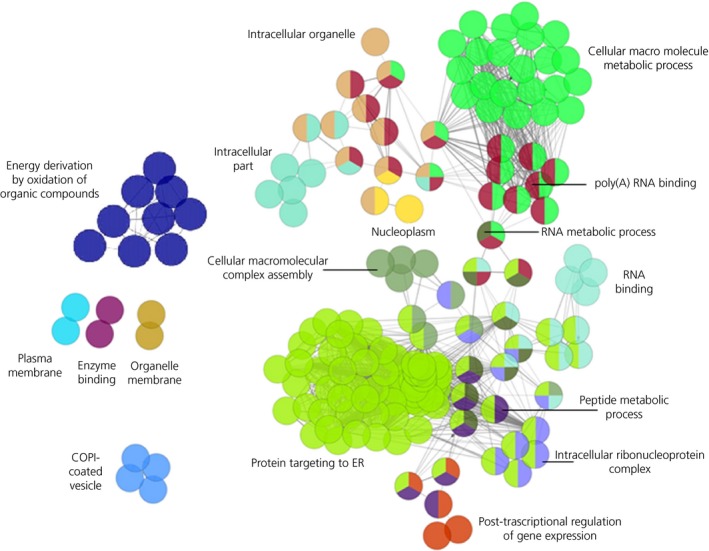
cytoscape analysis showing networks formed by principal significant terms. The number of nodes is proportional to the number of the term‐forming genes; nodes are colour‐coded according to functional annotations and can have more than 1 colour because genes may belong to more than 1 term. The 2 major terms are shown in bold. COPI, coat protein I; ER, endoplasmic reticulum
3.2. Clinical data analysis
Gene expression profiles led to the grouping of patients in 3 separate clusters (ie, A, B and C). Analyses of differences in clinical variables among clusters revealed a higher proportion of macroadenomas and extrasellar tumours in cluster A (Table 1); indeed, 91% of macroadenomas and 92% of extrasellar tumours from the entire series fell in cluster A, with the remainder in cluster B. With respect to a quantitative comparison of tumour size, adenomas in cluster A were bigger than those in clusters B and C, although this failed to reach statistical significance (F = 1.88, not significant). Patients in cluster A were also older (F = 3.258, P < 0.05) (Table 1), although age and tumour diameter were not associated with each other (r = 0.01, not significant). No differences regarding hormonal values, pathological findings and surgical outcomes were detected among clusters (Table 1).
Table 1.
Hormonal and clinical variables in individual patient clusters
| Cluster A | Cluster B | Cluster C | Significance | |
|---|---|---|---|---|
| Demographical features | ||||
| Sex distribution (male/female) | 5/15 | 2/14 | 2/2 | NS |
| Age (years) | 45.1 ± 3.01 | 36.7 ± 3.18 | 30.3 ± 3.74 | P < 0.05 |
| Hormonal parameters | ||||
| Urinary free cortisol (μg 24 h‐1) | 547.4 ± 153.4 | 690.9 ± 120.8 | 474.7 ± 179.2 | NS |
| Plasma ACTH (pg mL‐1) | 89.2 ± 13.9 | 63.1 ± 6.15 | 45.3 ± 3.22 | NS |
| Morning serum cortisol (μg dL‐1) | 23.5 ± 1.28 | 19.6 ± 1.60 | 18.5 ± 2.20 | NS |
| Midnight serum cortisol (μg dL‐1) | 20.5 ± 1.48 | 19.8 ± 2.52 | 18.2 ± 6.00 | NS |
| Cortisol after 1 mg of dexamethasone (μg dL‐1) | 16.5 ± 1.98 | 15.5 ± 2.83 | 7.95 ± 4.55 | NS |
| Cortisol suppression after 8 mg dexamethasone (% baseline) | 34.9 ± 9.44 | 43.0 ± 13.51 | 11.6 ± 1.81 | NS |
| ACTH peak after corticotrophin‐releasing hormone (% baseline) | 341.9 ± 93.8 | 271.5 ± 79.4 | 392.1 ± 57.4 | NS |
| Macroadenoma (%) | 50% | 6.2% | 0% | P < 0.005 |
| Extrasellar extension (%) | 55% | 6.2% | 0% | P < 0.005 |
| Pathology findings | ||||
| Tumour diameter (mm) | 10.1 ± 1.52 | 7.0 ± 0.64 | 6.7 ± 0.25 | NS |
| ACTH immunoreactive cells (%) | 86.0 ± 2.48 | 84.1 ± 3.54 | 93.7 ± 1.25 | NS |
| Crooke cells (absent/slight/moderate) | 45%/45%/10% | 69%/25%/6% | 75%/25%/0% | NS |
| Cellular atypia (absent/slight) | 55%/45% | 75%/25% | 100%/0% | NS |
| Surgical outcomes | ||||
| Immediate remission (%) | 85% | 87.5% | 100% | NS |
| Relapse (%) | 6% | 0% | 0% | NS |
| Length on steroid replacement therapy (months) | 19.3 ± 5.9 | 16.9 ± 3.5 | 23.2 ± 12.1 | NS |
Data are the mean ± SEM or percentage. ACTH, adrenocorticotrophic hormone; NS, not significant.
3.3. USP8 sequencing
Five patients presented USP8 variants: 2 patients showed missense variant rs672601311 (c.2159C>G, p.720Pro>Arg), 1 patient showed missense variant rs672601307 (c.2152T>C, p.718Ser>Pro), 1 patient showed the 3‐bp deletion variant rs672601306 (c.2155_2157delTCC, p.Ser718del) and 1 patient showed a novel, 15‐bp deletion (c.2157_2171delCCCAGATATAACCCA, p.Pro720_Gln724del). Gene expression profiles in specimens bearing USP8 variants did not group together, except for the 2 specimens with the same missense variant, which clustered next to each other (Figure 1).
