Abstract
The spatiotemporal dynamics of proteins or organelles plays a vital role in controlling diverse cellular processes. However, acute control of activity at distinct locations within a cell is challenging. A versatile multidirectional activity control (MAC) approach is presented, which employs a photoactivatable system that may be dimerized upon chemical inducement. The system comprises second‐generation SLF*‐TMP (S*T) and photocaged NvocTMP‐Cl dimerizers; where, SLF*‐TMP features a synthetic ligand of the FKBP(F36V) binding protein, Nvoc is a caging group, and TMP is the antibiotic trimethoprim. Two MAC strategies are demonstrated to spatiotemporally control cellular signaling and intracellular cargo transport. The novel platform enables tunable, reversible, and rapid control of activity at multiple compartments in living cells.
Keywords: chemo-optogenetics, intracellular transport, multidirectional activity, photoactivated, signaling
The spatiotemporal organization of cellular processes is often controlled by the subcellular distribution of molecules or organelles. As individual proteins can perform different functions depending on their local environment, or when tethered to different effectors, subcellular localization is essential for the functional diversity of proteins.1 A growing number of so‐called “moonlighting proteins” have been identified, which play distinct roles when located at different subcellular regions.2 For instance, at the plasma membrane, the Wnt‐signaling protein β‐catenin controls cell–cell adhesion, while it can also regulate gene transcription if it is localized in the nucleus.3 Aberrant localizations of proteins has been implicated in pathogenesis of diverse human diseases and the modulation of subcellular protein localization has emerged as a target for therapeutic intervention.4 Recent evidence suggests a broad role of organelle distribution in the spatial organization of several cellular processes, including cell signaling, cell polarization, and neurite outgrowth.5 For example, the bidirectional transport along axonal microtubules plays a central role in the proper subcellular distribution of cellular cargos and its misregulation is thought to play an important role in neurodegenerative diseases.6
To unravel the complex and dynamic nature of such systems that orchestrate cell function in space and time, acute perturbations with high spatial and temporal resolution are required. Conventional genetic manipulations, such as protein overexpression or siRNA‐mediated knockdown, are limited as they act very slowly on the entire cell. Chemically induced dimerization (CID) is a powerful tool to modulate protein function by bringing two proteins in close proximity to control their function and to perturb associated cellular processes.7 Optogenetic and optochemical approaches, such as light‐induced dimerization, confer superior spatiotemporal control to such perturbations.8
In the past, such approaches were successfully applied to perform a single layer of activity control, such as turning on/off by targeting a protein to a specific cellular compartment to study relatively simple cellular processes (Figure 1 A). However, the analysis of more complex processes that involve cycling, trafficking or shuttling of signal molecules between different cell compartments, such as growth factor or Wnt signaling, could benefit from multiple layers of activity control; for example, by subsequent targeting of proteins to distinct locations within a single cell (Figure 1 B).9 Herein, we report a versatile photoactivatable, dual chemically induced dimerization (pdCID) system to enable acute, switchable, and multidirectional activity control (MAC) in living cells.
Figure 1.
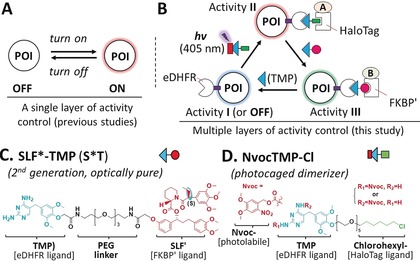
A) Previously developed systems only enable a single layer of activity control of cellular processes. B) MAC of protein function and complex cellular processes. C) The newly introduced second‐generation SLF*‐TMP dimerizer and D) the photocaged NvocTMP‐Cl dimerizer used in the pdCID system.
