Abstract
We present a catalog of common and well‐documented (CWD) alleles of the German population for the six HLA loci A, B, C, DRB1, DQB1, and DPB1. This study is based on a sample of over 5 million volunteer adult hematopoietic stem cell donors from the 26 German donor centers. To establish the catalog, allele and haplotype frequencies were estimated with a validated implementation of the expectation‐maximization algorithm. CWD criteria similar to existing CWD catalogs were applied in order to be able to put our findings into the context of relevant existing references. Overall, 2155 HLA‐A, ‐B, ‐C, ‐DRB1, ‐DQB1, and ‐DPB1 alleles were identified as CWD in the German donor population representing about 20% of the HLA alleles at two‐field resolution in the IPD‐IMGT/HLA Database release v3.25.0 from July 2016 for these six loci. We found a substantial concordance of CWD alleles between the three catalogs and showed the contribution of the German donor population to the CWD alleles domain. In conclusion, the definition of CWD criteria that allow interoperability, scalability, and flexibility will be crucial for the development of a worldwide CWD catalog.
Keywords: allele frequency, common and well‐documented allele catalog, haplotype frequency estimation, hematopoietic stem cell donor, HLA
1. INTRODUCTION
Many genes of the human major histocompatibility complex are well studied and show a high degree of polymorphism. It is also known that the distribution of the HLA alleles within and between populations is not equal. Therefore, the exact knowledge of a population's HLA composition is of great interest for a wide range of applications and is a long‐standing subject of research. One of the main driving forces behind this research is to understand and to minimize the transplant barrier between genetically different individuals.1, 2, 3, 4, 5, 6, 7, 8
The growing number of alleles and the subsequent problems in excluding ambiguous allele combinations motivated the American Society for Histocompatibility and Immunogenetics (ASHI) in 2007 to analyze the genetic HLA diversity of various populations. The aim was to establish criteria for alleles that need to be resolved when reporting results for external proficiency testing.9 The resulting catalog of common and well‐documented (CWD) alleles soon became an important tool even for applications outside its initial scope. This fact, together with scientific and technological progress, led to an updated and expanded version 2.0.0 of the CWD catalog in 2012,10 henceforth named ASHI CWD. Other groups applied the concept for individual populations,11, 12, 13 and only recently the European Federation for Immunogenetics (EFI) published a CWD catalog for Europe,14 designated as EFI CWD hereafter.
For this study, we examined a sample of over 5 million individuals from the German population for the six loci HLA‐A, ‐B, ‐C, ‐DRB1, ‐DQB1, and ‐DPB1. The six HLA loci investigated are considered to be the most relevant for the selection of an unrelated hematopoietic cell donor for transplantation.15 We resort to the first two fields of the HLA allele nomenclature because this level of resolution is regarded as sufficiently detailed for the assessment of the histocompatibility between a patient and a potential donor16 and most widely used in the documentation of HLA genotypes of volunteer donors.
At the end of 2015, about 57% of the total German population of 82 million were between 18 und 60 years old that is, generally eligible as hematopoietic stem cell (HSC) donors.17 Of the approximately 9 million foreigners (persons who do not have German citizenship), 73% are in this age group,18 and about one‐fifth of the German population has a migrant background (persons who themselves or one parent were not born of German nationality). The immigration is mainly from Turkey and other European countries.19 All these population subgroups are represented in the German donor file,20 and regional differences in Germany have been previously described.21 These specific characteristics justify a detailed analysis of CWD alleles targeted in our particular setting.
We investigated the prevalence of alleles for the 6 HLA loci currently regarded as primarily relevant for the success of an unrelated hematopoetic stem cell transplantation (HSCT). The aim of this study was to elaborate to which extent the large German donor pool contributes to the knowledge about the CWD alleles worldwide. We have applied criteria similar to the ASHI and EFI CWD catalogs in order to be able to compare our findings with relevant existing references. In that process, we also assessed the prevalence of exon‐defined null alleles in the German population.
2. MATERIAL AND METHODS
Our sample is composed of volunteer adult HSC donors from the 26 German donor centers. The HLA typing methodology of the donor sample is not individually documented but is a mixture of SSO, SSP, sequence‐based typing, and next‐generation sequencing methodologies as they were used over time. This heterogeneity with regard to typing techniques, typing resolution, and loci typed is typical for a long existing registry. In order to cope with this situation and to avoid a major selection bias, we did not exclude most donors with limited HLA typing which would be necessary to apply a pure counting approach. Instead, we used a validated implementation of the expectation‐maximization algorithm for allele and haplotype frequency estimation (HFE) capable of dealing with ambiguous allele assignments and missing data.22 This approach not only allows for inclusion of many more donors but inherently provides the haplotypical context for the alleles. Therefore, we could use the 5 104 477 individuals from the German donor pool as of September 30, 2016 with molecular typings for at least the three loci HLA‐A, ‐B, and ‐DRB1 for the HFE of the HLA‐A~B~C and the HLA‐DRB1~DQB1~DPB1 3‐locus haplotypes. For this work, all allele frequencies (AFs) are calculated as the marginal frequencies of the estimated 3‐locus haplotype frequencies (HFs) mentioned above.
The HF and AF obtained were further analyzed with Perl, R, and Excel.