3.4. Differentially expressed genes
Comparisons of gene profiles from the 3 different clusters allowed identification of over‐ or under‐expressed genes between clusters. A Volcano plot (Figure 3) shows genes up‐regulated with Diff Score ≥13 and genes down‐regulated with Diff Score ≤−13 between cluster A vs clusters B and C (Figure 3A), cluster B vs clusters A and C (Figure 3B), and cluster C vs clusters A and B (Figure 3C).
Figure 3.
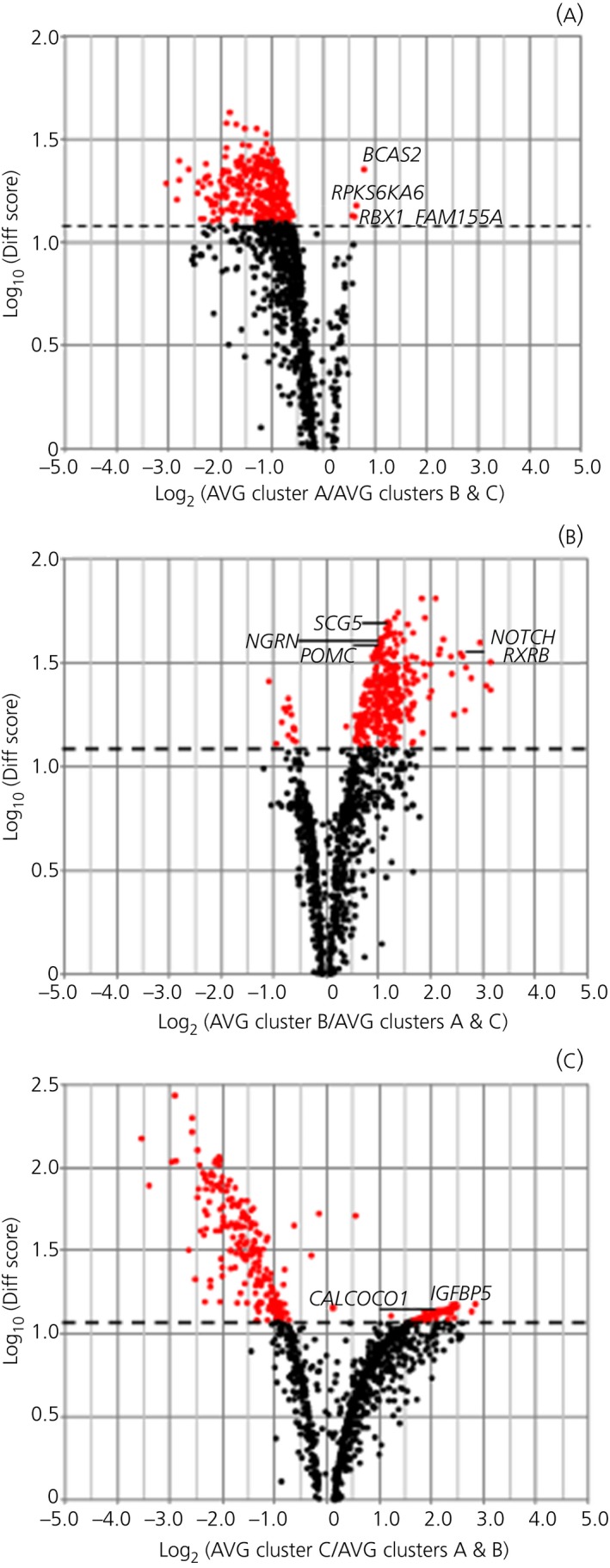
Volcano plot. Genes up‐ and down‐regulated in the 3 clusters. Effect (average signal, AVG) is shown on the x‐axis and significance (Diff Score) is shown on the y‐axis. Up‐regulated genes appear to the right and down‐regulated genes appear to the left of the x‐axis. (A) Comparison between cluster A and clusters B and C. (B) Comparison between cluster A and clusters A and C. (C) Comparison between cluster C and clusters A and B. Significant genes are highlighted in red (Diff Score >13) and selected genes are identified by name
Of 1259 significantly expressed genes, 4 genes were uniquely overexpressed in cluster A, 313 genes were overexpressed only in cluster B and 29 genes overexpressed only in cluster C (Table 2). Among the 4 genes uniquely overexpressed in cluster A, 3 genes were associated with tumoral processes (ie, BCAS2 [breast carcinoma amplified sequence 2] and RPS6KA6 [ribosomal protein S6 kinase 6] with breast cancer27, 28 and RBX1 [ringbox 1] with gastric and lung cancer). With regard to genes overexpressed in cluster B compared to the other 2 clusters, we observed overexpression of the POMC gene itself, as well as of SCG5, which encodes for secretogranin 5 (also known as neuroendocrine protein 7B2) associated with prohormone convertase 2 activation.29 Furthermore, genes involved in neuroendocrine tumours (eg, NOTCH3, LGALS3, PITX1 and NGRN) were also overexpressed in cluster B compared to clusters A and C. Additional genes of interest to corticotroph adenoma pathophysiology were RXRB and TIMP1 in view of their links to retinoic acid sensitivity30, 31 and matrix metalloproteinase, respectively.32, 33 Given the number of genes overexpressed in this cluster, we could perform functional analysis and detected significant enrichment for 123 genes. Highest enrichment scores were relative to ribosomal function and translation (enrichment 4.89), ribosome assembly (enrichment 1.97), protein biosynthesis (enrichment 1.67) and mRNA processing (enrichment 1.60), thus comprising items recurring with features identified for functional analysis of all corticotroph adenomas (see Supporting information, Table S1). The remaining 190 genes did not fall into specific functional annotation groups. Gene expression profiles in 4 patients differed markedly from all others and clustered separately (cluster C). Amongst genes uniquely overexpressed in this cluster (Table 2), IGFBP5 (insulin‐like growth factor binding protein 5) and genes associated with intracellular calcium flux (ie, CALCOCO1, CAMTA2) appeared to be considerably overexpressed as measured by fold‐change vs other clusters and as of interest to tumoral corticotroph pathophysiology.