This system combines either the newly introduced second‐generation SLF*‐TMP (S*T) dimerizer or the first‐generation SLF′‐TMP (ST)10 with the NvocTMP‐Cl photocaged dimerizer,11 which were recently established in our lab (Figure 1; Supporting Information, Figure S1). NvocTMP‐Cl consists of caged trimethoprim (TMP), a HaloTag ligand (chlorohexyl group) and a polyethylene glycol (PEG) linker. TMP selectively binds to Escherichia coli dihydrofolate reductase (eDHFR) with a K d of 1 nm and the chlorohexyl moiety can form a covalent bond with a bacterial alkyl dehalogenase mutant (HaloTag). NvocTMP‐Cl can covalently pre‐localize to a protein fused with a HaloTag. The dimerization between HaloTag and eDHFR does not occur until the nitroveratroyloxycarbonyl (Nvoc) caging group is removed by illumination using a 405 nm light pulse (a process called photolysis; Supporting Information, Figure S1). On the other hand, SLF*‐TMP and SLF′‐TMP feature a synthetic ligand of FKBP(F36V) (SLF′) and a TMP moiety. SLF*‐TMP (S*T) is an optically pure diastereomer while SLF′‐TMP (ST) is a diastereomeric mixture. SLF*/SLF′ binds with high affinity (subnanomolar) to the FKBP(F36V) mutant (FKBP′) with a 1000‐fold selectivity over wild‐type FKBP. Both dimerizers induce dimerization between FKBP′‐ and eDHFR‐fused proteins with nanomolar affinity (Supporting Information, Figure S1E,F).10 The NvocTMP‐Cl and S*T/ST‐induced dimerization is stable and essentially irreversible even after wash‐out, but can be disrupted by the competitor TMP to make both systems reversible (Supporting Information, Figures S1–S3). The NvocTMP‐Cl system is tunable with doses of illumination (Supporting Information, Figure S4) and the induced dimerization is very rapid (t 1/2=0.55–1.69 s; Supporting Information, Figure S5). Among many available dimerizer systems, the combination of two is challenging and problematic, because four dimerization modules are required and two individual dimerization processes are difficult to connect with each other. Herein, because the NvocTMP‐Cl system shares a common eDHFR dimerizing module with the S*T (or ST) system, it is possible to combine these modular systems to enable multidirectional control over protein or organelle activity in a single cell.
A major challenge in all dimerization systems is basal activity. Even in the absence of dimerization, the protein of interest (POI) can still reach the site of activation by diffusion through the cytosol, resulting in background activity.7b–7d A previous approach to reduce background activity utilized a Golgi‐binding fusion protein to sequester the POI away from its target localization at the plasma membrane. Upon CID, the POI was translocated from the Golgi to the plasma membrane.12 However, binding of the POI to Golgi was difficult to control, as there was competition between binding to the Golgi and to the plasma membrane. We sought to solve this issue with a novel parallel MAC approach (Figure 2 A; Supporting Information, Movie S1). Because the ST‐induced dimerization is stable and tunable (Supporting Information, Figures S2 and S6), ST could be used to tune the POI level in the cytosol. After wash‐out of ST, photoactivation of NvocTMP‐Cl induces dimerization. Since these two events are independent of each other, we termed this approach parallel MAC.
Figure 2.
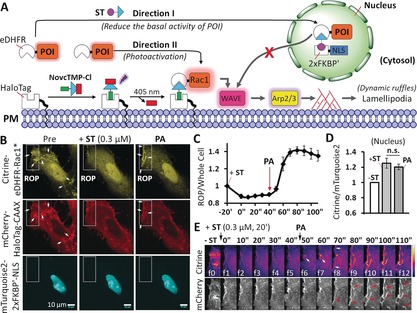
Parallel MAC of Rac1 in live cells. A) Representation of the principle of the parallel MAC approach. B) Confocal images of a HeLa cell co‐expressing Citrine‐eDHFR‐Rac1* (cytosol), mCherry‐HaloTag‐CAAX (plasma membrane), and mTurquoise2‐2×FKBP′‐NLS (nucleus). The rectangular area indicates the region of photoactivation (ROP) by 405 nm light. Arrows indicate ruffles. Scale bar=10 μm. C) Normalized fluorescence intensity ratio of Citrine‐eDHFR‐Rac1* at the ROP versus the entire cell (n=4 cells). The Citrine‐eDHFR‐Rac1* level in the cytosol decreased after adding 0.3 μm ST, and was then recruited to the ROP upon photoactivation (PA). D) Normalized fluorescence intensity ratio of Citrine‐eDHFR‐Rac1* versus mTurquoise2‐2×FKBP′‐NLS in the nucleus before (−ST) and after the addition of 0.3 μm ST for 20 min (+ST), and after PA. Not significant (n.s.; p=0.46) as determined from a paired Student's t‐test. E) Time series of images of the ROP shows the recruitment of Citrine‐eDHFR‐Rac1* (upper panel, intensity coded) and ruffle formation (arrows) at the plasma membrane (lower panel, mCherry‐HaloTag‐CAAX). For (C) and (D), mean values and the standard error of the mean (SEM) are shown (Supporting Information, Movie S1).
