Summary
Seafloor microorganisms impact global carbon cycling by mineralizing vast quantities of organic matter (OM) from pelagic primary production, which is predicted to increase in the Arctic because of diminishing sea ice cover. We studied microbial interspecies‐carbon‐flow during anaerobic OM degradation in arctic marine sediment using stable isotope probing. We supplemented sediment incubations with 13C‐labeled cyanobacterial necromass (spirulina), mimicking fresh OM input, or acetate, an important OM degradation intermediate and monitored sulfate reduction rates and concentrations of volatile fatty acids (VFAs) during substrate degradation. Sequential 16S rRNA gene and transcript amplicon sequencing and fluorescence in situ hybridization combined with Raman microspectroscopy revealed that only few bacterial species were the main degraders of 13C‐spirulina necromass. Psychrilyobacter, Psychromonas, Marinifilum, Colwellia, Marinilabiaceae and Clostridiales species were likely involved in the primary hydrolysis and fermentation of spirulina. VFAs, mainly acetate, produced from spirulina degradation were mineralized by sulfate‐reducing bacteria and an Arcobacter species. Cellular activity of Desulfobacteraceae and Desulfobulbaceae species during acetoclastic sulfate reduction was largely decoupled from relative 16S rRNA gene abundance shifts. Our findings provide new insights into the identities and physiological constraints that determine the population dynamics of key microorganisms during complex OM degradation in arctic marine sediments.© 2018 Society for Applied Microbiology and John Wiley & Sons Ltd
Introduction
The Earth's seafloor is habitat for more than half of the microbial cells in the marine environment (Kallmeyer et al., 2012). Microorganisms in marine sediments are highly dependent on energy and carbon that is derived from the degradation of detrital organic matter (OM) (Jørgensen and Boetius, 2007), which is produced by marine phytoplankton (Arndt et al., 2013). Although only 1% of the OM exported from surface waters reaches the seafloor on a global scale (Hedges and Keil, 1995), regional differences are high and sediments in coastal regions receive approximately 6–12% of pelagic primary production (Dunne et al., 2007). The labile fraction of OM that reaches the seafloor is gradually degraded and mineralized by the concerted activity of diverse anaerobic microorganisms (Arndt et al., 2013; Orsi et al., 2018). Cellular macromolecules such as proteins, nucleic acids, lipids and polysaccharides that make‐up the majority of phytoplankton debris (Burdige, 2007) are degraded by mostly unknown microorganisms into oligomers and monomers, which are subsequently fermented to a range of products, including volatile fatty acids (VFAs), H2 and CO2 (Burdige, 2007; Muyzer and Stams, 2008). In continental shelf sediments, the most favorable electron acceptors (O2, Fe(III), Mn(IV), ) are depleted first (Froelich et al., 1979) and sulfate reduction is the predominant process that accounts for the oxidation of half of the VFAs that are produced during anaerobic OM breakdown (Jørgensen, 1982). Thus, on a global scale sulfate‐reducing microorganisms (SRM) facilitate the remineralization of approximately 29% of the total OM flux to the entire seafloor (Bowles et al., 2014).
The Arctic is severely impacted by climate change, resulting in a constant decline of the arctic sea ice cover (Vaughan et al. 2013). This allows marine primary production to prevail (Arrigo et al., 2008) and predictions suggest this will initially cause an increased export of OM to the seafloor (Lalande et al., 2009; Sørensen et al., 2015). OM degradation rates and pathways in arctic marine sediments have been intensively studied in Svalbard fjords with a focus on enzymatic hydrolysis, sulfate reduction, iron reduction and denitrification (Arnosti and Jørgensen, 2006; Finke et al., 2007; Canion et al., 2014). Intriguingly, OM‐degrading microorganisms in permanently cold sediments are relatively active at temperatures close to the freezing point (Kostka et al., 1999; Arnosti and Jørgensen, 2003; Robador et al., 2016), because of psychrophilic adaptations (Knoblauch and Jørgensen, 1999; Cavicchioli, 2016). The microbial communities in Svalbard fjord sediments are highly diverse (Teske et al., 2011), and dominated by representatives of the classes Deltaproteobacteria, Gammaproteobacteria and the phylum Bacteroidetes (Ravenschlag et al., 1999, 2000, 2001). Even though a variety of specific members of this arctic sediment microbial community could be isolated (Knoblauch et al., 1999; Sahm et al., 1999; Wang et al., 2015), the identity and population dynamics of most microorganisms that are responsible for the degradation of complex OM under cold, anoxic conditions in marine sediments is unknown.
In this study, we conducted microcosm experiments with 13C‐labelled substrates in order to directly link OM degradation processes in arctic marine sediments with specific microorganisms (Supporting Information Fig. S1). We used sediment samples from a fjord in Svalbard (Smeerenburgfjorden, station J) where sulfate reduction is the predominant terminal remineralization process (Finke et al., 2007). Anoxic sediment incubations were either supplemented with a low dose (LD) or a high dose (HD) of 13C‐labelled cyanobacterial necromass (spirulina) in order to mimic a pulse of bulk OM input to the seabed or 13C‐labelled acetate, an important organic carbon degradation intermediate and electron donor for anaerobic respiration in marine sediments. We investigated bacterial community responses to substrate supplementation by amplicon‐sequencing of 16S rRNA genes and transcripts in light of changes in VFA concentrations and sulfate reduction rates, and by single‐cell Raman microspectroscopy. This allowed us to (i) identify species‐level phylotypes involved in the degradation cascade of cyanobacterial necromass and in the terminal oxidation of acetate, (ii) reveal main products of cyanobacterial necromass degradation and provide evidence for their consumption by SRM and microorganisms that use alternative electron acceptors for energy conservation and (iii) confirm the specific incorporation of substrate‐derived 13C into individual target cells via a combination of catalyzed reporter deposition fluorescence in situ hybridization (CARD‐FISH) and Raman microspectroscopy.
Results
Enhanced sulfate reduction rates in spirulina necromass‐ and acetate‐amended microcosms
Sulfate reduction rates in the unamended controls ranged from 9 to 18 nmol cm−3 d−1 (Fig. 1 A and B). The addition of cyanobacterial necromass significantly enhanced sulfate reduction rates to 18–40 nmol cm−3 d−1 (P < 0.001) and to 45–64 nmol cm−3 d−1 (P < 0.001) in LD and HD 13C‐spirulina incubations respectively (Fig. 1A). Amendment of 13C‐acetate also significantly increased sulfate reduction rates to 14–34 nmol cm−3 d−1 (P < 0.001) and to 22–40 nmol cm−3 d−1 (P < 0.001) in LD and HD incubations respectively (Fig. 1B). Sulfate reduction rates were below the detection limit in incubations with molybdate as inhibitor (Fig. 1A and B).
Figure 1.
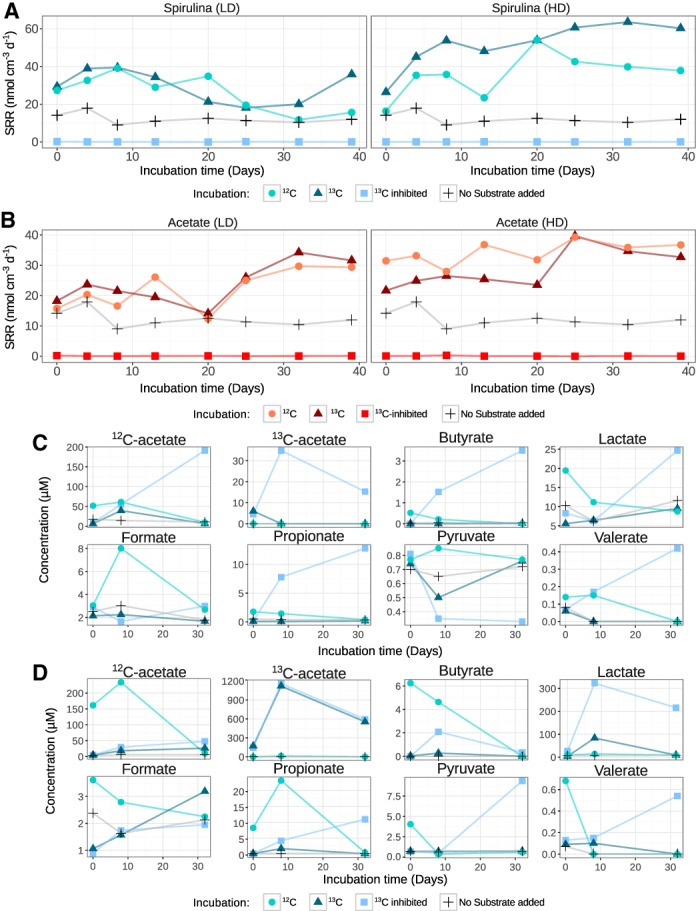
Sulfate reduction rates and volatile fatty acid concentrations in substrate‐supplemented sediment incubations. Sulfate reduction rates (nmol cm−1 d−1) are shown for incubations supplemented with spirulina (A) and acetate (B). Concentrations of volatile fatty acids are indicated for LD (C) and HD (D) incubations with spirulina. 13C‐inhibited, sediment incubations with 13C‐substrate and molybdate. LD, low substrate dose (50 µg ml−1 spirulina or 50 μM acetate). HD, high substrate dose (1 mg ml−1 spirulina or 1 mM acetate). Note that the scales are different for each treatment and VFA.