Table 2.
Genes up‐regulated in individual clusters
| Symbol | Diff Score | Definition |
|---|---|---|
| Cluster A | ||
| BCAS2 | 22,57 | Homo sapiens breast carcinoma amplified sequence 2, mRNA |
| RPS6KA6 | 15,18 | Homo sapiens ribosomal protein S6 kinase, 90 kDa, polypeptide 6, mRNA |
| RBX1 | 13,5 | Homo sapiens ringbox 1, mRNA |
| FAM155A | 13,2 | Homo sapiens family with sequence similarity 155, member A, mRNA |
| Cluster B (first 20 genes with highest Diff Scores listed here, the entire list is given in the Supporting information, Table S2) | ||
| RAB33B | 64,25 | Homo sapiens RAB33B, member RAS oncogene family, mRNA |
| RAX2 | 64,25 | Homo sapiens retina and anterior neural fold homeobox 2, mRNA |
| EIF2AK4 | 54,75 | Homo sapiens eukaryotic translation initiation factor 2 alpha kinase 4, mRNA |
| RBM25 | 51,91 | Homo sapiens RNA binding motif protein 25, mRNA |
| ZNF358 | 51,62 | Homo sapiens zinc finger protein 358, mRNA |
| ACTR1A | 49,43 | Homo sapiens ARP1 actin‐related protein 1 homolog A, mRNA |
| SCG5 | 49,18 | Homo sapiens secretogranin V (7B2 protein), mRNA |
| NT5C2 | 48,13 | Homo sapiens 5′‐nucleotidase, cytosolic II, mRNA |
| BTBD2 | 47,99 | Homo sapiens BTB (POZ) domain containing 2, mRNA |
| TNPO2 | 46,04 | Homo sapiens transportin 2, mRNA |
| UBXN2A | 45,06 | Homo sapiens UBX domain protein 2A, mRNA |
| SH3YL1 | 43,87 | Homo sapiens SH3 and SYLF domain containing 1 (SH3YL1), mRNA |
| HNRNPUL2 | 43,84 | Homo sapiens heterogeneous nuclear ribonucleoprotein U‐like 2, mRNA |
| HARS2 | 43,51 | Homo sapiens histidyl‐tRNA synthetase 2, mitochondrial, mRNA |
| DARS | 43,26 | Homo sapiens aspartyl‐tRNA synthetase, mRNA |
| RNGTT | 43,15 | Homo sapiens RNA guanylyltransferase and 5′‐phosphatase, mRNA |
| NDUFS8 | 42,47 | Homo sapiens NADH dehydrogenase (ubiquinone) Fe‐S protein 8, 23 kDa (NADH‐coenzyme Q reductase), mRNA |
| THBS3 | 42,47 | Homo sapiens thrombospondin 3, mRNA |
| FOXO4 | 41,59 | Homo sapiens forkhead box O4 (FOXO4), mRNA |
| GCN1 | 41,08 | Homo sapiens eIF2 alpha kinase activator homolog, also known as general control of amino‐acid synthesis 1‐like 1 (GCN1L1), mRNA |
| Cluster C | ||
| SNX29P1 | 15,12 | Homo sapiens sorting nexin 29 pseudogene 1, also known as RUN domain containing 2B, mRNA (RUNDC2B) |
| IGFBP5 | 14,78 | Homo sapiens insulin‐like growth factor binding protein 5, mRNA |
| PLD2 | 14,78 | Homo sapiens phospholipase D2, mRNA |
| SNORA54 | 14,34 | Homo sapiens small nucleolar RNA, H/ACA box 54, small nucleolar RNA |
| SNORD1141 | 14,30 | Homo sapiens small nucleolar RNA, C/D box 114‐1, small nucleolar RNA |
| ACIN1 | 14,28 | Homo sapiens apoptotic chromatin condensation inducer 1, mRNA |
| SNORA10 | 14,18 | Homo sapiens small nucleolar RNA, H/ACA box 10, small nucleolar RNA |
| SNORD104 | 14,05 | Homo sapiens small nucleolar RNA, C/D box 104, small nucleolar RNA |
| SNORD41 | 13,86 | Homo sapiens small nucleolar RNA, C/D box 41, small nuclear RNA |
| SNORA73B | 13,74 | Homo sapiens RNA, U105A small nucleolar, small nucleolar RNA a.k.a RNU105A |
| CALCOCO1 | 13,71 | Homo sapiens calcium binding and coiled‐coil domain 1, mRNA |
| SCAF1 | 13,69 | Homo sapiens SR‐related CTD‐associated factor 1, mRNA |
| SNORA64 | 13,65 | Homo sapiens small nucleolar RNA, H/ACA box 64, small nucleolar RNA |
| CAMTA2 | 13,49 | Homo sapiens calmodulin binding transcription activator 2, mRNA |
| SNORA73B | 13,45 | Homo sapiens small nucleolar RNA, H/ACA box 73B, small nucleolar RNA |
| ARSG | 13,42 | Homo sapiens arylsulfatase G, mRNA |
| SNORA32 | 13,36 | Homo sapiens small nucleolar RNA, H/ACA box 32, small nucleolar RNA |
| RNU5A | 13,31 | Homo sapiens RNA, U5A small nuclear, small nuclear RNA |
| SNORD12 | 13,30 | Homo sapiens small nucleolar RNA, C/D box 12, small nucleolar RNA |
| RNU1G2 | 13,24 | Homo sapiens RNA, U1G2 small nuclear, small nuclear RNA |
| PPID | 13,23 | Homo sapiens peptidylprolyl isomerase D, mRNA |
| SNORD3D | 13,19 | Homo sapiens small nucleolar RNA, C/D box 3D, small nucleolar RNA |
| SNORD33 | 13,16 | Homo sapiens small nucleolar RNA, C/D box 33, small nucleolar RNA |
| SNORD68 | 13,14 | Homo sapiens small nucleolar RNA, C/D box 68, small nucleolar RNA |
| SNORD110 | 13,11 | Homo sapiens small nucleolar RNA, C/D box 110, small nucleolar RNA |
| SCARNA3 | 13,05 | Homo sapiens small Cajal body‐specific RNA 3, guide RNA |
| KMT5A | 13,05 | Homo sapiens lysine methyltransferase 5A, also known as SET domain containing 8 (SETD8), mRNA |
| SNORD66 | 13,02 | Homo sapiens small nucleolar RNA, C/D box 66, small nucleolar RNA |
| SNORA71A | 13,01 | Homo sapiens small nucleolar RNA, H/ACA box 71A, small nucleolar RNA |
Real‐time PCR confirmed overexpression of BCAS2 in cluster A, SCG5 in cluster B and IGFBP5 in cluster C (Figure 4), in support of the differential expression observed with respect to microarray data.