About three quarters of the donors in this study came from one donor center (DKMS). The fourth quarter was added from the other donor centers, of which the majority is organized as “Stiftung Knochenmarkspende Deutschland” (SKD). This part of the donor data is summarized as SKD hereafter. Because DKMS also contributed data to the EFI CWD, there is some overlap with this study, which has been assessed for the six HLA loci under consideration in Table 1. The potential overlap with the EFI study is under 26% for HLA class I loci, however, up to 75% for HLA class II loci.
Table 1.
DE CWD study population characteristics and potential overlap (p.o.) with the EFI CWD
| Number of donors | ||||||
|---|---|---|---|---|---|---|
| HLA‐A | HLA‐B | HLA‐C | HLA‐DRB1 | HLA‐DQB1 | HLA‐DPB1 | |
| DE | 5 104 477 | 5 104 477 | 4 257 003 | 5 104 477 | 3 664 597 | 2 495 957 |
| DKMS | 76% | 76% | 84% | 76% | 85% | 91% |
| EFI | 639 416 | 1 371 921 | 798 159 | 3 966 984 | 2 700 141 | 1 708 550 |
| DKMS | 89% | 95% | 91% | 97% | 99% | 99% |
| p.o. | 11% | 26% | 17% | 75% | 73% | 68% |
Abbreviations: CWD, common and well‐documented; DE CWD, CWD catalog of the German population; EFI, European Federation for Immunogenetics.
The AF and HF, their analysis, and the comparison with the ASHI and EFI CWD catalogs are based on the first two fields of HLA allele names in the IMGT/HLA database release 3.25.0 (2016‐07). If necessary, allele designations were collapsed to two fields. At that level, we were able to additionally provide information about alleles that are categorized in P‐groups, that is, alleles that have an identical protein structure in the antigen recognition domain (ARD) excluding null alleles (see Appendices S1 and S2).
Current HFE methods assume that the study population is in Hardy‐Weinberg equilibrium (HWE). As a test for deviation from HWE is always significant because of the sample size analyzed in this study, the amount of deviation from HWE was assessed by the effect size statistic Wn according to23, 24 and the comparison of observed and expected homozygosity rates. Both calculations were performed on the allele groups (first field) for the six individual loci, because of the mixed resolution in the data set.
Our first reference data set, the CWD 2.0.0 catalog (hereafter called ASHI CWD), is based on 139 961 SBT observations from several populations and the IMGT/HLA database release 3.9.0 (2012‐07). The ASHI CWD catalog uses the following allele categorizations:
A1. Common alleles (C) are observed at frequencies greater than 1 in 1000 in several populations of at least 1500 individuals.
A2. Well‐documented (WD) alleles are observed either at least
five times in unrelated individuals or
three times in a specific haplotype in unrelated individuals.
Our second reference data set, the EFI CWD catalog, is based on between 639 416 (HLA‐A) and 3 966 984 (HLA‐DRB1) individuals of European origin. The EFI CWD catalog uses the following allele categorizations:
E1. Common alleles (C) are observed more than three times in at least three different populations.
E2. WD alleles are observed at least five times in the total set of populations.
For the CWD catalog of the German population (DE CWD hereafter), we have established the following allele categorizations:
D1. Common alleles are observed at frequencies greater than 1 in 1000, that is, more than about 10 000 times in the sample.
D2. We distinguish four groups of WD alleles comprising
those observed with a frequency of at least 1 in 100 000 (WD1), that is, about 100 times in the sample;
those observed at least five times (WD3), that is, about 1 per 2 000 000 in the sample;
those observed three times in a specific haplotype (WD4), that is, about 1 in 3 400 000 in the sample;
alleles which satisfy conditions b) and c) are categorized as WD2.
The definition for common is identical for ASHI CWD (A1) and DE CWD (D1), and the original definition of WD (A2) is divided into four subcategories for DE CWD (D2a‐D2d) to allow for a more differentiated view on the properties of this large group of alleles. We will present the rationale for our refined approach in the discussion.
3. RESULTS
We identified 2155 HLA‐A, ‐B, ‐C, ‐DRB1, ‐DQB1, and ‐DPB1 two‐field alleles as CWD in the German donor population representing approximately 20% of the HLA alleles at two‐field resolution in the IPD‐IMGT/HLA Database release v3.25.0 for these loci. HLA‐DPB1 is an outlier with a particularly high proportion of about 34%. Table 2 summarizes the number of CWD alleles in Germany (column C) and their proportions with regard to all alleles observed in the German donor pool (column O) and the total number of alleles described in the official nomenclature (column A).
Table 2.
Number and proportion of common and well‐documented alleles in Germany (DE CWD)
| Locus | Number of two‐field alleles in IMGT/HLA 3.25.0 | Number of two‐field alleles estimated from DE sample | Number of two‐field DE CWD | Percentage of described two‐field alleles that are DE CWD | Percentage of described two‐field alleles that have a positive frequency in DE | Percentage of two‐field alleles in DE that are DE CWD |
|---|---|---|---|---|---|---|
| A | O | C | C/A(%) | O/A(%) | C/O(%) | |
| A | 2629 | 934 | 437 | 16.6 | 35.5 | 46.8 |
| B | 3354 | 1215 | 618 | 18.4 | 36.2 | 50.9 |
| C | 2310 | 853 | 417 | 18.1 | 36.9 | 48.9 |
| DRB1 | 1452 | 619 | 345 | 23.8 | 42.6 | 55.7 |
| DQB1 | 672 | 243 | 142 | 21.1 | 36.2 | 58.4 |
| DPB1 | 570 | 262 | 196 | 34.4 | 46.0 | 74.8 |
| Total | 10 987 | 4126 | 2155 | 19.6 | 37.6 | 52.2 |
Abbreviations: CWD, common and well‐documented; DE CWD, CWD catalog of the German population.