To this end, three constructs were co‐expressed in HeLa cells: 1) cytosolic Citrine‐eDHFR‐Rac1*, 2) mCherry‐HaloTag‐CAAX at the plasma membrane, and 3) mTurquoise2‐2×FKBP′‐NLS (nucleus localization signal) in the nucleus (Figures 2 A,B). Herein Rac1* (that is, Rac1Q61LΔCAAX) stands for a constitutively active Q61L mutant that lacks plasma membrane localization. Targeting of Rac1* to the plasma membrane will activate WAVE, and the Arp2/3 complex, which will stimulate actin polymerization near the plasma membrane, resulting in cell protrusion (Figure 2 A). In HeLa cells, those localized protrusions mainly take the shape of dynamic ruffles at the plasma membrane.13 Citrine‐eDHFR‐Rac1* can freely diffuse in the cytosol, which can already cause significant membrane ruffling prior to light‐induced dimerization (Figure 2 B left panel, indicated by arrows). To control the amount of Rac1* in the cytosol, we added the first dimerizer ST to recruit cytosolic Rac1* into the nucleus. Titration of ST revealed a dose‐dependent decrease of the cytosol/nucleus ratio with a half maximal effective concentration (EC50) of 0.53±0.04 μm, showing that cytosolic Rac1* concentration can be fine‐tuned by the addition of ST (Supporting Information, Figure S6). We found that partial sequestration of Rac1* to the nucleus by addition of 0.3 μm ST is sufficient to minimize basal Rac1*‐induced ruffle formation (Figures 2 B–E). Following wash‐out of ST, illumination at the edge of the cell rapidly targeted Rac1* to the region of photoactivation (ROP) at the plasma membrane (Figures 2 B,E) without alteration of Rac1* in the nucleus (Figure 2 D). This suggests that ST promotes stable dimerization to sequester Rac1* in the nucleus, which is not reversible without the competitor (Supporting Information, Figure S2). Light‐induced plasma‐membrane‐targeting leads to significant formation of ruffles (observed with the plasma membrane marker mCherry‐HaloTag‐CAAX) only at the ROP (Figure 2 B, right panel; Figure 2 E). Therefore, the parallel MAC approach, using the pdCID system, is well‐suited to tune background activity before light‐induced control of protein function in cells.
Another challenge in chemical genetic and optogenetic systems is to achieve multiple layers of functional control in a single cell. To address this problem, we developed the so‐called competitive MAC approach to enable multilayered control over protein or organelle function. In this approach, S*T can competitively inhibit the NvocTMP‐Cl‐induced dimerization of eDHFR and HaloTag while triggering the S*T‐induced dimerization of eDHFR and FKBP′. To enhance the efficiency of eDHFR/FKBP′ dimerization, we introduced the second‐generation S*T dimerizer that is optically pure (Figure 1; Supporting Information, Scheme S1). Compared to the first‐generation ST (EC50=0.53±0.04 μm; Supporting Information, Figure S6), which is a diastereomeric mixture, S*T not only shows eightfold higher efficacy (EC50=0.065±0.005 μm), but also displays a higher extent of protein dimerization in cells (Supporting Information, Figure S7). First, we demonstrated that S*T can disrupt the dimerization between eDHFR and HaloTag that was initially induced by TMP‐Cl (the uncaged form of NvocTMP‐Cl) with an EC50 of 3.2±0.2 μm (Supporting Information, Figure S8), suggesting that the competitive MAC approach is feasible.