Acetate is the main volatile fatty acid produced from spirulina degradation
Average background concentrations of VFAs in the unamended control microcosms were low (12 µM acetate, 9 µM formate, 4 µM lactate and < 1 µM propionate, pyruvate, butyrate and valerate) (Fig. 1 and Supporting Information Fig. S2). The concentrations of VFAs differed substantially between the incubations with 13C‐spirulina and 12C‐spirulina (Fig. 1C and D), which suggests considerable differences in the composition of the purchased spirulina products (Supporting Information Results and Discussion). Only results from the 13C‐spirulina incubations are discussed in the main text. A pulse of 13C‐spirulina was added at the beginning of the experiment and its degradation led to the production of mainly 13C‐acetate and several other VFAs (Fig. 1C and D). Compared to uninhibited incubations, concentrations of VFAs increased more over time in incubations with the sulfate‐reduction inhibitor molybdate. Exceptions were HD 13C‐spirulina incubations where 13C‐acetate increased to similarly high concentrations ( > 1.1 mM) with and without molybdate (Fig. 1D). In LD 13C‐spirulina incubations, 13C‐acetate only accumulated in the presence of molybdate and reached concentrations of 35 µM (Fig. 1C). Other VFAs that consistently accumulated in molybdate‐inhibited 13C‐spirulina microcosms were formate, propionate, butyrate and valerate, which reached concentrations of 25 µM, 11 µM, 4 µM and 0.42 µM in LD incubations and 324 µM, 13 µM, 2 µM and 0.52 µM in HD incubations. Pyruvate increased up to 9 µM but only in HD 13C‐spirulina incubations (Fig. 1D) and not in LD 13C‐spirulina incubations (Fig. 1C). Lactate concentrations were always comparable to the unamended controls (Fig. 1C and D). Intriguingly, low amounts of 12C‐acetate accumulated in molybdate‐inhibited 13C‐spirulina incubations (up to 191 and 48 µM in LD and HD incubations respectively), indicating that addition of fresh cyanobacterial necromass additionally stimulated the breakdown of endogenous OM (van Nugteren et al., 2009).
Concentrations of acetate, formate, propionate, butyrate and valerate remained mostly unchanged in incubations with repeated amendments of acetate (Supporting Information Fig. S2). Similar to spirulina incubations, inhibition with molybdate led to an accumulation of acetate, butyrate, propionate and valerate (Supporting Information Fig. S2). However, the concentrations of these VFAs were consistently lower in molybdate‐inhibited incubations with 13C‐acetate than with 13C‐spirulina. This further suggests that a substantial part of the VFAs that accumulated in the inhibited 13C‐spirulina incubations came from breakdown of the supplemented substrate.
An assemblage of eleven phylotypes from diverse taxa is associated with degradation of spirulina necromass under sulfidic conditions
We monitored changes in the relative abundance and ribosomal activity of bacterial community members during the microcosm experiments by pyrosequencing of 16S rRNA gene and transcript amplicons. The sediment community at the start of the experiment (day 0) was dominated by Deltaproteobacteria, Bacteroidetes, Gammaproteobacteria and Planctomycetes (Fig. 2); these taxa also harboured 21 of the 25 abundant phylotypes ( ≥ 1% 16S rRNA gene and/or transcript abundance) (Supporting Information Table S1). The other four abundant phylotypes were affiliated with Fusobacteria, Betaproteobacteria, candidate group Milano‐WF1B‐44 and Cyanobacteria‐Chloroplast (Supporting Information Table S1). Amendments with acetate did not trigger considerable changes in 16S rRNA gene or transcript alpha‐ (Supporting Information Table S2) and beta‐diversity (Supporting Information Fig. S3). In contrast, incubations with 13C‐spirulina led to LD‐ and HD‐dependent shifts in 16S rRNA gene and transcript composition (Supporting Information Figs S3 and S4).
Figure 2.
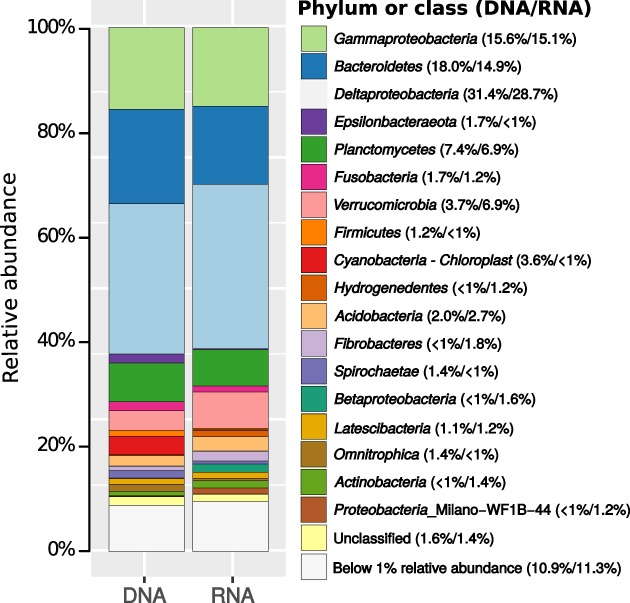
Bacterial community composition in sulfidic Smeerenburgfjorden surface sediment at the start of the incubation experiments (day 0). Average (n = 4) relative 16S rRNA gene (DNA) and transcript (RNA) abundance of bacterial phyla or proteobacterial classes. Only phyla/classes with a relative abundance ≥ 1% on DNA or RNA level are indicated.
We identified individual phylotypes that responded to the added substrates by comparing their relative 16S rRNA gene and transcript abundances in incubations with substrate versus unamended controls. Overall, eleven responsive phylotypes of the phyla/classes Gammaproteobacteria, Deltaproteobacteria, Epsilonbacteraeota, Bacteroidetes, Firmicutes and Fusobacteria were identified by two‐proportion T‐tests (P ≤ 0.01); of which seven were only associated with spirulina amendments, one with acetate amendment alone and three with both substrate amendments (Fig. 3). Ten of these phylotypes had a relative 16S rRNA gene abundance of ≥ 0.1% at the start of the incubations (including four phylotypes with ≥ 1%) (Supporting Information Fig. S5), demonstrating that the majority of the responsive consortium are abundant microbiota members in the sulfidic zone of the arctic marine sediment in situ. Five phylotypes (1452, 4400, 4749, 7435, 10615) responded by significant increases in relative abundances of both 16S rRNA genes and transcripts, while six (2011, 4982, 7234, 7300, 9869, 10263) responded only by increases in relative 16S rRNA transcript abundance, including three deltaproteobacterial phylotypes that were stimulated by the amendment of acetate. The fourth, putative acetate‐utilizing Arcobacter phylotype 10615 was low in abundance and ribosomal activity ( < 0.1%) at day 0, but became dominant in spirulina‐ and acetate‐amended incubations (Fig. 3). Four deltaproteobacterial phylotypes, including Desulfobacteraceae phylotype 2011 and [Desulfobacterium] phylotype 4982 that responded to acetate, were considered as putative SRM because they were inhibited by molybdate. The other two SRM phylotypes (Desulfobacteraceae phylotype 11380 and Desulfobulbaceae phylotype 3944) did not respond significantly to spirulina or acetate amendments (Fig. 3).