The characteristics of the DE CWD catalog are shown in Tables 3 and 4. Only between 18 (HLA‐DQB1) and 40 (HLA‐B) alleles were found to be common for the six HLA loci investigated but they comprise between 98.3% (HLA‐B) and 99.9% (HLA‐DQB1) of the cumulated AFs. This leads to the observation, that only between 1 (HLA‐DQB1) and 284 (HLA‐B) of a million individuals carries no common allele at a specific locus. Adding the WD alleles, the cumulated frequency is above 99.99% for all loci, thereby placing the number of individuals with no DE CWD allele at a specific locus at below 1 per million.
Table 3.
Number (#) and cumulated frequencies of CWD alleles in CWD catalog of the German population
| Locus | Common | Well documented | CWD | #Individual with no common allele per million | |||
|---|---|---|---|---|---|---|---|
| # | Cum. AF(%) | # | Cum. AF(%) | # | Cum. AF(%) | ||
| A | 26 | 99.152 | 411 | 0.839 | 437 | 99.991 | 72 |
| B | 40 | 98.316 | 578 | 1.673 | 618 | 99.990 | 284 |
| C | 21 | 99.309 | 396 | 0.682 | 417 | 99.992 | 48 |
| DRB1 | 33 | 99.362 | 312 | 0.633 | 345 | 99.996 | 41 |
| DQB1 | 18 | 99.906 | 124 | 0.092 | 142 | 99.998 | 1 |
| DPB1 | 22 | 99.521 | 174 | 0.477 | 196 | 99.998 | 23 |
| Total | 160 | 1995 | 2155 | ||||
Abbreviations: AF, allele frequency; CWD, common and well documented.
Table 4.
Number (#) and cumulated frequencies for the different well‐documented alleles categories in CWD catalog of the German population
| Locus | # | WD1 | WD2 | WD3 | WD4 | |||
|---|---|---|---|---|---|---|---|---|
| Cum. AF(%) | # | Cum. AF(%) | # | Cum. AF(%) | # | Cum. AF(%) | ||
| A | 66 | 0.762 | 254 | 0.071 | 50 | 0.005 | 41 | 0.002 |
| B | 108 | 1.580 | 364 | 0.088 | 39 | 0.003 | 67 | 0.003 |
| C | 64 | 0.622 | 245 | 0.056 | 27 | 0.002 | 60 | 0.003 |
| DRB1 | 66 | 0.585 | 182 | 0.045 | 23 | 0.001 | 41 | 0.001 |
| DQB1 | 16 | 0.070 | 90 | 0.021 | 12 | 0.001 | 6 | 0.000 |
| DPB1 | 50 | 0.441 | 106 | 0.035 | 10 | 0.001 | 8 | 0.000 |
| Total | 370 | 1241 | 161 | 223 | ||||
Abbreviations: AF, allele frequency; CWD, common and well documented.
Of the four WD categories, only WD1 (greater than 1 in 100 000) has a substantial frequency contribution (ie, >0.1%). The categories WD2‐WD4 contribute the majority of alleles but less than one per thousand of the cumulated frequencies (see Table 4). The relationship among allele numbers, AFs, and cumulated frequencies is illustrated in Figure 1 for the HLA class I and class II loci.
Figure 1.

Individual and cumulated allele frequencies of common and well‐documented catalog of the German population. HLA class I alleles on top, HLA class II alleles at the bottom. The color coded x‐axis ticks designate the number of alleles for each locus needed for a cumulated frequency of 0.99
Table 5 gives a comparison of the three catalogs' CWD numbers. The numbers for common alleles in any catalog are in bold face, and the numbers of alleles for which all three catalogs are in concordance are in red. Table 5 provides the same information but compares the SKD part of our sample with ASHI and EFI CWD.
Table 5.
(A) Comparison of the three catalogs' CWD numbers. (B) Comparison of the SKD part of our sample with the ASHI and EFI catalogs' CWD numbers
| ASHI CWD | EFI CWD | C | WD | Not CWD |
|---|---|---|---|---|
| (A) CWD catalog of the German population | ||||
| C | C | 153 | 52 | 0 |
| WD | 5 | 89 | 0 | |
| Not CWD | 0 | 60 | 4 | |
| WD | C | 0 | 12 | 0 |
| WD | 0 | 207 | 2 | |
| Not CWD | 1 | 244 | 185 | |
| Not CWD | C | 1 | 7 | 0 |
| WD | 0 | 494 | 6 | |
| Not CWD | 0 | 830 | n.a. | |
| 160 | 1995 | 197 | ||
| (B) SKD CWD | ||||
| C | C | 152 | 53 | 0 |
| WD | 5 | 86 | 3 | |
| Not CWD | 1 | 52 | 11 | |
| WD | C | 0 | 12 | 0 |
| WD | 0 | 170 | 39 | |
| Not CWD | 1 | 182 | 247 | |
| Not CWD | C | 0 | 8 | 0 |
| WD | 0 | 231 | 269 | |
| Not CWD | 0 | 317 | n.a. | |
| 159 | 1111 | 569 | ||
Abbreviations: ASHI, American Society for Histocompatibility and Immunogenetics; CWD, common and well documented; EFI, European Federation for Immunogenetics; SKD, Stiftung Knochenmarkspende Deutschland; WD, well documented.