Figure 4.
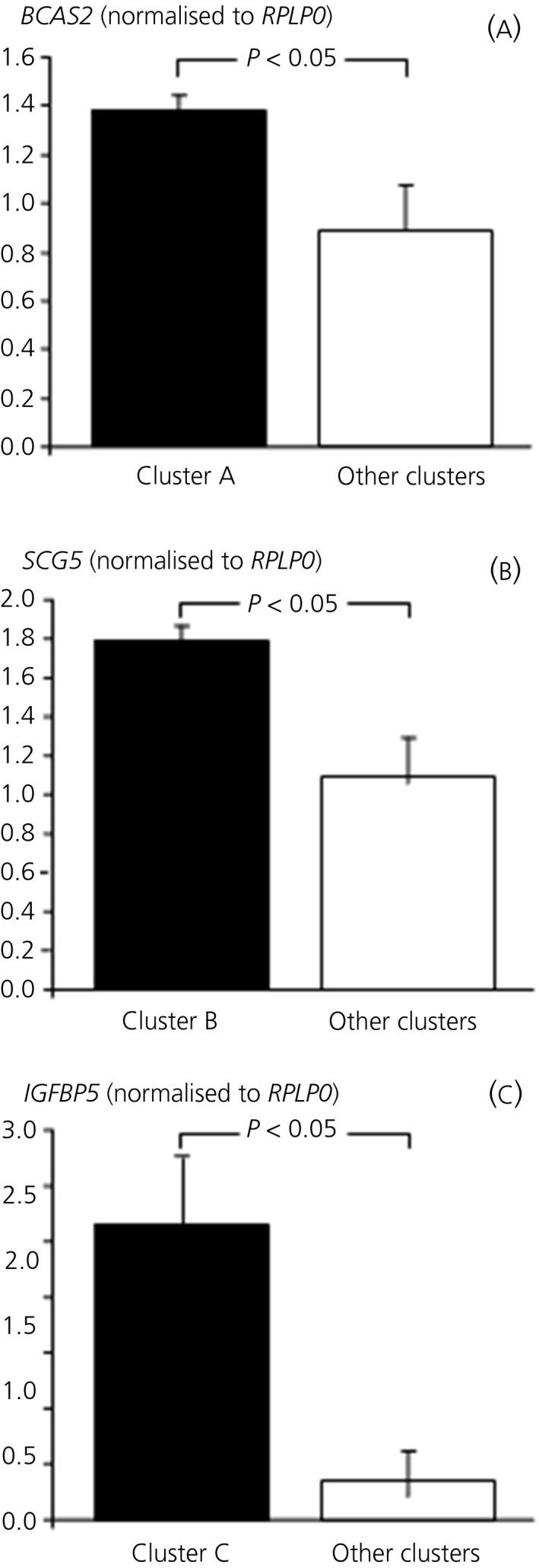
Quantitative expression of selected genes in different clusters. (A) Expression of BCAS2 in cluster A vs other clusters. (B) Expression of SCG5 in cluster B vs other clusters. (C) Expression of IGFBP5 in cluster C vs other clusters
4. DISCUSSION
ACTH‐secreting pituitary tumours are a rare and poorly understood disease. The studies performed so far in adenomatous specimens have indicated the involvement of individual genes (ie, mutations in USP8)3, 4 and the variable expression of factors that mediate sensitivity to glucocorticoids, such as HSP90, BRG‐1 and HSD11B2.5, 34, 35 Furthermore, cellular models derived from rat or dog tumoral corticotrophs called EGF36 and testicular orphan nuclear receptor 4 (TR4)37 into play. Studies in zebrafish also linked the development of pro‐opiomelanocortin (POMC)‐cell tumours to pituitary tumour‐transforming gene (PTTG) overexpression.38 Altogether, several mechanisms appear to be involved in corticotroph tumorigenesis, although a clear‐cut picture of the changes occurring in corticotroph tumours is as yet missing.
The present study had 2 main aims: identifying gene expression patterns characteristic to tumoral corticotrophs and evaluating differences in among corticotroph adenoma subgroups. Over recent decades, it has become increasingly evident that ACTH‐secreting adenomas differ considerably among themselves, in terms of secretory parameters and responses to medical therapy. Thus, studies investigating the variability of these tumours are necessary to identify the features associated with tumour subtypes and to aid in the development of targeted therapeutical approaches.
Heatmap profiling revealed the significant expression of 1259 of 29 285 probes in corticotroph adenoma specimens. Obviously, genes associated with the corticotroph phenotype (ie, POMC, TBX19) were among the highly expressed genes39, 40 Other genes associated with POMC‐processing (ie, proprotein convertase inhibitor [PCSKIN], secretogranin [SCG5], cathepsin L1 [CTSL1]), with drug action on corticotrophs (ie, retinoid X receptors [RXRB], GABA receptors [GABARAPL2, GABRB3]), or with steroid feedback (ie, 11β‐hydroxysteroid dehydrogenase [HSD11BIL]), were also significantly expressed. As regards the individual genes noted above, the expression of HDAC2, SMARCA4 (ie, formerly BRG‐1) TR4, FGFR4 and EGF or EGFR was not included among the significantly expressed genes in the corticotroph adenoma set; PPTG1‐interacting protein (PTTG11P) was significantly expressed, although PTTG itself was not. Likewise, several members of the heat shock protein family A (HSP70) (eg, HSPA8, HSPA9) were detected, although there were none related to heat shock protein alpha family (HSP90). Furthermore, given that the response to vasopressin, somatostatin and dopamine receptor agonists in Cushing's disease has been linked to specific receptor expression,8, 10, 11 we sought evidence for the significant expression of these receptors, however none proved significant.
Several genes detected at normal human pituitary expression profiling41 were also found in tumoral corticotrophs, such as PUM1 (pumilio homolog 1), TPT1 (tumour protein translationally controlled 1) and CDK2AP1 (cyclin‐dependent kinase 2‐associated protein 1), as were some genes detected in other pituitary tumours,42, 43, 44 such as AKT2 (v‐akt murine thymoma viral oncogene homolog 2), NOTCH3 (Notch homolog 3) and PITX1 (paired‐like homeodomain transcription factor 1). Our data confirmed some of the findings obtained in 7 corticotroph macroadenomas with the UniGEM microarray probe on some 7000 genes25 (ie, overexpression of HSPH1, SH3BGRL2 and RGS2, with the latter being of particular interest because it increases during corticotrophin‐releasing hormone incubation in murine tumoral corticotrophs).45 Moreover, we confirmed the results obtained in 12 corticotroph adenomas reporting the expression of several genes (eg, SEZ6L, CALY, CD200, TCF7L2, NISCH, RUNDC3A and NGFRAP1).46 We could also confirm increased expression of cold‐inducible RNA binding protein (CIRP), recently proven to stimulate murine corticotroph tumour growth47 and KRT8 (ie, keratin 8 or CAM 5.2), which, together with significant expression of LGALS3, represents a immunophenotype marker for corticotroph tumours.48, 49, 50 Furthermore, our results corroborate those obtained in 9 adenomas by RNA sequencing on 32 186 protein‐coding genes,51 which revealed the significant expression of a variety of genes, some with clear links to neuroendocrine or pituitary tumoral phenotype (eg, CGA, FILIP1, PITX1, IGFBP5) and others with as yet unknown links to corticotroph tumour development (eg, FBOXO31, FZD7, GPX3, MYH6). Our data also matched the results obtained in ectopic ACTH‐secreting tumours52 (ie, significant expression of MAX dimerisation protein 4 [MXD4], neugrin [NGRN] and SHC transforming protein 1 [SHC1]).
Most recent studies have revealed that up to 60% of human pituitary ACTH‐secreting adenomas may harbour somatic mutations in the ubiquitin‐specific protease 8 (ie, USP8) gene3, 4 and, indeed, 5 specimens in our series presented USP8 variants. The detected mutations were either missense or deletion variants and fell within the 14‐3‐3 binding motif of USP8 suggesting enhanced USP8 activity, as most frequently occurs.3, 4 Interestingly, the 2 specimens carrying the same variant clustered next to each other, indicating highly similar gene expression profiles, whereas the 3 remaining variants were associated with differing expression profiles and fell into separate clusters. It appears that, although variants appear to lead to a defect in USP8 activity, the resulting gene expression profile may diverge; indeed, catalytic activity of USP8 mutants reportedly differs.3
As regards enrichment analysis, pathways involved in protein biosynthesis, macromolecule metabolic processes and RNA processing appeared to be the most significant, in keeping with a neuroendocrine, secretory phenotype. Pathways associated with cell death and DNA repair ranked amongst the low enrichment scores, as expected given the benign nature of these tumours. Of note, there was little overlap with pathways involved in prolactinomas53 and nonsecreting pituitary adenomas.54
Lastly, differential expression analysis was able to identify genes uniquely expressed in each of the 3 clusters formed by unsupervised clustering analysis. Interestingly, although POMC is obviously significantly expressed in all specimens, it proved to be overexpressed together with the POMC‐processing co‐adjuvator SCG5 in specimens grouping in cluster B compared to those in clusters A and C. Increased expression of genes involved in other pituitary tumours or ACTH‐secreting neuroendocrine tumours, most notably NOTCH3, PITX1, LGALS3 and NGRN,38, 43, 44, 52 was also observed in cluster B. Furthermore, adenomas in this cluster overexpressed the retinoid X receptor beta gene (RXRB), thus confirming possibly greater sensitivity to RXR agonists30, 31 in selected adenomas. TIMP1 (tissue metallopeptidase inhibitor 1) was also overexpressed in cluster B adenomas, which is an interesting result given the recent reports on matrix metalloproteinase‐932 and TIMP1 itself33 in corticotroph adenomas.
Four adenomas presented a markedly different gene expression phenotype and clustered separately (cluster C). IGFBP5 was among the genes highly overexpressed in this cluster, which may prove to be of particular interest given its role in both carcinogenesis and normal cell growth.55 Indeed, IGFBP5 is known to be expressed in the pituitary itself, independently from growth hormone‐producing cells,56 and, furthermore, IGFBP5 expression and protein synthesis was increased in a subset of nonfunctioning pituitary adenomas.57 Lastly, overexpression of BCAS2 (breast carcinoma amplified sequence 2) and RPS6KA6 (ribosomal protein S6 kinase 6), both associated with the tumour suppressor p53,27, 28 as well as RBX1 (ringbox 1), a gene linked to cullin‐modulated cell cycle progression58, was detected in cluster A; p53 itself has been shown to accumulate in a subset of pituitary ACTH‐secreting lesions, in particular invasive adenomas.59 Accordingly, we observed that patients in cluster A presented the highest prevalence of macroadenomas and invasive tumours and both p53 and cullin‐RING pathways represent attractive therapeutic targets.58 A recent study aimed at identifying the genetic signature of invasive macroadenomas reported an increased expression of CCND2 (cyclin 2) and ZNF676 (zinc‐finger protein 676) in 3 invasive adenomas compared to non‐invasive corticotroph adenomas.46 Not surprisingly, neither gene was significantly expressed in our series, which comprised 12 invasive adenomas, because our study aim differed and required an unforced approach to gene expression analysis (ie, unsupervised clustering without a priori classification); additional studies into this as well as other group‐derived signatures will allow the above results to be extended and confirmed.
In conclusion, the present study describes the gene expression profile of human pituitary ACTH‐secreting adenomas and has identified genes associated with distinct expression clusters. Overall, it appears that these tumours present a neuroendocrine cell profile but, at the same time, there are clearly distinct gene expression patterns in individual subgroups. This evidence provides the basis for future studies into the molecular pathophysiology of these adenomas and, possibly, paves the way to target therapy.
CONFLICT OF INTERESTS
The authors declare that they have no conflicts of interest that could be perceived as prejudicing the impartiality of the research reported.
Supporting information
Cassarino MF, Ambrogio AG, Cassarino A, et al. Gene expression profiling in human corticotroph tumours reveals distinct, neuroendocrine profiles. J Neuroendocrinol. 2018;30:e12628 10.1111/jne.12628
Secure Array data link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=itsvwwkuzjyvpsj&acc=GSE93825
REFERENCES
- 1. Graversen D, Vestergaard P, Stochholm K, Gravholt CH, Jørgensen JOL. Mortality in Cushing's syndrome: a systematic review and meta‐analysis. Eur J Intern Med. 2012;23:278‐282. [DOI] [PubMed] [Google Scholar]
- 2. Theodoropoulou M, Arzberger T, Gruebler Y, et al. Expression of epidermal growth factor receptor in neoplastic pituitary cells: evidence for a role in corticotropinoma cells. J Endocrinol. 2004;183:385‐394. [DOI] [PubMed] [Google Scholar]
- 3. Reincke M, Sbiera S, Hayakawa A, et al. Mutations in the deubiquitinase gene USP8 cause Cushing's disease. Nat Genet. 2015;47:31‐38. [DOI] [PubMed] [Google Scholar]
- 4. Ma ZY, Song ZJ, Chen JH, et al. Recurrent gain‐of‐function USP8 mutations in Cushing's disease. Cell Res. 2015;25:306‐317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bilodeau S, Vallette‐Kasic S, Gauthier Y, et al. Role of Brg1 and HDAC2 in GR trans‐repression of the pituitary POMC gene and misexpression in Cushing disease. Genes Dev. 2006;20:2871‐2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pecori Giraldi F, Pagliardini L, Cassarino MF, Losa M, Lasio G, Cavagnini F. Responses to CRH and dexamethasone in a large series of human ACTH‐secreting pituitary adenomas in vitro reveal manifold corticotroph tumoural phenotypes. J Neuroendocrinol. 2011;23:1214‐1221. [DOI] [PubMed] [Google Scholar]
- 7. Suda T, Tozawa F, Yamada M, et al. Effects of corticotropin‐releasing hormone and dexamethasone on proopiomelanocortin messenger RNA levels in human corticotroph adenoma cells in vitro. J Clin Invest. 1988;82:110‐114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pecori Giraldi F, Marini E, Torchiana E, Mortini P, Dubini A, Cavagnini F. Corticotrophin‐releasing activity of desmopressin in Cushing's disease. Lack of correlation between in vivo and in vitro responsiveness. J Endocrinol. 2003;177:373‐379. [DOI] [PubMed] [Google Scholar]
- 9. Senovilla L, Núñez L, De Campos JM, et al. Multifunctional cells in human pituitary adenomas: implications for paradoxical secretion and tumorigenesis. J Clin Endocrinol Metab. 2004;89:4545‐4552. [DOI] [PubMed] [Google Scholar]
- 10. Hofland LJ, van de Hoek J, Feelders RA, et al. The multi‐ligand somatostatin analogue SOM230 inhibits ACTH secretion by cultured human corticotroph adenomas via somatostatin receptor type 5. Eur J Endocrinol. 2005;152:645‐654. [DOI] [PubMed] [Google Scholar]
- 11. Pivonello R, Ferone D, De Herder WW, et al. Dopamine receptor expression and function in corticotroph pituitary tumors. J Clin Endocrinol Metab. 2004;89:2452‐2462. [DOI] [PubMed] [Google Scholar]
- 12. Nakano‐Tateno T, Tateno T, Hlaing MM, et al. FGFR4 polymorphic variants modulate phenotypic features of Cushing disease. Mol Endocrinol. 2014;28:525‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perez‐Rivas LG, Theodoropoulou M, Ferraù F, et al. The gene of the ubiquitin‐specific protease 8 is frequently mutated in adenomas causing Cushing's disease. J Clin Endocrinol Metab. 2015;100:E997‐E1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ravo M, Mutarelli M, Ferraro L, et al. Quantitative expression profiling of highly degraded RNA from formalin‐fixed, paraffin‐embedded breast tumor biopsies by oligonucleotide microarrays. Lab Invest. 2008;88:430‐440. [DOI] [PubMed] [Google Scholar]
- 15. Raitoharju E, Seppäla I, Lyytikäinen LP, et al. A comparison of the accuracy of Illumina HumanHT‐12 v3 expression BeadChip and TaqMan qRT‐PCR gene expression results in patient samples from the Tampere Vascular study. Atherosclerosis. 2013;226:149‐152. [DOI] [PubMed] [Google Scholar]
- 16. Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354:246‐2472. [DOI] [PubMed] [Google Scholar]
- 17. Chu TT, Fink MY, Mong JA, Auger AP, Ge Y, Sealfon SC. Effective use of microarrays in neuroendocrine research. J Neuroendocrinol. 2007;19:145‐161. [DOI] [PubMed] [Google Scholar]
- 18. White NJ, Contaifer D Jr, Martin EJ, et al. Early hemostatic responses to trauma identified with hierarchical clustering analysis. J Thromb Haemost. 2015;13:978‐988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li W. Volcano plots in analyzing differential expressions with mRNA microarrays. J Bioinform Comput Biol. 2012;10:1231003. [DOI] [PubMed] [Google Scholar]
- 20. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44‐57. [DOI] [PubMed] [Google Scholar]
- 21. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498‐2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug‐in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091‐1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module‐centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183‐R199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morris DG, Musat M, Czirják S, et al. Differential gene expression in pituitary adenomas by oligonucleotide array analysis. Eur J Endocrinol. 2005;153:143‐151. [DOI] [PubMed] [Google Scholar]
- 25. Evans CO, Young AN, Brown MR, et al. Novel patterns of gene expression in pituitary adenomas identified by complementary deoxyribunucleic acid microarrays and quantitative reverse transcription‐polymerase chain reaction. J Clin Endocrinol Metab. 2001;86:3097‐3107. [DOI] [PubMed] [Google Scholar]
- 26. Hu J, Song H, Wang X, et al. Gene expression profiling in human null cell pituitary adenoma tissue. Pituitary. 2007;10:47‐52. [DOI] [PubMed] [Google Scholar]
- 27. Kuo PC, Tsao YP, Chang HW, et al. Breast cancer amplified sequence 2, a novel negative regulator of the p53 tumor suppressor. Cancer Res. 2009;69:8877‐8885. [DOI] [PubMed] [Google Scholar]
- 28. Lleonart ME, Vidal F, Gallardo D, et al. New p53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol Rep. 2006;16:603‐608. [DOI] [PubMed] [Google Scholar]
- 29. Westphal CH, Muller L, Zhou A, et al. The neuroendocrine protein 7B2 is required for peptide hormone processing in vivo and provides a novel mechanism for pituitary Cushing's disease. Cell. 1999;96:689‐700. [DOI] [PubMed] [Google Scholar]
- 30. Saito‐Hakoda A, Uruno A, Yokoyama A, et al. Effects of RXR agonists on cell proliferation/apoptosis and ACTH secretion/Pomc expression. PLoS ONE. 2015;10:e0141960‐e0141981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pecori Giraldi F, Ambrogio AG, Andrioli M, et al. Potential role for retinoic acid in patients with Cushing's disease. J Clin Endocrinol Metab. 2012;97:3577‐3583. [DOI] [PubMed] [Google Scholar]
- 32. Liu X, Feng M, Zhang Y, et al. Expression of matrix metalloproteinase‐9, pituitary tumor transforming gene, high mobility group A 2, and Ki‐67 in adrenocorticotropic hormone‐secreting pituitary tumors and their association with tumor recurrence. World Neurosurg. 2018;113:e213‐e221. [DOI] [PubMed] [Google Scholar]
- 33. Tofrizal A, Fuhiwara K, Azuma M, et al. Tissue inhibitors of metalloproteinase‐expressing cells in human anterior pituitary and pituitary adenoma. Med Mol Morphol. 2017;50:145‐154. [DOI] [PubMed] [Google Scholar]
- 34. Riebold M, Kozany C, Freiburger L, et al. A C‐terminal HSP90 inhibitor restores glucocorticoid sensitivity and relieves a mouse allograft model of Cushing disease. Nat Med. 2015;21:276‐280. [DOI] [PubMed] [Google Scholar]
- 35. Korbonits M, Bujalska IK, Shimojo M, et al. Expression of the 11b‐hydroxysteroid dehydrogenase isoenzymes in the human pituitary: induction of the type 2 enzyme in corticotropinomas and other pituitary tumors. J Clin Endocrinol Metab. 2001;86:2728‐2733. [DOI] [PubMed] [Google Scholar]