As a proof of principle, we used the competitive MAC approach to cycle the positioning of proteins among multiple sites within a cell. Recent studies show that, in addition to the cytosol and plasma membrane, Rac1 also localizes in the nucleus and regulates nuclear membrane organization and morphology. The nucleocytoplasmic shuttling of Rac1 plays an important role in tumor invasion.14 Using the pdCID system, we manipulated the shuttling of Rac1 among the cytosol, plasma membrane, and nucleus (Supporting Information, Figure S9, Movie S2). We also performed multiple cycles of Rac1 shuttling among the cytosol, mitochondria, and nucleus (Supporting Information, Figure S10).
We further used the competitive MAC approach to control bidirectional transport of organelles by targeting two microtubule motors with opposite directionality (Figure 3 A). To this end, three constructs, PEX3‐mCherry‐eDHFR that anchors at the peroxisome through the PEX3 peptide motif, KIF5BN‐Citrine‐HaloTag that contains the N‐terminal motor domain (1–560) of the kinesin KIF5B (that is, KIF5BN), and BicD2N‐mTurquoise2‐2×FKBP′ that contains the N‐terminal dynein binding domain (1–594) of the dynein adaptor protein Bicaudal D2 (that is, BicD2N), were co‐expressed in HeLa cells. Recruitment of kinesin (KIF5BN) or the dynein complex (via BicD2N) to cargos induces microtubule plus‐end‐or minus‐end‐directed transport to the cell periphery or to the cell body, respectively. NvocTMP‐Cl was pre‐bound to KIF5BN via the HaloTag. Photoactivation (PA) led to the recruitment of KIF5BN to peroxisomes (Figures 3 B,C), which induced their anterograde transport to the cell periphery (Figure 3 B; Pre, PA 2′, and PA 5′). Subsequently, addition of S*T led to the displacement of KIF5BN and recruitment of BicD2N to peroxisomes (Figure 3 B; S*T 80′′; Figure 3 C), which stimulated retrograde transport of peroxisomes to the cell center (Figure 3 B; S*T 9′; Figures 3 D,E; Supporting Information, Movie S3). This bidirectional peroxisome transport was readily visualized within a subcellular region. Upon PA, dispersed peroxisome vesicles moved to the cell periphery, where they formed a cluster. Addition of S*T triggered the retrograde transport of peroxisomes toward the cell body (Figure 3 F; Supporting Information, Figure S11, Movie S4). Finally, TMP dissociated BicD2N from peroxisomes (cyan channels of Figure 3 B; TMP 80′′ TMP 3′; Figure 3 D) and disrupted the directional movement of peroxisomes (red channels of Figure 3 B; TMP 80′′ TMP 3′; Figure 3 E). By swapping KIF5BN and BicD2N, BicD2N was first recruited to cargos to induce retrograde motility, followed by replacing BicD2N with KIF5BN to trigger anterograde transport (Supporting Information, Figure S12, Movie S5). Hence, the competitive MAC approach can be flexibly implemented to control the direction of cargo transport.
Figure 3.