The numbers for common alleles in any catalog are bold, and the numbers of alleles for which all three catalogs are in concordance are red.
The complete two‐field allele DE CWD catalogs including the null alleles and the P‐group catalog are provided in the supporting information (Appendix S1 and S2). An example is given in Table 6, which shows the classification of the allele for DE, ASHI, and EFI CWD, the AF, the most frequent 3‐locus haplotype with the accompanying HF and the ratio of HF/AF as linkage indicator, all the latter for DE CWD only. Remarkably, in this example, only one of the 14 German CWD DRB1*07 alleles has been identified in the EFI study too.
Table 6.
HLA‐DRB1*07 example for DE CWD table
| Allele | DE CWD | American Society for Histocompatibility and Immunogenetics CWD | European Federation for Immunogenetics CWD | AF | Most frequent DRB1~DQB1~DPB1 haplotype | HF | HF/AF(%) |
|---|---|---|---|---|---|---|---|
| DRB1*07:01 | C | C | C | 1.21E‐01 | 07:01~02:02~04:01 | 2.54E‐02 | 21 |
| DRB1*07:03 | WD2 | WD | ‐ | 2.26E‐06 | 07:03~02:02~04:02 | 1.55E‐06 | 68 |
| DRB1*07:04 | WD4 | ‐ | ‐ | 4.17E‐07 | 07:04~02:04~02:01 | 4.17E‐07 | 100 |
| DRB1*07:05 | WD1 | WD | ‐ | 1.26E‐05 | 07:05~02:02~11:01 | 1.10E‐05 | 87 |
| DRB1*07:07 | WD1 | WD | ‐ | 2.23E‐05 | 07:07~03:03~04:01 | 1.82E‐05 | 81 |
| DRB1*07:10 N | WD1 | ‐ | ‐ | 1.05E‐05 | 07:10 N~02:02~04:02 | 8.87E‐06 | 85 |
| DRB1*07:11 | WD2 | WD | ‐ | 2.17E‐06 | 07:11~02:02~02:01 | 1.03E‐06 | 48 |
| DRB1*07:14 | WD2 | ‐ | ‐ | 8.10E‐07 | 07:14~03:03~04:01 | 7.12E‐07 | 88 |
| DRB1*07:21 | WD4 | ‐ | ‐ | 3.06E‐07 | 07:21~03:03~09:01 | 3.06E‐07 | 100 |
| DRB1*07:23 | WD2 | ‐ | ‐ | 6.07E‐07 | 07:23~02:02~02:01 | 5.17E‐07 | 85 |
| DRB1*07:26N | WD4 | ‐ | ‐ | 4.05E‐07 | 07:26 N~02:02~04:01 | 4.01E‐07 | 99 |
| DRB1*07:29 | WD2 | ‐ | ‐ | 6.07E‐07 | 07:29~02:02~04:02 | 4.86E‐07 | 80 |
| DRB1*07:32 | WD2 | ‐ | ‐ | 5.06E‐07 | 07:32~02:02~11:01 | 5.06E‐07 | 100 |
| DRB1*07:34 | WD2 | ‐ | ‐ | 9.09E‐06 | 07:34~02:02~11:01 | 8.54E‐06 | 94 |
Abbreviations: AF, allele frequency; CWD, common and well documented; DE CWD, CWD catalog of the German population; HF, haplotype frequency; WD, well documented.
Observed alleles that did not qualify for the ASHI and EFI CWD catalogs were not published and are therefore not available for analysis here.
The frequency of null alleles defined by exon variations was also analyzed. Approximately, 10% to 15% of all null alleles described for HLA‐A, ‐B, ‐C, ‐DRB1, ‐DQB1, and ‐DPB1 were actually found to be CWD in the German sample (20 of 158 for HLA‐A, 13 of 137 for HLA‐B, 10 of 115 for HLA‐C, 5 of 46 for HLA‐DRB1, 3 of 25 for HLA‐DQB1, and 3 of 18 for HLA‐DPB1). Evidently, HLA‐C*04:09N is the most frequent null allele with an AF of over 1 in 10 000. In Germany, it accounts for almost two‐thirds of the occurrences of the 10 null alleles defined by variations outside the ARD defining region (Appendix S1). Together the 10 non‐ARD defined null alleles account for more than half of the occurrences of the 54 HLA‐A, ‐B, ‐C, ‐DRB1, ‐DQB1, and ‐DPB1 null alleles found as CWD in Germany. The frequency distribution and the categories of the 54 DE CWD null alleles are shown in Figure 2.
Figure 2.