- 36. Fukuoka H, Cooper O, Ben‐Shlomo A, et al. EGFR as a therapeutic target for human, canine, and mouse ACTH‐secreting pituitary adenomas. J Clin Invest. 2011;121:4712‐4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du L, Bergsneider M, Mirsadraei L, et al. Evidence for orphan nuclear receptor TR4 in the etiology of Cushing disease. Proc Natl Acad Sci USA. 2013;21:8555‐8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu NA, Jiang H, Ben‐Shlomo A, et al. Targeting zebrafish and murine pituitary corticotroph tumors with a cyclin‐dependent kinase (CDK) inhibitor. Proc Natl Acad Sci USA. 2011;108:8414‐8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pascual‐Le Tallec L, Dulmet E, Bertagna X, De Keyzer Y. Identification of genes associated with the corticotroph phenotype in bronchial tumors. J Clin Endocrinol Metab. 2002;87:5015‐5022. [DOI] [PubMed] [Google Scholar]
- 40. Tateno T, Izumiyama H, Doi M, et al. Differential gene expression in ACTH‐secreting and non‐functioning pituitary tumors. Eur J Endocrinol. 2007;157:717‐724. [DOI] [PubMed] [Google Scholar]
- 41. Tanaka S, Tatsumi K, Okubo K, et al. Expression profile of active genes in the human pituitary gland. J Mol Endocrinol. 2002;28:33‐44. [DOI] [PubMed] [Google Scholar]
- 42. Musat M, Korbonits M, Kola B, et al. Enhanced protein kinase B/Akt signalling in pituitary tumors. Endocr Relat Cancer. 2005;12:423‐433. [DOI] [PubMed] [Google Scholar]
- 43. Evans CO, Moreno CS, Zhan X, et al. Molecular pathogenesis of human prolactinomas identified by gene expression profiling, RT‐qPCR, and proteomic analysis. Pituitary. 2008;11:231‐245. [DOI] [PubMed] [Google Scholar]
- 44. Moreno CS, Evans CO, Zhan X, Okor M, Desiderio DM, Oyesiku NM. Novel molecular signaling and classification of human clinically nonfunctional pituitary adenomas identified by gene expression profiling and proteomic analyses. Cancer Res. 2005;65:10214‐10222. [DOI] [PubMed] [Google Scholar]
- 45. Peeters PJ, Gohlmann HW, Van den Wyngaert I, et al. Transcriptional response to corticotropin‐releasing factor in AtT‐20 cells. Mol Pharmacol. 2004;66:1083‐1092. [DOI] [PubMed] [Google Scholar]
- 46. de Araujo LJT, Lerario AM, de Castro M, et al. Transcriptome analysis showed a differential signature between invasive and non‐invasive corticotrophinomas. Front Endocrinol. 2017;8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jian FF, Chen YF, Ning G, et al. Cold inducible RNA binding protein upregulation in pituitary corticotroph adenoma induces corticotroph cell proliferation via Erk signaling pathway. Oncotarget. 2016;7:9175‐9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jin L, Riss D, Ruebel KH, et al. Galectin‐3 expression in functioning and silent ACTH‐producing adenomas. Endocr Pathol. 2005;16:107‐114. [DOI] [PubMed] [Google Scholar]
- 49. Ruebel KH, Leontovich AA, Jin L, et al. Patterns of gene expression profiles in pituitary carcinomas and adenomas analyzed by high‐density oligonucleotide arrays, reverse transcriptase‐quantitative PCR, and protein expression. Endocrine. 2006;29:435‐444. [DOI] [PubMed] [Google Scholar]
- 50. Höfler H, Denk H, Walter GF. Immunohistochemical demonstration of cytokeratins in endocrine cells of the human pituitary gland and in pituitary adenomas. Virchows Arch A Pathol Anat Histopathol. 1984;404:359‐368. [DOI] [PubMed] [Google Scholar]
- 51. Wang R, Yang Y, Sheng M, et al. Phenotype‐genotype association analysis of ACTH‐secreting pituitary adenoma and its molecular link to patient osteoporosis. Int J Mol Sci. 2016;17:E1654‐E1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bi YF, Liu RX, Ye L, et al. Gene expression profiles of thymic neuroendocrine tumors (carcinoids) with ectopic ACTH syndrome reveal novel molecular mechanism. Endocr Relat Cancer. 2009;16:1273‐1282. [DOI] [PubMed] [Google Scholar]
- 53. Jiang Z, Gui S, Zhang Y. Analysis of differential gene expression by fiber‐optic BeadArray and pathway in prolactinomas. Endocrine. 2010;35:360‐368. [DOI] [PubMed] [Google Scholar]
- 54. Jiang Z, Gui S, Zhang Y. Analysis of differential gene expression by bead‐based fiber‐optic array in nonfunctioning pituitary adenomas. Horm Metab Res. 2011;43:325‐330. [DOI] [PubMed] [Google Scholar]
- 55. Güllü G, Karabulut S, Akkiprik M. Functional roles and clinical values of insulin‐like growth factor‐binding protein‐5 in different types of cancers. Chin J Cancer. 2012;31:266‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bach MA, Bondy CA. Anatomy of the pituitary insulin‐like growth factor system. Endocrinology. 1992;131:2588‐2594. [DOI] [PubMed] [Google Scholar]
- 57. Galland F, Lacroix L, Saulnier P, et al. Differential gene expression profiles of invasive and non‐invasive non‐functioning pituitary adenomas based on microarray analysis. Endocr Relat Cancer. 2010;17:361‐371. [DOI] [PubMed] [Google Scholar]
- 58. Shafique S, Ali W, Kanwal S, Rashid S. Structural basis for cullins and RING component inhibition: Targeting E3 ubiquitin pathway conductors for cancer therapeutics. Int J Biol Macromol. 2018;106:532‐543. [DOI] [PubMed] [Google Scholar]
- 59. Buckley N, Bates AS, Broome JC, et al. p53 Protein accumulates in Cushing's adenomas and invasive non‐functional adenomas. J Clin Endocrinol Metab. 1994;79:1513‐1516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