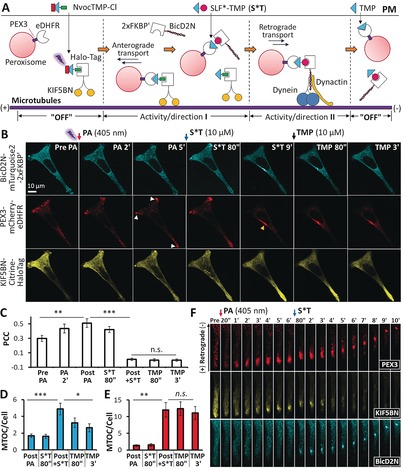
Competitive MAC of peroxisome transport. A) Representation of the principle of competitive MAC. B) Representative confocal images. Cherry channel: the arrows indicate that peroxisomes cluster at the cell periphery; the arrow head indicates that peroxisomes accumulate at the microtubule organization center (MTOC) at the cell center. C) Pearson's correlation coefficient (PCC) analysis of the colocalization between KIF5BN‐Citrine‐eDHFR and PEX3‐mCherry‐HaloTag showed that KIF5BN is recruited to peroxisomes after PA and then dissociates from peroxisomes after adding S*T. D) The fluorescence intensity ratio of BicD2N at the MTOC versus the entire cell increases after adding S*T and decreases after adding TMP. E) The fluorescence intensity ratio of PEX3 (peroxisome marker) at the MTOC versus the entire cell increases after S*T, but does not change significantly after adding TMP for 3 min. Scale bar=25 μm. For (C), (D), and (E), mean values and SEM are shown; data were based on n=6 cells from three independent experiments; p values are determined by a paired Student's t‐test (***: p<0.001; **: p<0.01; *: p<0.05; n.s.: not significant); “Post PA” indicates 5–10 min after PA and just before adding S*T, while “Post+S*T” means 5–10 min after the addition of S*T and just before adding TMP. F) Time series of an enlarged region within the cell as shown in Figure S11 (Supporting Information; see also Movies S3 and S4).
Most cargos simultaneously bind to kinesin and cytoplasmic dynein motors, which move in opposite directions along microtubules. A model of a stochastic “tug‐of‐war” that describes the mechanical competition between antagonistic motors bound to the same cargo has been proposed.15 Small changes in the number of active motors lead to significant changes in the net direction of cargo motility.16 In our study, the extra force generated by the recruitment of additional kinesin or dynein motors outcompetes the force generated by the endogenous motors, thereby enforcing cargo transport toward the cell periphery or toward the cell center (Supporting Information, Figure S13). Our competitive MAC experiments provide direct experimental support for the “tug‐of‐war” model, regardless of the manner in which dynein and kinesin motors coordinate with each other at the molecular level.
Herein, we have shown that the pdCID system can be flexibly implemented in distinct perturbation strategies to achieve multilayered activity control in a single cell. Another advantage of the pdCID system is its selectivity and full bioorthogonality. NvocTMP‐Cl and ST enable the combination of rapid, light‐induced (typically t 1/2<1 s), and chemically induced (t 1/2<1 min) dimerization, respectively. Simultaneous combination of all these desirable features has not been achieved in other CID systems.
In summary, using the pdCID system, we developed a multidirectional activity control (MAC) strategy. The parallel MAC approach was used to address the inherent problem of undesirable background activity in chemical/optogenetic systems through fine‐tuning of the cytosolic level of Rac1. The competitive MAC approach enables reversible, and multiple layers of functional control, which is exemplified by multidirectional control of the positioning of organelles and proteins in a single cell. We envision that these versatile MAC strategies open up new avenues for spatiotemporal control of proteins or organelles, which cannot be addressed with existing techniques. These new tools are particularly useful to study how protein or organelle function is modulated by their local, subcellular environment, and, conversely, how proteins or organelles reciprocally affect their local environment and eventually cellular function. The MAC approach could also be used to emulate or interfere with disease conditions involving protein/organelle positioning to study pathogenic mechanisms, and ultimately aid the development of therapeutic interventions.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Acknowledgements
We thank Sven Müller for technical microscopy support and Hui Ma for helpful discussions. This work was supported by the Deutsche Forschungsgemeinschaft, DFG (grant No.: SPP 1623), Behrens Weise Stiftung, European Research Council, ERC (ChemBioAP), and the Knut and Alice Wallenberg Foundation (to Y.‐W.W.). L.D. is supported by a DFG Heisenberg Stipend.
X. Chen, M. Venkatachalapathy, L. Dehmelt, Y.-W. Wu, Angew. Chem. Int. Ed. 2018, 57, 11993.
References
- 1. Butler G. S., Overall C. M., Nat. Rev. Drug Discovery 2009, 8, 935–948. [DOI] [PubMed] [Google Scholar]
- 2. Jeffery C. J., Mol. Biosyst. 2009, 5, 345–350. [DOI] [PubMed] [Google Scholar]
- 3. MacDonald B. T., Tamai K., He X., Dev. Cell 2009, 17, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hung M. C., Link W., J. Cell Sci. 2011, 124, 3381–3392. [DOI] [PubMed] [Google Scholar]
- 5.