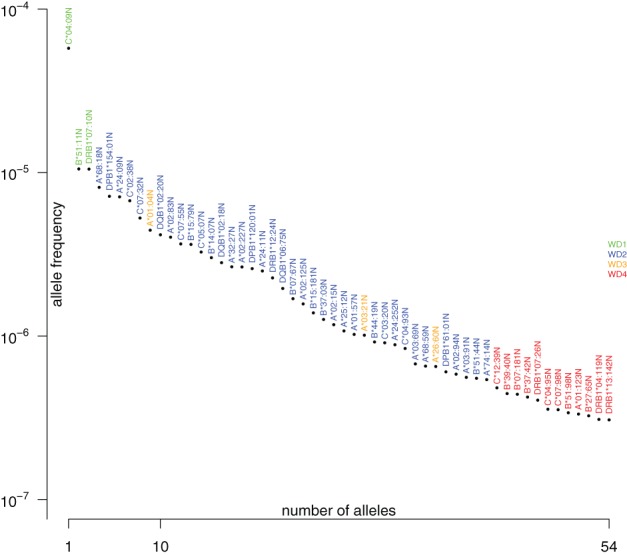
Frequency of common and well‐documented null alleles in Germany
The effect size statistic Wn was close to zero for all six loci (between 0.006 and 0.02, see column Wn in Table 7), which does not indicate a relevant deviation from HWE and is in accordance with findings from previous studies.25, 26 The deviation observed is consistently small and toward homozygosity for all six loci (see Table 7 columns HoObs and HoExp), which does not affect the accuracy of the HFE.27 Both subsamples of our population (DKMS and SKD) cover all geographic regions and are therefore representative for the general donor population in Germany.
Table 7.
Effect size statistic Wn, observed (HoObs) and expected homozygosity (HoExp) rates
| Locus | Wn | HoObs | HoExp |
|---|---|---|---|
| A | 0.006 | 0.155 | 0.153 |
| B | 0.011 | 0.081 | 0.077 |
| C | 0.009 | 0.161 | 0.156 |
| DRB1 | 0.006 | 0.116 | 0.113 |
| DQB1 | 0.004 | 0.255 | 0.253 |
| DPB1 | 0.021 | 0.335 | 0.331 |
4. DISCUSSION
As shown in the introduction, our sample comprises almost 11% of the population theoretically available as HSC donors and about 6% of the total population in Germany. Although this sample may not exactly reflect the age and ethnic structure of the total population, it is sufficiently large to be regarded as representative for the German population as a whole. More than one‐third of all two‐field HLA‐A, ‐B, ‐C, ‐DRB1, ‐DQB1, and ‐DPB1 alleles described in the IMGT/HLA database release 3.25.0 are found in our sample and slightly more than half of these are actually CWD (Table 2). Almost all individuals in the sample carry at least one CWD allele on every investigated HLA locus.
An important design decision of our study was the choice of criteria for “common” and “well‐defined.” Actually, the criteria of our reference studies were so divergent that the common name “CWD” for their outcome is slightly misleading. ASHI used an absolute frequency (1:1000) with a quorum sample size of 1500 for common, whereas EFI required three occurrences in each of three different populations. The latter criteria make the result of the study quite dependent on the sample sizes and on the notion used to delineate populations among the 196 individual samples of the study. The EFI CWD catalog removed the relative frequency part probably because over 80% of the population samples considered were under the quorum of 1500 individuals. However, given our sample size, there was no need to abandon this criterion and, in particular, our decision was not motivated by the lack of complete and reliable data on ethnic or geographic background of the individuals tested.
In contrast to the ASHI CWD definition of “common,” both previous catalogs only used an occurrence count criterion for “well‐defined” alleles, which makes comparing classifications based on different sample sizes difficult. This made us include a size‐independent, frequency‐based criterion WD1 (>1:100000) as a part of our “well‐defined” definition, and we included the other variants (WD2‐WD4) for reasons of completeness and comparability. However, it can clearly be seen from Table 4 that although the groups WD2‐WD4 contribute over 80% of the WD allele list their contribution to the actual occurrences is under 10% clearly limiting their practical relevance.
Generally, there is a substantial concordance of CWD alleles between the three catalogs. Altogether, 40% of the 380 different HLA‐A, ‐B, ‐C, ‐DRB1, ‐DQB1, and ‐DPB1 common alleles are classified as common in all three catalogs. This is not surprising in view of the dominant contribution of Europeans to all the samples processed.
81% of the ASHI CWD catalog is shared with DE CWD and about half is shared with EFI CWD. However, ASHI CWD contains a higher number of common alleles, reflecting the higher diversity of the global sample, whereas DE and EFI CWD have more WD alleles, reflecting their larger samples.
Overall, it is quite striking that in spite of a much narrower ethnic and geographic coverage our study reports substantially greater details of the allelic polymorphism than both other studies if all allele categorizations are applied (ie, down to a minimum of three copies in the sample). DE CWD contains about twice as many alleles as ASHI and EFI CWD. Only 830 alleles are included in DE CWD, whereas just 197 are found in the EFI CWD or ASHI CWD catalog only.
However, if the WD1 criterion is applied for DE CWD alone, 67 DE CWD alleles are not part of the ASHI CWD catalog. Figure 3 shows the different allele counts of the three catalogs per locus and illustrates the higher population diversity of ASHI and EFI CWD. The high proportion of two‐field HLA‐DPB1 alleles found (46%) and classified as CWD (34%) in the German population illustrates the contribution of specific donor typing efforts on the revelation of the HLA‐DPB1 polymorphism after HLA‐DBP1 had been identified as a relevant transplant antigen.28, 29
Figure 3.