- 5a. van Bergeijk P., Hoogenraad C. C., Kapitein L. C., Trends Cell Biol. 2016, 26, 121–134; [DOI] [PubMed] [Google Scholar]
- 5b. van Bergeijk P., Adrian M., Hoogenraad C. C., Kapitein L. C., Nature 2015, 518, 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Hancock W. O., Nat. Rev. Mol. Cell Biol. 2014, 15, 615–628; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b. De Vos K. J., Grierson A. J., Ackerley S., Miller C. C., Annu. Rev. Neurosci. 2008, 31, 151–173. [DOI] [PubMed] [Google Scholar]
- 7.
- 7a. Fegan A., White B., Carlson J. C. T., Wagner C. R., Chem. Rev. 2010, 110, 3315–3336; [DOI] [PubMed] [Google Scholar]
- 7b. Voss S., Klewer L., Wu Y. W., Curr. Opin. Chem. Biol. 2015, 28, 194–201; [DOI] [PubMed] [Google Scholar]
- 7c. DeRose R., Miyamoto T., Inoue T., Pflug. Arch. Eur. J. Phy. 2013, 465, 409–417; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7d. Putyrski M., Schultz C., FEBS Lett. 2012, 586, 2097–2105. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Tischer D., Weiner O. D., Nat. Rev. Mol. Cell Biol. 2014, 15, 551–558; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Weitzman M., Hahn K. M., Curr. Opin. Cell Biol. 2014, 30, 112–120; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8c. Chen X., Wu Y. W., Angew. Chem. Int. Ed. 2018, 57, 6796–6799; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 6912–6915; [Google Scholar]
- 8d. Ankenbruck N., Courtney T., Naro Y., Deiters A., Angew. Chem. Int. Ed. 2018, 57, 2768–2798; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 2816–2848. [Google Scholar]
- 9.
- 9a. Villaseñor R., Kalaidzidis Y., Zerial M., Curr. Opin. Cell Biol. 2016, 39, 53–60; [DOI] [PubMed] [Google Scholar]
- 9b. Schmick M., Vartak N., Papke B., Kovacevic M., Truxius D. C., Rossmannek L., Bastiaens P. I. H., Cell 2014, 157, 459–471; [DOI] [PubMed] [Google Scholar]
- 9c. Niehrs C., Nat. Rev. Mol. Cell Biol. 2012, 13, 767–779. [DOI] [PubMed] [Google Scholar]
- 10. Liu P., Calderon A., Konstantinidis G., Hou J., Voss S., Chen X., Li F., Banerjee S., Hoffmann J. E., Theiss C., Dehmelt L., Wu Y. W., Angew. Chem. Int. Ed. 2014, 53, 10049–10055; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 10213–10219. [Google Scholar]
- 11. Chen X., Venkatachalapathy M., Kamps D., Weigel S., Kumar R., Orlich M., Garrecht R., Hirtz M., Niemeyer C. M., Wu Y.-W., Dehmelt L., Angew. Chem. Int. Ed. 2017, 56, 5916–5920; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 6010–6014. [Google Scholar]
- 12. Phua S. C., Pohlmeyer C., Inoue T., ACS Chem. Biol. 2012, 7, 1950–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu Y. I., Frey D., Lungu O. I., Jaehrig A., Schlichting I., Kuhlman B., Hahn K. M., Nature 2009, 461, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Navarro-Lérida I., Pellinen T., Sanchez S. A., Guadamillas M. C., Wang Y. H., Mirtti T., Calvo E., Del Pozo M. A., Dev. Cell 2015, 32, 318–334. [DOI] [PubMed] [Google Scholar]
- 15. Müller M. J. I., Klumpp S., Lipowsky R., Proc. Natl. Acad. Sci. USA 2008, 105, 4609–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hendricks A. G., Perlson E., Ross J. L., Schroeder, 3rd H. W., Tokito M., Holzbaur E. L., Curr. Biol. 2010, 20, 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary