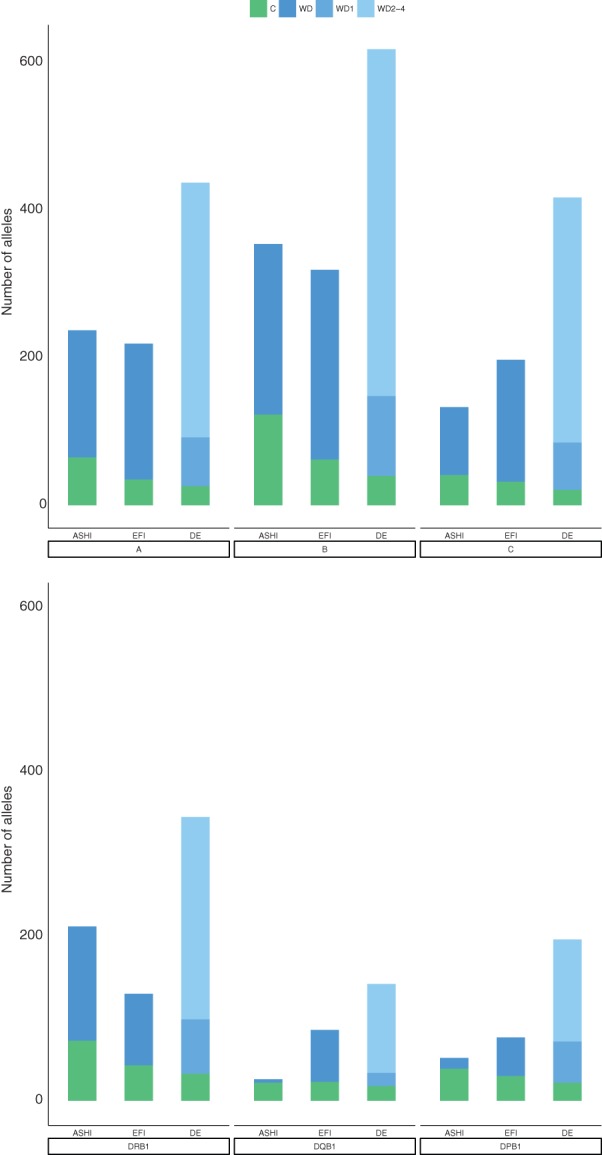
Comparison of common and well‐documented numbers per locus
For the EFI CWD, the DKMS sample accounts for between 89% and 99% of the HLA data for HLA‐A, ‐B, ‐C, ‐DRB1, ‐DQB1, and ‐DPB1 and for a total of 76% of the donors considered in this study. This explains why DE CWD covers 99% of the alleles reported in EFI CWD. The over 1000 additional alleles of DE CWD compared with EFI CWD can be attributed to methodological refinements and a substantially larger sample. For HLA class I loci, the sample was between 3.7 (HLA‐B) and 8 (HLA‐A) times larger mainly because our approach allowed for inclusion of many more DKMS donors than the EFI study leading to a low overlap between 11% (HLA‐A) and 26% (HLA‐B). For HLA class II loci, on the other hand, the sample increase between 25% and 32% was mainly because of the inclusion of the SKD donor centers. Overall, our study based on the German donor pool gives new insight into the CWD alleles on the worldwide level, in particular for the HLA class I loci.
The six WD alleles only found in the EFI catalog are present in our sample, but their frequencies did not qualify for CWD. The absence of 60 ASHI and DE CWD alleles in the EFI CWD is probably explained by the varying number of population criteria. The relationships among the three catalogs are illustrated in Figure 4, pointing out the level of detail found in the individual CWD catalogs.
Figure 4.
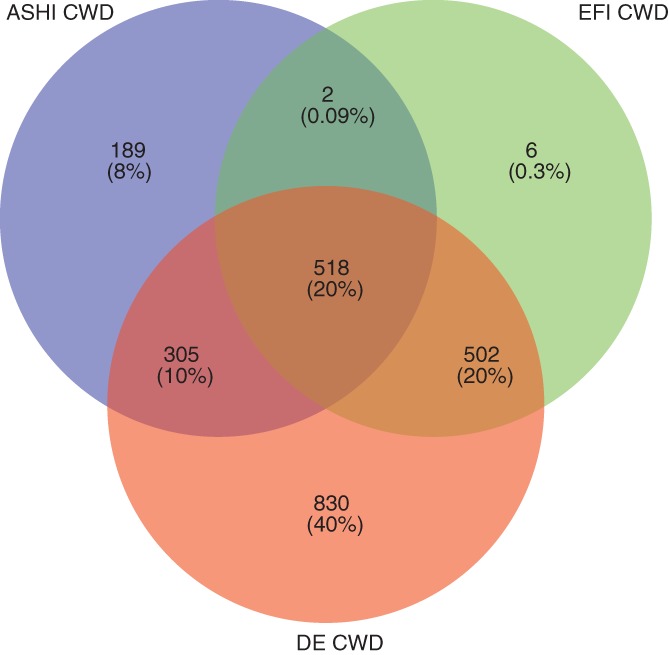
Venn diagram showing all possible logical relationships between the American Society for Histocompatibility and Immunogenetics catalog, European Federation for Immunogenetics catalog, and common and well‐documented catalog of the German population
Both the ASHI and EFI catalogs partly require different populations for their categories, whereas here we used a single donor sample with some limited population admixture. The combination of the number of populations and the sample size per population for a category seems to be unsuitable for standardized and comprehensible criteria. The allele HLA‐DRB1*07:05 for example is seen 117 times in our complete sample (95 times in the DKMS subsample) by counting alone and categorized as WD1 and as WD in the ASHI CWD (Table 6). EFI CWD, however, although definitely seeing a substantial number of HLA‐DRB1*07:05 alleles because of the overlap of donors in both studies, can only report one HLA‐DRB1*07 allele (DRB1*07:01), perhaps because of the number of population criteria. In summary, globally comprehensive and scalable categories are required to make the different catalogs more consistent.
The vast majority of known null alleles is defined by sequence variations within the region coding the ARD and is therefore reliably detected by most typing techniques. Only six depend on variations outside all exons and are rarely considered. However, the identification of null alleles defined in non‐ARD exons is handled inconsistently30, 31 because of the lack of an efficient general typing strategy.32 Therefore, the AFs of those null alleles are most likely underestimated.
In this study, we did not observe the AFs directly rather estimated them from heterogeneous donor data as it is typically found in donor registries. The concordance between the three catalogs shows the applicability of our approach. The estimation procedure has inherently larger estimation variance than direct counting procedures,33 particularly for HLA‐C, ‐DQB1, and ‐DPB1, where 17%, 28%, and 51% of the individuals in this study are not typed (Table 1). However, this is more than compensated for by the large sample size, reducing bias, and allowing the description of rare alleles (three copies) on a sound statistical basis and providing the haplotypical context for the alleles automatically.
Finally, we would like to advocate once more for the usage of relative frequency CWD criteria because they allow better interoperability, scalability, and flexibility. Such CWD criteria do also need a standardized geographic and/or population dimension.
Supporting information
Appendix S1. Two‐field allele common and well‐documented catalog of the German population including null alleles (Excel 2016 file).
Appendix S2. P‐group common and well‐documented catalog of the German population (Excel 2016 file).
ACKNOWLEDGMENTS
This work was supported in part by a grant from the Deutsche José Carreras Leukämie Stiftung (DJCLS R15/02) to K.F. We are indebted to all donors and their managing donor centers contributing to scientific progress to help more patients better. We would like to acknowledge Ingrid Tistl for proofreading, Sandra Frank, and Caroline Räther for R‐support.
Conflict of interest
The authors have declared no conflicting interests.
Eberhard H‐P, Schmidt AH, Mytilineos J, Fleischhauer K, Müller CR. Common and well‐documented HLA alleles of German stem cell donors by haplotype frequency estimation. HLA. 2018;92:206–214. 10.1111/tan.13378
Funding information Deutsche Jose Carreras Leukämie Stiftung, Grant/Award Number: DJLCS R15/02
REFERENCES
- 1. Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genom Hum Genet. 2013;14(1):301‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gragert L, Eapen M, Williams E, et al. HLA match likelihoods for hematopoietic stem‐cell grafts in the U.S. registry. N Engl J Med. 2014;371(4):339‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beatty PG, Mori M, Milford EL. Impact of racial genetic polymorphism on the probability of finding an HLA‐matched donor. Transplantation. 1995;60(8):778‐783. [PubMed] [Google Scholar]
- 4. Klein JP, Sato A. The HLA system—first of two parts. N Engl J Med. 2000;343(10):702‐709. [DOI] [PubMed] [Google Scholar]
- 5. Klein JP, Sato A. The HLA system. Second of two parts. N Engl J Med. 2000;343(11):782‐786. [DOI] [PubMed] [Google Scholar]
- 6. Thomas ED. Stem cell transplantation: past, present and future. Stem Cells. 1994;12(6):539‐544. [DOI] [PubMed] [Google Scholar]
- 7. Alper CA, Larsen CE, Dubey DP, Awdeh ZL, Fici DA, Yunis EJ. The haplotype structure of the human major histocompatibility complex. Hum Immunol. 2006;67(1–2):73‐84. [DOI] [PubMed] [Google Scholar]
- 8. Blake JT, McTaggart K, Killeen D. Modelling the optimal ethnic composition of an adult stem cell registry. Eur J Oper Res. 2018;264(3):870‐883. [Google Scholar]
- 9. Cano P, Klitz W, Mack SJ, et al. Common and well‐documented HLA alleles: report of the ad‐hoc committee of the american society for histocompatibility and immunogenetics. Hum Immunol. 2007;68(5):392‐417. [DOI] [PubMed] [Google Scholar]
- 10. Mack SJ, Cano P, Hollenbach JA, et al. Common and well‐documented HLA alleles: 2012 update to the CWD catalogue. Tissue Antigens. 2013;81(4):194‐203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He J, Li Y, Bao X, et al. Common and well‐documented (CWD) alleles of human leukocyte antigen‐A, ‐B, ‐C, ‐DRB1, and ‐DQB1 loci for the Chinese Han population do not quite correlate with the ASHI CWD alleles. Hum Immunol. 2012;73(1):61‐66. [DOI] [PubMed] [Google Scholar]
- 12. Grubic Z, Burek Kamenaric M, Maskalan M, Stingl Jankovic K, Zunec R. Nonfrequent but well‐documented, rare and very rare HLA alleles observed in the Croatian population. Tissue Antigens. 2014;84(6):560‐564. [DOI] [PubMed] [Google Scholar]
- 13. Sanchez‐Mazas A, Buhler S, Nunes JM. A new HLA map of Europe: regional genetic variation and its implication for peopling history, disease‐association studies and tissue transplantation. Hum Hered. 2013;76(3–4):162‐177. [DOI] [PubMed] [Google Scholar]
- 14. Sanchez‐Mazas A, Nunes JM, Middleton D, et al. Common and well‐documented HLA alleles over all of Europe and within European sub‐regions: a catalogue from the European Federation for Immunogenetics. HLA. 2017;89(2):104‐113. [DOI] [PubMed] [Google Scholar]
- 15. Shaw BE, Mayor NP, Szydlo RM, et al. Recipient/donor HLA and CMV matching in recipients of T‐cell‐depleted unrelated donor haematopoietic cell transplants. Bone Marrow Transplant. 2017;52(5):717‐725. [DOI] [PubMed] [Google Scholar]
- 16. Howard CA, Fernandez‐Vina MA, Appelbaum FR, et al. Recommendations for donor human leukocyte antigen assessment and matching for allogeneic stem cell transplantation: consensus opinion of the blood and marrow transplant clinical trials network (BMT CTN). Biol Blood Marrow Transplant. 2015;21(1):4‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.State and Society—Current population—Federal Statistical Office (Destatis) [Internet]. destatis.de. https://www.destatis.de/EN/FactsFigures/SocietyState/Population/CurrentPopulation/CurrentPopulation.html. Accessed November 8, 2016.
- 18.Das Bundesamt in Zahlen 2015 [Internet]. bamf.de. https://www.bamf.de/SharedDocs/Anlagen/DE/Publikationen/Broschueren/bundesamt-in-zahlen-2015.pdf?__blob=publicationFile. Accessed November 8, 2016.
- 19.Demographics of Germany—Wikipedia [Internet]. en.http://wikipedia.org. https://en.wikipedia.org/wiki/Demographics_of_Germany#Population. Accessed November 8, 2016.
- 20. Pingel J, Solloch UV, Hofmann JA, Lange V, Ehninger G, Schmidt AH. High‐resolution HLA haplotype frequencies of stem cell donors in Germany with foreign parentage: how can they be used to improve unrelated donor searches? Hum Immunol. 2013;74(3):330‐340. [DOI] [PubMed] [Google Scholar]
- 21. Schmidt AH, Solloch UV, Baier DM, et al. Regional differences in HLA antigen and haplotype frequency distributions in Germany and their relevance to the optimization of hematopoietic stem cell donor recruitment. Tissue Antigens. 2010;76(5):362‐379. [DOI] [PubMed] [Google Scholar]
- 22. Bochtler W, Gragert L, Patel ZI, et al. A comparative reference study for the validation of HLA‐matching algorithms in the search for allogeneic hematopoietic stem cell donors and cord blood units. HLA. 2016;87(6):439‐448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klitz W, Stephens JC, Grote MN, Carrington M. Discordant patterns of linkage disequilibrium of the peptide‐transporter loci within the HLA class II region. Am J Hum Genet. 1995;57(6):1436‐1444. [PMC free article] [PubMed] [Google Scholar]
- 24. Maiers M, Gragert L, Klitz W. High‐resolution HLA alleles and haplotypes in the United States population. Hum Immunol. 2007;68(9):779‐788. [DOI] [PubMed] [Google Scholar]
- 25. Eberhard H‐P, Feldmann U, Bochtler W, et al. Estimating unbiased haplotype frequencies from stem cell donor samples typed at heterogeneous resolutions: a practical study based on over 1 million German donors. Tissue Antigens. 2010;76(5):352‐361. [DOI] [PubMed] [Google Scholar]
- 26. Müller CR, Ehninger G, Goldmann SF. Gene and haplotype frequencies for the loci HLA‐A, HLA‐B, and HLA‐DR based on over 13,000 german blood donors. Hum Immunol. 2003;64(1):137‐151. [DOI] [PubMed] [Google Scholar]
- 27. Fallin D, Schork NJ. Accuracy of haplotype frequency estimation for biallelic loci, via the expectation‐maximization algorithm for unphased diploid genotype data. Am J Hum Genet. 2000;67(4):947‐959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fleischhauer K. Immunogenetics of HLA‐DP—A new view of permissible mismatches. N Engl J Med. 2015;373(7):669‐672. [DOI] [PubMed] [Google Scholar]
- 29. Crocchiolo R, Zino E, Vago L, et al. Nonpermissive HLA‐DPB1 disparity is a significant independent risk factor for mortality after unrelated hematopoietic stem cell transplantation. Blood. 2009;114(7):1437‐1444. [DOI] [PubMed] [Google Scholar]
- 30. NMDP . NMDP policy for adult donor and patient HLA confirmatory [Internet]. https://bioinformatics.bethematchclinical.org/HLA-Resources. 2015. https://bioinformatics.bethematchclinical.org/workarea/downloadasset.aspx?id=10528. Accessed January 31, 2017.
- 31. Müller CR, Mytilineos J, Ottinger HD, et al. Deutscher Konsensus 2013 zur immungenetischen Spenderauswahl für die allogene Stammzelltransplantation. Transfusionsmedizin. 2014;4(4):190‐196. [Google Scholar]
- 32. Eberhard H‐P, Mytilineos J, Fleischhauer K, Schmidt AH, Müller CR. Effectiveness of linkage disequilibrium for the identification of HLA null alleles. Bone Marrow Transplant. 2016;51(S1):S552. [Google Scholar]
- 33. Eberhard H‐P, Madbouly AS, Gourraud P‐A, et al. Comparative validation of computer programs for haplotype frequency estimation from donor registry data. Tissue Antigens. 2013;82(2):93‐105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Two‐field allele common and well‐documented catalog of the German population including null alleles (Excel 2016 file).
Appendix S2. P‐group common and well‐documented catalog of the German population (Excel 2016 file).