Figure 3.

Relative abundance heat‐maps of substrate‐responsive and molybdate‐inhibited 16S rRNA phylotypes. Depicted are phylotypes that responded to substrate amendment and/or molybdate inhibition in spirulina (A) and acetate (B) incubations with significant relative abundance changes in at least two 16S rRNA gene or transcript libraries as compared to unamended or uninhibited incubations. SRM (i.e., molybdate‐inhibited) phylotypes are highlighted in bold. Phylotypes that responded to both spirulina and acetate amendments are indicated with a section sign (§). Phylotypes that were analyzed by CARD‐FISH‐Raman microspectroscopy are indicated with an asterisk (*). Substrate + , phylotypes that responded to substrate additions. Only inhib., phylotypes that did not respond to substrate addition but were inhibited by molybdate. No Sub., no‐substrate control incubation. Mol., Molybdate.
Single cell stable isotope probing revealed 13C‐incorporation in phylotypes that responded to substrate supplementation
Raman microspectroscopy of randomly selected cells from day 32 of the HD 13C‐spirulina incubations showed that only 4% of cells displayed a low level of 13C labelling, which supports the finding that only a small part of the native sediment community is involved in the degradation of spirulina necromass (Fig. 4). We further combined CARD‐FISH and Raman microspectroscopy to confirm 13C‐substrate utilization and assimilation by individual cells of selected taxa that responded to the substrate additions. The used or newly developed CARD‐FISH probes covered four putative hydrolytic/fermenting phylotypes, i.e., Clostridiales phylotype 1452, Marinifilum phylotype 4400, Psychrilyobacter phylotype 4749 and Psychromonas phylotype 7435 and two putative acetate‐utilizing phylotypes, i.e., Desulfobacteraceae phylotype 2011 and Arcobacter phylotype 10615 (Supporting Information Fig. S6). All phylotypes that responded to 13C‐spirulina, including Arcobacter phylotype 10615 that responded to 13C‐spirulina and 13C‐acetate, showed significant cellular 13C‐incorporation at day 32 of the HD 13C‐spirulina incubations (Fig. 4). Desulfobacteraceae phylotype 2011 was also significantly enriched in comparison to the 12C‐spirulina control but only a small fraction (11%) of the cells were labelled at low level (Fig. 4). The fraction of 13C‐labeled Desulfobacteraceae phylotype 2011 cells was larger (37%) in HD 13C‐acetate incubations but individual cells had low 13C/12C phenylalanine peak height ratios, indicating only minor 13C‐incorporation.
Figure 4.

13 C‐labelling of individual cells of substrate responsive phylotypes. CARD‐FISH‐Raman microspectroscopy analysis of 13C‐labelled probe‐targeted cell populations (including selected substrate‐responsive phylotypes) at day 32 in high dose (HD) 13C‐spirulina (1 mg ml−1) and 13C‐acetate (1 mM) incubations. Stars (***) indicate significant (P < 0.001) 13C‐labelling, as revealed by comparing 13C‐labelled target populations to the respective negative control using Wilcoxon Rank Sum tests. Negative 12C‐spirulina, randomly selected cells from incubations with 12C‐spirulina at day 0. Negative 12C‐acetate, cells of Desulfobacteraceae phylotype 2011 (Deltaproteobacteria) from incubations with 12C‐acetate at day 0. The red dashed line shows the threshold for detection of 13C‐labelling in an individual cell.
Discussion
Different cellular responses of members of a bacterial consortium involved in mineralization of organic matter in an arctic marine sediment
OM in anoxic, sulfidic marine sediments is mainly degraded by the joint activity of hydrolysers, fermenters and SRM (Arndt et al., 2013). In order to investigate the microorganisms involved in sequential OM degradation, we performed anoxic incubation experiments at 0°C with arctic marine sediments from Smeerenburgfjorden (Svalbard). In these sediments sulfate reduction is the dominant carbon mineralization process, with important contributions by iron reduction (Finke et al., 2007). Sulfate reduction rates in sediment incubations without external substrates (9–18 nmol cm−3 d−1) (Fig. 1A and B) were in the range of previously determined mean in situ rates of 15–20 nmol cm−3 d−1 in the sulfate reduction zone of Smeerenburgfjorden sediments (Jørgensen et al., 2010; Sawicka et al., 2012; Robador et al., 2016). Supplementation of spirulina and acetate, each at two different doses, to individual sediment microcosms immediately triggered significant increases in sulfate reduction rates, which were more pronounced in spirulina incubations (Fig. 1A). Interestingly, a 20‐fold increase in substrate concentration resulted in only 55% and 38% increased sulfate reduction rates in spirulina and acetate incubations respectively. This indicated that the SRM community had almost reached maximal physiological activity at the lower substrate dose. Hydrolysis and fermentation of spirulina biomass predominantly generated acetate and smaller amounts of butyrate, propionate, valerate, formate and pyruvate (Fig. 1C and D). Accumulation of these VFAs in sulfate reduction‐inhibited incubations confirmed that mineralization of OM and utilization of VFAs in this sediment is largely dependent on the activity of SRM.
Smeerenburgfjorden sediments harbor a cold‐adapted microbial community that is primarily composed of Deltaproteobacteria, Gammaproteobacteria and representatives of the phylum Bacteroidetes (Ravenschlag et al., 1999, 2000, 2001). These classes/phyla were also dominant in the sediments analyzed in the present study (Fig. 2). Here, we show that only eleven species‐level phylotypes of this complex sediment microbiota responded significantly to spirulina or acetate supplementation during a 38 days incubation period at 0°C. Responsive phylotypes either only increased in relative 16S rRNA transcript abundance, which suggests response by increased ribosomal activity, or additionally in relative 16S rRNA gene abundance, which suggests response by growth (Hausmann et al., 2016). Acetate mainly triggered increases in ribosomal activity in SRM phylotypes 2011 and 4982, whereas four (phylotypes 4400, 1452, 4749, 7435) of the six phylotypes involved in the fermentation of spirulina necromass and the acetate‐utilizing, non‐sulfate‐reducing Arcobacter phylotype 10615, also responded by increased relative 16S rRNA gene abundance (Fig. 3). Differences in spirulina‐ and acetate‐based 13C‐labeling of individual cells confirmed different cellular response patterns of the diverse functional guilds in anaerobic carbon decomposition (Fig. 4). This difference in cellular response could relate to frequently observed higher process rates of hydrolysis as compared to the mineralization of VFAs (Arnosti, 2011). Hydrolytic microorganisms in marine sediments can respond to increased substrate availability by synthesis of extracellular enzymes (Brüchert and Arnosti, 2003), which might even persist in sediments and enable a fast response to new OM input (Fabiano and Danovaro, 1998). It has also been speculated that glucose‐utilizing fermenters in arctic marine sediments have substantially higher growth yields than acetate‐utilizing SRM (Kirchman et al., 2014). SRM in marine sediments require several days to upregulate their VFA turnover (Arnosti et al., 2005). The calculated energy yield from acetoclastic sulfate reduction at in situ conditions in Smeerenburgfjorden sediment (Supporting Information Material and Methods; Table S3) was −93.3 kJ mol−1, which is much higher than the energetic limit of this process of −30 kJ mol−1 (Jin and Bethke, 2009; Glombitza et al., 2015). Thus, SRM in our sediment microcosms seemed not to be energy‐limited even at low in situ acetate concentrations. However, most of the substrate might be funnelled through catabolic pathways for energy conservation as opposed to also supplying elements and energy for anabolism and thus growth and cell division (Rabus et al., 2013). Additionally, doubling times of previously isolated, psychrophilic SRM at 0°C are considerably lower than at their optimal growth temperatures, i.e., 30–170 h at 7–18°C (Knoblauch and Jørgensen, 1999). Cell physiological constraints of SRM might further limit the uptake of substrate from the sediment and intracellular activation (Schauder et al., 1986; Glombitza et al., 2015), and also adsorption of VFAs by the sediment matrix can significantly reduce substrate uptake and mineralization (Shaw et al., 1984; Finke et al., 2007). Increase in relative 16S rRNA gene abundance, i.e., putative growth of the non‐sulfate‐reducing Arcobacter phylotype in acetate incubations (Fig. 3) is conflicting. However, this may be explained by the use of the energetically more favourable electron acceptors manganese, nitrate or iron, which result in higher energy yields during OM oxidation than the use of sulfate (Froelich et al., 1979; LaRowe and Amend, 2014).
Psychrophilic fermenters of cyanobacterial necromass
Six phylotypes were considered hydrolysers/fermenters, because they showed a significant response to spirulina amendments, did not respond to the amendment of acetate, were not negatively impacted by amendment of the sulfate reduction inhibitor molybdate, and were related to described species with hydrolytic and/or fermentative metabolic capabilities (Fig. 5). Four of the phylotypes (Psychromonas phylotype 7435, Gammaproteobacteria; Psychrilyobacter phylotype 4749, Fusobacteria; Marinifilum phylotype 4400, Bacteroidetes; Clostridiales phylotype 1452, Firmicutes) grew (Fig. 3A) and incorporated spirulina‐derived 13C into their cells (Fig. 4). These phylotypes thus seemed to be primarily r‐selected (Lynch and Neufeld, 2015) and thus able to quickly take advantage and grow upon availability of larger amounts of OM that, for example, are available during spring bloom‐like events of primary producers that deposit to the seafloor upon cell death (Hebbeln and Wefer, 1991). In contrast, Colwellia phylotype 7234 (Gammaproteobacteria) and Marinilabiliaceae phylotype 9869 (Bacteroidetes) only responded by relative increase in ribosomal activity (Fig. 5). Colwellia phylotype 7234 was of high relative abundance in the native sediment (day 0) (Supporting Information Table S1), which suggests that it retains a more stable population size in periods of low nutrient supply.
Figure 5.
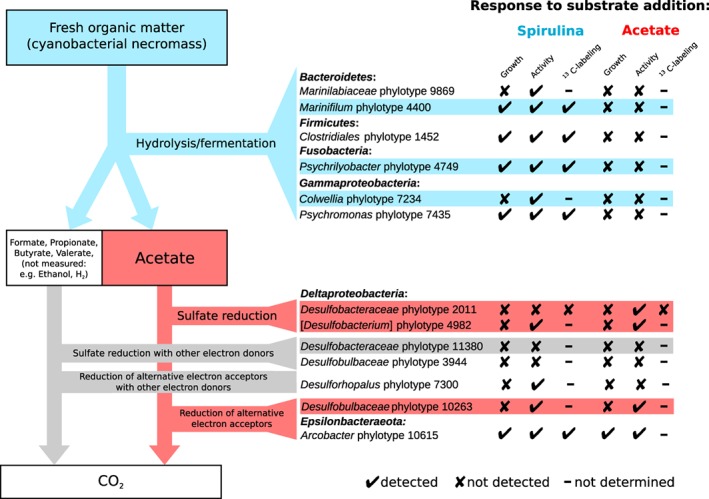
A bacterial interaction model for organic matter degradation in sulfidic arctic marine sediments of Smeerenburgfjorden, Svalbard. Fresh biomass that is introduced into the system by sedimentation is utilized by hydrolytic and fermentative bacteria and degraded to VFAs, mainly acetate, which are further utilized by SRM or microorganisms that use alternative electron acceptors. Putative roles in organic carbon degradation are shown for identified phylotypes (Fig. 3). Growth, significant response in relative 16S rRNA gene abundance. Activity, significant response in relative 16S rRNA transcript abundance. 13C‐labelling, significant 13C‐incorporation by > 50% of phylotype cells from 13C‐spirulina or 13C‐acetate as revealed by single‐cell CARD‐FISH‐Raman (Fig. 4). Responsive phylotypes that are highly abundant and/or ribosomally active in native sediment, i.e., ≥ 1% abundance in 16S rRNA gene or transcript libraries at day 0 (Supporting Information Table S1), are shaded.
Bacterial cellular biomass is composed of thousands of individual molecules and metabolites (Engelking, 2015), but amendment of spirulina necromass activated only very few members of the complex sediment microbiota, as observed in another study (Graue et al., 2012). Dry spirulina cell extract is mostly composed of crude protein (64‐74%), carbohydrates (12‐20%) and lipids (9‐14%) (Ciferri, 1983), which thus represent the dominant food sources for the responsive bacteria and is in agreement with the various fermentative capabilities of their phylogenetically most closely related species (Supporting Information Results and Discussion).
SRM utilizing acetate and other electron donors
All four phylotypes that were inhibited by molybdate, and thus identified as SRM (Fig. 5), are members of the class Deltaproteobacteria (Fig. 3B), which contains the majority of ecologically relevant SRM in marine sediments (Wasmund et al., 2017). The abundant Desulfobacteraceae phylotype 2011 belongs to the globally distributed Sva0081 group of yet uncultured sediment bacteria (Supporting Information Fig. S5). Phylotype 4982 (Desulfobulbaceae) is very closely related to [Desulfobacterium] catecholicum (99% 16S rRNA identity) (Supporting Information Fig. S5), a complete oxidizer of acetate and other VFAs (Szewzyk and Pfennig, 1987). Both phylotypes responded to acetate amendment but mainly by increased ribosomal activity (Fig. 3B). Additionally, Desulfobacteraceae phylotype 2011 incorporated only small amounts of 13C from acetate into its biomass (Fig. 4). Our results suggest that cellular activity of acetate‐utilizing SRM in arctic marine sediments is largely decoupled from an increase in their population size. We postulate that populations of SRM in marine surface sediments with constantly cold temperatures are rather stable (Robador et al., 2009), and only change slowly in response to long‐term changes in substrate availability.
Besides the two acetate‐utilizing SRM‐phylotypes, we also identified Desulfobulbaceae phylotype 3944 and Desulfobacteraceae phylotype 11380 to be suppressed in incubations with molybdate (Fig. 3). Both phylotypes did not respond significantly to either spirulina or acetate amendments, which suggests that they utilize other, endogenous electron donors for sulfate reduction (Fig. 5). Alternatively, they might have other energy metabolisms that are inhibited by molybdate, e.g., utilization of taurine (Lie et al., 1999) or disproportionation of thiosulfate or elemental sulfur (Finster et al., 1998).
Bacteria that utilize VFAs but are not strictly dependent on sulfate reduction
Three VFA‐utilizing phylotypes were not suppressed by molybdate (Fig. 5); Arcobacter phylotype 10615, Desulfobulbaceae phylotype 10263 and Desulforhopalus phylotype 7300. The first two responded to spirulina and acetate amendments (Figs 3 and 4), which indicated that both phylotypes primarily utilized the spirulina fermentation product acetate. In contrast Desulforhopalus phylotype 7300 only responded to spirulina incubations (Fig. 3A), potentially by consuming spirulina degradation intermediates other than acetate. Given that Arcobacter phylotype 10615 responded to acetate with particularly massive relative abundance increases (Fig. 3B), we hypothesize that it used electron acceptors more energetically favorable than sulfate for the oxidation of VFAs (Fig. 5). Arcobacter‐related microorganisms use manganese, iron or nitrate as electron acceptors (Thamdrup et al., 2000; Vandieken et al., 2012; Canion et al., 2013; Vandieken and Thamdrup, 2013). Also, the SRM Desulfobulbus propionicus, which is related to Desulfobulbaceae phylotype 10263 (94.4% 16S rRNA identity) (Supporting Information Fig. S5), has similar metabolic capabilities in the absence of sulfate (Dannenberg et al., 1992; Lovley and Phillips, 1994; Holmes et al., 2004). Desulforhopalus phylotype 7300 was closely related to Desulforhopalus singaporensis (Supporting Information Fig. S5), which besides sulfate can also use nitrate, sulfite and thiosulfate as electron acceptor (Lie et al., 1999). Iron and nitrate reduction contribute to organic carbon oxidation in Svalbard fjord sediments (Kostka et al., 1999; Finke et al., 2007; Canion et al., 2014) and many SRM use intermediate sulfur species for respiration (Wasmund et al., 2017). Thus, iron, nitrate or intermediate sulfur species might have been utilized by the phylotypes that were not inhibited by molybdate.
Conclusions
We investigated degradation of complex OM in sulfidic Smeerenburgfjorden sediment from the arctic archipelago of Svalbard. We propose a metabolic and species interaction model for dead biomass mineralization in the conditions studied that may be found in permanently cold and sulfate‐reduction‐dominated sediments (Fig. 5). Deposited organic biomass was primarily degraded by a limited number of species from diverse phyla. Initial hydrolysis and fermentation of mainly proteins, carbohydrates and lipids to diverse VFAs were catalyzed by Psychrilyobacter (Fusobacteria), Marinifilum and Marinilabiaceae (Bacteroidetes), Colwellia and Psychromonas (Gammaproteobacteria) and Clostridiales species (Firmicutes). Acetate, the main fermentation product, was mineralized by sulfate‐reducing species of the families Desulfobacteraceae and Desulfobulbaceae (Deltaproteobacteria) and by Arcobacter (Epsilonbacteraeota) and Desulfobulbaceae (Deltaproteobacteria) species that presumably used other electron acceptors than sulfate. Interestingly, cellular activation (i.e., increases in relative 16S rRNA transcript abundance and in single‐cell 13C‐labelling) upon availability of new substrate was coupled to relative population growth (i.e., increases in relative 16S rRNA gene abundance) in microorganisms able to directly hydrolyze/ferment cyanobacterial necromass and in the acetate‐utilizing Arcobacter species, but not in acetate‐utilizing SRM. These different cellular responses of each species have important implications for the population dynamics of the different functional guild members in marine sediments. It is tempting to speculate that the taxa identified here will play increasing roles in the future mineralization of the predicted increased phyto‐detrital organic material in arctic benthic systems (Lalande et al., 2009; Sørensen et al., 2015), and in determining the extent of burial of organic material into the subsurface.
Experimental procedures
Marine sediment sampling
Arctic marine sediment samples were collected from Smeerenburgfjorden, Svalbard (station J; 79°43′N, 11°05′E; water depth 216 m) in August 2011. Sediment from 5 to 10 cm depth was collected from several HAPS cores (KC Denmark A/S, Silkeborg, Denmark) (Kanneworff and Nicolaisen, 1972), transferred into gas‐tight plastic bags (Hansen et al., 2000) and stored on ice at 0°C for up to 2.5 months prior to incubations.
Sediment incubations
Approximately 2 l of anoxic sediment slurry was prepared per set of substrate and concentrations (Supporting Information Fig. S1A) by homogenizing sediment with anoxic bottom water from station J at a 1:1 (w/w) ratio under constant flow of N2 gas. 200 ml of slurry was distributed into 250 ml serum bottles, amended with different substrates, sealed with butyl rubber stoppers (Geo‐Microbial Technologies) and incubated at 0°C (Supporting Information Fig. S1A). Incubations were performed with either acetate or spirulina (Sigma‐Aldrich, Steinheim, Germany). Spirulina consists of freeze‐dried cells of the cyanobacterium Arthrospira platensis. Incubations were performed with two different substrate concentrations; i.e., a low dose (LD) and a high dose (HD) with 50 µg ml−1 and 1 mg ml−1 of spirulina and with 50 µM and 1 mM of acetate respectively (Supporting Information Fig. S1A). Spirulina was added as a one‐time pulse at the beginning of the experiment, whereas acetate was continuously added every four to seven days (Supporting Information Fig. S1B) in order to compensate for the supposedly high acetate turnover (Finke et al., 2007). LD acetate supplementation was in the range of concentrations measured in situ in Svalbard sediments (Finke et al., 2007). Parallel incubations were performed with 13C‐labeled acetate or 13C‐labelled spirulina (both Sigma Aldrich, 99 atom% 13C), 12C‐substrates (unlabeled control), 13C‐labelled substrate in combination with 5 mM molybdate as a sulfate‐reduction inhibitor (inhibited control) and an unamended control incubation (no‐substrate control) (Supporting Information Fig. S1A). Molybdate was added at day 0 and after 3 weeks of incubation. At days 0, 4, 8, 13, 20, 25, 32 and 39 after the start of the incubations, subsamples were taken through the rubber stopper with a sterile 16‐gauge needle (Braun) attached to a 10 ml Omnifix syringe (Braun). At each sampling time point, a subsample of 6.5 ml was taken and stored either at −80°C for VFA and nucleic acid analyses or fixed for CARD‐FISH analyses as described below.
Biogeochemical analyses
Concentrations of the VFAs acetate, formate, propionate, butyrate, valerate, lactate and pyruvate were measured in the supernatant by 2‐dimensional ion chromatography‐mass spectrometry (IC‐IC‐MS; Dionex ICS‐3000 coupled to an MSQ Plus™, both Thermo Scientific), equipped with an Ion Pack™ AS 24 as the first column to separate the VFAs from chloride, and an Ion Pack™ AS 11 HC as the second column) (Glombitza et al., 2014). Prior to the analyses, samples were thawed and filtered through disposable syringe filters (Acrodisc™ 13 mm, IC grade, polyethersulfone (PES) membrane with 0.2 µM pore size), that were previously cleaned by rinsing with 10 ml Milli‐Q water (Utrapure, Type 1) (Glombitza et al., 2014). Both isotopic variants of acetate were analyzed in parallel in separate selected ion monitoring (SIM) channels, i.e., 12C‐acetate (m/z 59) and 13C‐acetate (m/z 61). The Gibbs Energy of acetoclastic sulfate reduction was calculated as described in Supporting Information Materials and Methods. Parallel incubations were set up with 35S‐labelled carrier‐free sulfate tracer and sulfate reduction rates were determined using a single‐step cold chromium distillation method (Kallmeyer et al., 2004). Significance of SRR differences between no substrate controls and substrate amended incubations were determined by Wilcoxon tests using the function wilcox.test() from the R statistical package (R Core Team, 2015).
16S rRNA gene and transcript sequence analyses
Nucleic acid extraction and preparation of 16S rRNA gene and transcript libraries is described in Supporting Information Material and Methods. Sequencing was performed on a GS FLX + instrument using Titanium chemistry (Roche, Mannheim, Germany) by Eurofins MWG Operon (Ebersberg, Germany). Sequencing reads were filtered using mothur's implementation of PyroNoise (Schloss et al., 2009; Quince et al., 2011) and clustered into phylotypes of approximate species‐level at 97% sequence identity using UCLUST (Edgar, 2010) as evaluated previously (Berry et al., 2012). Representative sequences (mean length of 220 bp) were aligned with mothur using default settings. Chimeras that were detected with Chimera Slayer (Haas et al., 2011) were excluded from further analysis. Altogether we obtained 344,867 high quality reads (on average 4732 reads per sample) that clustered into 11,548 species‐level phylotypes. Phylum/class‐level classification was performed using mothur's implementation of the Ribosomal Database Project naïve Bayesian classifier (Wang et al., 2007; Schloss et al., 2009) with default settings. Note that reclassification of the class Epsilonproteobacteria into the new phylum Epsilonbacteraeota was recently proposed (Waite et al., 2017). Alpha diversity metrics were calculated (Caporaso et al., 2010) with re‐sampling (100 re‐samples) at 3250 reads to avoid sample based artifacts (Lozupone et al., 2011). Principal coordinates analysis (PCoA) was performed in the R software environment (R Core Team, 2015) on the re‐sampled phylotype table based on Bray‐Curtis dissimilarities using the software package phyloseq (McMurdie and Holmes, 2013). Phylotypes with a relative abundance of ≥ 1% of the community in at least one sample were aligned against the SILVA database SSU_Ref_NR99_128 (Quast et al., 2013) using SINA (Pruesse et al., 2012). A phylogenetic tree was calculated with FastTree (Price et al., 2009) using an alignment of closely related sequences that were selected from the non‐redundant SILVA database SSU_Ref_NR99_128 (Quast et al., 2013). Phylotype sequences were subsequently added to the resulting tree using the EPA algorithm (Berger et al., 2011) in RAxML (Stamatakis, 2014). Trees were visualized using iTOL (Letunic and Bork, 2016). The sequence similarity between phylotypes and representative strains in the non‐redundant SILVA database SSU_Ref_NR99_128 (Quast et al., 2013) was calculated using t_coffe (Notredame et al., 2000). Phylotypes associated with the utilization of the added substrate at a given dose or with sulfate reduction had to be significantly enriched in at least two 16S rRNA gene and/or transcript libraries (n = 4 for substrate utilization, n = 8 for sulfate reduction) compared to the respective control (no substrate control or inhibited control). Significant enrichment of phylotypes was determined using a two‐proportion T‐test (Müller et al., 2014; Robador et al., 2016). P‐values were corrected for multiple comparisons using the Benjamini‐Hochberg false discovery rate method with p.adjust() from the R statistical package (R Core Team, 2015). Corrected P‐values of ≤ 0.01 were considered significant.
Sequence data
16S rRNA gene and transcript sequence data is available in the NCBI Short Read Archive under accession number SRP133170.
Catalyzed reporter deposition‐fluorescence in situ hybridization coupled with raman microspectroscopy (CARD‐FISH‐raman)
Microbial cells were extracted from arctic marine sediment as described in Supporting Information Materials and Methods. The extracted cells were collected by filtration on polycarbonate filters (GTTP type, pore size 0.2 µm, Millipore) and identified by CARD‐FISH or two‐step CARD‐FISH (Supporting Information Materials and Methods). Hybridized cells were extracted from the filters (Supporting Information Materials and Methods) and spotted onto an aluminum‐coated slide (A1136; EMF Corporation) for analysis on a LabRAM HR800 confocal Raman microscope (Horiba Jobin‐Yvon) as outlined previously (Huang et al., 2007). The obtained single cell spectra were baseline‐corrected using a polynomial function. Spectra were then aligned according to the phenylalanine peak region and normalized by dividing the spectral intensity at each wavelength by the total spectral intensity. Ratios between the height of the 13C‐phenylalanine peak (wavelength of 960–970 cm−1) and the height of the 12C‐phenylalanine peak (wavelength of 1000–1005 cm−1) were calculated. Significance of the peak height ratio differences between 13C‐labeled probe‐targeted cell populations and negative controls were determined in the R software environment (R Core Team, 2015) with wilcoxon.test(). Testing for spirulina uptake was performed between probe‐targeted cells in 13C‐spirulina incubations and populations of cells randomly selected from 12C‐spirulina incubations. Testing for acetate uptake by Desulfobacteraceae phylotype 2011 was performed between populations from 13C‐acetate and 12C‐acetate incubations respectively. The threshold for detection of 13C‐labeled cells was calculated by adding three times the standard deviation of the negative (12C) control to the mean of the negative control.
Supporting information
Fig. S1. Experimental overview.
A. Setup of anoxic sediment incubations.
B. Timeline for substrate additions and sampling. LD, low substrate dose (50 μg ml‐1 spirulina and 50 μM acetate). HD, high substrate dose (1 mg ml‐1 spirulina and 1 mM acetate). SR‐inhibitor, the sulfate reduction inhibitor molybdate.
Fig. S2. Volatile fatty acids concentrations in acetate incubations. Concentrations of volatile fatty acids in (A) LD and (B) HD incubations with acetate. 13C‐inhibited, sediment incubations with 13C‐substrate and molybdate. Note that acetate was periodically added (black arrows) right before the measurement. LD, low dose (50 μM) of acetate. HD, high dose (1 mM) of acetate. Note that the scales are different for each treatment and each VFA.
Fig. S3. Principal coordinates analysis of bacterial beta‐diversity in anoxic sediment incubations. The analyses were based on Bray‐Curtis dissimilarities of relative 16S rRNA gene (DNA) and transcript (RNA) abundances of bacterial phylotypes. Sample colour indicates type and concentration of added substrate. Sample shape indicates the day of sampling.
Fig. S4. Bacterial community dynamics in spirulina and acetate amended anoxic sediment incubations. Only phyla/classes with a relative 16S rRNA gene (DNA) and/or transcript (RNA) abundance of ≥ 1% at day 0 are indicated. 13C., sediment incubations with 13C‐substrate. 13Ci., sediment incubations with 13C‐substrate and molybdate. 12C., sediment incubations with 12C‐substrate. None., no‐substrate control. LD, low substrate dose (50 μg ml−1 spirulina or 50 μm acetate). HD, high substrate dose (1 mg ml−1 spirulina or 1 mM acetate).
Fig. S5. Phylogeny of abundant phylotypes. Only phylotypes with ≥ 1% relative 16S rRNA gene or transcript abundance in at least one incubation sample are shown. The tree was calculated using FastTree (Price et al., 2009) and an alignment of close relatives of phylotypes selected from the SILVA database SSU_Ref_NR99_128 (Quast et al., 2013). Short amplicon sequences were subsequently aligned with SINA (Pruesse et al., 2012) and added to the reference tree without changing its topology using the EPA algorithm (Berger et al., 2011) in RAxML (Stamatakis, 2014). Numbers in parentheses indicate average relative 16S rRNA gene/transcript abundances at day 0. Blue and red squares indicate significant enrichment (P ≤ 0.01) in spirulina and acetate incubations respectively, compared to the no substrate control. Numbers within the squares give the number of samples (of 24 spirulina and 22 acetate incubation samples in total) in which the phylotype was significantly enriched. Yellow squares indicate sulfate‐reduction‐associated phylotypes, numbers in the square indicate numbers of samples (of 15 in total) in which the phylotype was significantly enriched in uninhibited incubations with substrate compared to molybdate‐inhibited controls.
Fig. S6. Raman spectra of responsive phylotypes and their relative sequence abundance.
A. Overlays of single cell Raman spectra are displayed for the CARD‐FISH‐ probe‐labelled phylotypes in 13C‐spirulina (1 mg ml−1) or 13C‐acetate (1 mM) incubations. Clostridiales phylotype 1452 (Firmicutes), Marinifilum phylotype 4400 (Bacteroidetes), Psychrilyobacter phylotype 4749 (Fusobacteria), Psychromonas phylotype 7435 (Gammaproteobacteria), Arcobacter phylotype 10615 (Epsilonbacteraeota) and Desulfobacteraceae phylotype 2011 (Deltaproteobacteria). Random, Raman spectra from randomly selected cells in the 13C‐spirulina (1 mg ml−1) incubations. Negative 13C‐spirulina, Raman spectra from randomly selected cells from the 12C‐spirulina incubations. Negative 13C‐ acetate, Raman spectra from cells of Desulfobacteraceae phylotype 2011 (Deltaproteobacteria) from incubations with 12C‐acetate at day 0. Insets show enlarged Raman spectrum regions containing 13C‐phenylalanine (wavelength of 960–970 cm−1) and 12C‐phenylalanine (wavelength of 1000–1005 cm−1) peaks.
B. Relative 16S rRNA gene and transcript abundance of CARD‐FISH targeted cell populations in the respective substrate supplemented sediment incubation.
Table S1. Relative abundances of phylotypes with ≥ 1% of all bacterial 16S rRNA genes or transcripts at day 0.
Table S2. Read number, coverage and alpha‐diversity of bacterial 16S rRNA gene and transcript libraries.
Table S3. Data for calculation of in situ Gibbs energy of acetoclastic sulfate reduction.
Table S4. Thermodynamic properties used for the calculation of standard state Gibbs energy change of reaction [DG0 (T, p)]. Using the SUPCRT92 software (Johnson et al., 1992). References: (a) Schock, (1995) and (b) Schock and Helgeson (1988).
Table S5. 16S rRNA‐targeted FISH probes.
Acknowledgements
We thank Laura Wehrmann for the excellent organization of the 2011 Svalbard cruise, Kristian Lund (captain) and Klaus Ryberg (first mate) of the MS Farm and cruise participants Carol Arnosti, Andy Canion, Patrick Chanton and Kolja Kindler for their assistance with sample collection. Furthermore, we thank Marc Mußmann for provide us with the CARD‐FISH probe DSS1431. This work was financially supported by the Austrian Science Fund (FWF, P29426‐B29 to KW; P25111‐B22 to AL). The authors declare no conflict of interest.
References
- Arndt, S. , Jørgensen, B.B. , LaRowe, D.E. , Middelburg, J.J. , Pancost, R.D. , and Regnier, P. (2013) Quantifying the degradation of organic matter in marine sediments: A review and synthesis. Earth‐Sci Rev 123: 53–86. [Google Scholar]
- Arnosti, C. (2011) Microbial extracellular enzymes and the marine carbon cycle. Ann Rev Mar Sci 3: 401–425. [DOI] [PubMed] [Google Scholar]
- Arnosti, C. , Finke, N. , Larsen, O. , and Ghobrial, S. (2005) Anoxic carbon degradation in Arctic sediments: Microbial transformations of complex substrates. Geochim Cosmochim Acta 69: 2309–2320. [Google Scholar]
- Arnosti, C. , and Jørgensen, B.B. (2003) High activity and low temperature optima of extracellular enzymes in Arctic sediments: Implications for carbon cycling by heterotrophic microbial communities. Mar Ecol Prog Ser 249: 15–24. [Google Scholar]
- Arnosti, C. , and Jørgensen, B.B. (2006) Organic carbon degradation in Arctic marine sediments, Svalbard: A comparison of initial and terminal steps. Geomicrobiol J 23: 551–563. [Google Scholar]
- Arrigo, K.R. , van Dijken, G. , and Pabi, S. (2008) Impact of a shrinking Arctic ice cover on marine primary production. Geophys Res Lett 35: L19603. [Google Scholar]
- Berger, S.A. , Krompass, D. , and Stamatakis, A. (2011) Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol 60: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, D. , Schwab, C. , Milinovich, G. , Reichert, J. , Ben Mahfoudh, K. , and Decker, T. (2012) Phylotype‐level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J 6: 2091–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles, M.W. , Mogollón, J.M. , Kasten, S. , Zabel, M. , and Hinrichs, K.‐U. (2014) Global rates of marine sulfate reduction and implications for sub‐sea‐floor metabolic activities. Science 344: 889–891. [DOI] [PubMed] [Google Scholar]
- Brüchert, V. , and Arnosti, C. (2003) Anaerobic carbon transformation: experimental studies with flow‐through cells. Mar Chem 80: 171–183. [Google Scholar]
- Burdige, D.J. (2007) Preservation of organic matter in marine sediments: Controls, mechanisms, and an imbalance in sediment organic carbon budgets? Chem Rev 107: 467–485. [DOI] [PubMed] [Google Scholar]
- Canion, A. , Overholt, W.A. , Kostka, J.E. , Huettel, M. , Lavik, G. , and Kuypers, M.M.M. (2014) Temperature response of denitrification and anaerobic ammonium oxidation rates and microbial community structure in Arctic fjord sediments. Environ Microbiol 16: 3331–3344. [DOI] [PubMed] [Google Scholar]
- Canion, A. , Prakash, O. , Green, S.J. , Jahnke, L. , Kuypers, M.M.M. , and Kostka, J.E. (2013) Isolation and physiological characterization of psychrophilic denitrifying bacteria from permanently cold Arctic fjord sediments (Svalbard, Norway). Environ Microbiol 15: 1606–1618. [DOI] [PubMed] [Google Scholar]
- Caporaso, J.G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F.D. , and Costello, E.K. (2010) QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavicchioli, R. (2016) On the concept of a psychrophile. ISME J 10: 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri, O. (1983) Spirulina, the edible microorganism. Microbiol Rev 47: 551–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg, S. , Kroder, M. , Dilling, W. , and Cypionka, H. (1992) Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate‐reducing bacteria. Arch Microbiol 158: 93–99. [Google Scholar]
- Dunne, J.P. , Sarmiento, J.L. , and Gnanadesikan, A. (2007) A synthesis of global particle export from the surface ocean and cycling through the ocean interior and on the seafloor. Global Biogeochem Cycles 21. [Google Scholar]
- Edgar, R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Engelking, L.R. (2015) Chemical composition of living cells In Textbook of Veterinary Physiological Chemistry. New York, USA: Elsevier, pp. 2–6. [Google Scholar]
- Fabiano, M. , and Danovaro, R. (1998) Enzymatic activity, bacterial distribution, and organic matter composition in sediments of the Ross Sea (Antarctica). Appl Environ Microbiol 64: 3838–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke, N. , Vandieken, V. , and Jã¸Rgensen, B.B. (2007) Acetate, lactate, propionate, and isobutyrate as electron donors for iron and sulfate reduction in Arctic marine sediments, Svalbard. FEMS Microbiol Ecol 59: 10–22. [DOI] [PubMed] [Google Scholar]
- Finster, K. , Liesack, W. , and Thamdrup, B. (1998) Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol 64: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich, P.N. , Klinkhammer, G.P. , Bender, M.L. , Luedtke, N.A. , Heath, G.R. , Cullen, D. , et al (1979) Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: Suboxic diagenesis. Geochim Cosmochim Acta 43: 1075–1090. [Google Scholar]
- Glombitza, C. , Jaussi, M. , Røy, H. , Seidenkrantz, M.‐S. , Lomstein, B.A. , and Jørgensen, B.B. (2015) Formate, acetate, and propionate as substrates for sulfate reduction in sub‐arctic sediments of Southwest Greenland. Front Microbiol 6: 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glombitza, C. , Pedersen, J. , Røy, H. , and Jørgensen, B.B. (2014) Direct analysis of volatile fatty acids in marine sediment porewater by two‐dimensional ion chromatography‐mass spectrometry. Limnol Oceanogr Methods 12: 455–468. [Google Scholar]
- Graue, J. , Engelen, B. , and Cypionka, H. (2012) Degradation of cyanobacterial biomass in anoxic tidal‐flat sediments: A microcosm study of metabolic processes and community changes. ISME J 6: 660–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Gevers, D. , Earl, A.M. , Feldgarden, M. , Ward, D.V. , Giannoukos, G. , et al (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454‐pyrosequenced PCR amplicons. Genome Res 21: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J.W. , Thamdrup, B. , and Jørgensen, B.B. (2000) Anoxic incubation of sediment in gas‐tight plastic bags: A method for biogeochemical process studies. Mar Ecol Prog Ser 208: 273–282. [Google Scholar]
- Hausmann, B. , Knorr, K.‐H. , Schreck, K. , Tringe, S.G. , Glavina Del Rio, T. , Loy, A. , and Pester, M. (2016) Consortia of low‐abundance bacteria drive sulfate reduction‐dependent degradation of fermentation products in peat soil microcosms. ISME J 10: 2365–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbeln, D. , and Wefer, G. (1991) Effects of ice coverage and ice‐rafted material on sedimentation in the Fram Strait. Nature 350: 409–411. [Google Scholar]
- Hedges, J.I. , and Keil, R.G. (1995) Sedimentary organic matter preservation: An assessment and speculative synthesis. Mar Chem 49: 81–115. [Google Scholar]
- Holmes, D.E. , Bond, D.R. , and Lovley, D.R. (2004) Electron transfer by Desulfobulbus propionicus to Fe(III) and graphite electrodes. Appl Environ Microbiol 70: 1234–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.E. , Stoecker, K. , Griffiths, R. , Newbold, L. , Daims, H. , Whiteley, A.S. , and Wagner, M. (2007) Raman‐FISH: Combining stable‐isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol 9: 1878–1889. [DOI] [PubMed] [Google Scholar]
- Jin, Q. , and Bethke, C.M. (2009) Cellular energy conservation and the rate of microbial sulfate reduction. Geology 37: 1027–1030. [Google Scholar]
- Jørgensen, B.B. (1982) Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 296: 643–645. [Google Scholar]
- Jørgensen, B.B. , and Boetius, A. (2007) Feast and famine–microbial life in the deep‐sea bed. Nat Rev Microbiol 5: 770–781. [DOI] [PubMed] [Google Scholar]
- Jørgensen, B.B. , Dunker, R. , Grünke, S. , and Røy, H. (2010) Filamentous sulfur bacteria, Beggiatoa spp., in arctic marine sediments (Svalbard, 79 degrees N). FEMS Microbiol Ecol 73: 500–513. [DOI] [PubMed] [Google Scholar]
- Kallmeyer, J. , Ferdelman, T.G. , Weber, A. , Fossing, H. , and Jørgensen, B.B. (2004) A cold chromium distillation procedure for radiolabeled sulfide applied to sulfate reduction measurements. Limnol Oceanogr Methods 2: 171–180. [Google Scholar]
- Kallmeyer, J. , Pockalny, R. , Adhikari, R.R. , Smith, D.C. , and D'Hondt, S. (2012) Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci U S A 109: 16213–16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneworff, E. , and Nicolaisen, W. (1972) The “Haps” a frame‐supported bottom corer. Ophelia 10: 119–128. [Google Scholar]
- Kirchman, D.L. , Hanson, T.E. , Cottrell, M.T. , and Hamdan, L.J. (2014) Metagenomic analysis of organic matter degradation in methane‐rich Arctic Ocean sediments. Limnol Oceanogr 59: 548–559. [Google Scholar]
- Knoblauch, C. , and Jørgensen, B.B. (1999) Effect of temperature on sulphate reduction, growth rate and growth yield in five psychrophilic sulphate‐reducing bacteria from Arctic sediments. Environ Microbiol 1: 457–467. [DOI] [PubMed] [Google Scholar]
- Knoblauch, C. , Sahm, K. , and Jørgensen, B.B. (1999) Psychrophilic sulfate‐reducing bacteria isolated from permanently cold arctic marine sediments: Description of Desulfofrigus oceanense gen. nov., sp. nov., Desulfofrigus fragile sp. nov., Desulfofaba gelida gen. nov., sp. nov., Desulfotalea psychrophila gen. nov., sp. nov. and Desulfotalea arctica sp. nov. Int J Syst Bacteriol 49: 1631–1643. [DOI] [PubMed] [Google Scholar]
- Kostka, J.E. , Thamdrup, B. , Glud, R.N. , and Canfield, D.E. (1999) Rates and pathways of carbon oxidation in permanently cold Arctic sediments. Mar Ecol Prog Ser 180: 7–21. [Google Scholar]
- Lalande, C. , Bélanger, S. , and Fortier, L. (2009) Impact of a decreasing sea ice cover on the vertical export of particulate organic carbon in the northern Laptev Sea, Siberian Arctic Ocean. Geophys Res Lett 36: 1–5. [Google Scholar]
- LaRowe, D. , and Amend, J. (2014) Energetic constraints on life in marine deep sediments In Life in Extreme Environments: Microbial Life of the Deep Biosphere. Kallmeyer J., and Wagner D. (eds). Berlin: De Gruyter, pp. 279–302. [Google Scholar]
- Letunic, I. , and Bork, P. (2016) Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie, T.J. , Clawson, M.L. , Godchaux, W. , and Leadbetter, E.R. (1999) Sulfidogenesis from 2‐aminoethanesulfonate (taurine) fermentation by a morphologically unusual sulfate‐reducing bacterium, Desulforhopalus singaporensis sp. nov. Appl Environ Microbiol 65: 3328–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley, D.R. , and Phillips, E.J. (1994) Novel processes for anaerobic sulfate production from elemental sulfur by sulfate‐reducing bacteria. Appl Environ Microbiol 60: 2394–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, C. , Lladser, M.E. , Knights, D. , Stombaugh, J. , and Knight, R. (2011) UniFrac: An effective distance metric for microbial community comparison. ISME J 5: 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M.D.J. , and Neufeld, J.D. (2015) Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13: 217–229. [DOI] [PubMed] [Google Scholar]
- McMurdie, P.J. , and Holmes, S. (2013) phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, A.L. , de Rezende, J.R. , Hubert, C.R.J. , Kjeldsen, K.U. , Lagkouvardos, I. , Berry, D. , et al (2014) Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents. ISME J 8: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer, G. , and Stams, A.J.M. (2008) The ecology and biotechnology of sulphate‐reducing bacteria. Nat Rev Microbiol 6: 441–454. [DOI] [PubMed] [Google Scholar]
- Notredame, C. , Higgins, D.G. , and Heringa, J. (2000) T‐Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217. [DOI] [PubMed] [Google Scholar]
- van Nugteren, P. , Moodley, L. , Brummer, G.‐J. , Heip, C.H.R. , Herman, P.M.J. , and Middelburg, J.J. (2009) Seafloor ecosystem functioning: The importance of organic matter priming. Mar Biol 156: 2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi, W.D. , Richards, T.A. , and Francis, W.R. (2018) Predicted microbial secretomes and their target substrates in marine sediment. Nat Microbiol 3: 32–37. [DOI] [PubMed] [Google Scholar]
- Price, M.N. , Dehal, P.S. , and Arkin, A.P. (2009) FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse, E. , Peplies, J. , and Glöckner, F.O. (2012) SINA: Accurate high‐throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , Peplies, J. , and Glöckner, F.O. (2013) The SILVA ribosomal RNA gene database project: Improved data processing and web‐based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince, C. , Lanzen, A. , Davenport, R.J. , and Turnbaugh, P.J. (2011) Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus, R. , Hansen, T.A. , and Widdel, F. (2013) Dissimilatory sulfate‐ and sulfur‐reducing prokaryotes In The Prokaryotes. Dworkin M., Falkow S., Rosenberg E., Schleifer K.‐H., Stackebrandt E. (eds). New York: Springer, pp. 309–404. [Google Scholar]
- Ravenschlag, K. , Sahm, K. , Pernthaler, J. , and Amann, R. (1999) High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol 65: 3982–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenschlag, K. , Sahm, K. , Knoblauch, C. , Jørgensen, B.B. , and Amann, R. (2000) Community structure, cellular rRNA content, and activity of sulfate‐reducing bacteria in marine arctic sediments. Appl Environ Microbiol 66: 3592–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenschlag, K. , Sahm, K. , and Amann, R. (2001) Quantitative molecular analysis of the microbial community in marine arctic sediments (Svalbard). Appl Environ Microbiol 67: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2015) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; URL: http://www.r-project.org/ [Google Scholar]
- Robador, A. , Brüchert, V. , and Jørgensen, B.B. (2009) The impact of temperature change on the activity and community composition of sulfate‐reducing bacteria in arctic versus temperate marine sediments. Environ Microbiol 11: 1692–1703. [DOI] [PubMed] [Google Scholar]
- Robador, A. , Müller, A.L. , Sawicka, J.E. , Berry, D. , Hubert, C.R.J. , Loy, A. , et al (2016) Activity and community structures of sulfate‐reducing microorganisms in polar, temperate and tropical marine sediments. ISME J 10: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm, K. , Knoblauch, C. , and Amann, R. (1999) Phylogenetic affiliation and quantification of psychrophilic sulfate‐reducing isolates in marine Arctic sediments. Appl Environ Microbiol 65: 3976–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka, J.E. , Jørgensen, B.B. , and Brüchert, V. (2012) Temperature characteristics of bacterial sulfate reduction in continental shelf and slope sediments. Biogeosci Discuss 9: 3425–3700. [Google Scholar]
- Schauder, R. , Eikmanns, B. , Thauer, R.K. , Widdel, F. , and Fuchs, G. (1986) Acetate oxidation to CO2 in anaerobic bacteria via a novel pathway not involving reactions of the citric acid cycle. Arch Microbiol 145: 162–172. [Google Scholar]
- Schloss, P.D. , Westcott, S.L. , Ryabin, T. , Hall, J.R. , Hartmann, M. , Hollister, E.B. , et al (2009) Introducing mothur: Open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, D.G. , Alperin, M.J. , Reeburgh, W.S. , and McIntosh, D.J. (1984) Biogeochemistry of acetate in anoxic sediments of Skan Bay, Alaska. Geochim Cosmochim Acta 48: 1819–1825. [Google Scholar]
- Sørensen, H.L. , Meire, L. , Juul‐Pedersen, T. , de Stigter, H.C. , Meysman, F. , Rysgaard, S. , et al (2015) Seasonal carbon cycling in a Greenlandic fjord: An integrated pelagic and benthic study. Mar Ecol Prog Ser 539: 1–17. [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewzyk, R. , and Pfennig, N. (1987) Complete oxidation of catechol by the strictly anaerobic sulfate‐reducing Desulfobacterium catecholicum sp. nov. Arch Microbiol 147: 163–168. [Google Scholar]
- Teske, A. , Durbin, A. , Ziervogel, K. , Cox, C. , and Arnosti, C. (2011) Microbial community composition and function in permanently cold seawater and sediments from an arctic fjord of svalbard. Appl Environ Microbiol 77: 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamdrup, B. , Rosselló‐Mora, R. , and Amann, R. (2000) Microbial manganese and sulfate reduction in Black Sea shelf sediments. Appl Environ Microbiol 66: 2888–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandieken, V. , Pester, M. , Finke, N. , Hyun, J.‐H. , Friedrich, M.W. , Loy, A. , and Thamdrup, B. (2012) Three manganese oxide‐rich marine sediments harbor similar communities of acetate‐oxidizing manganese‐reducing bacteria. ISME J 6: 2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandieken, V. , and Thamdrup, B. (2013) Identification of acetate‐oxidizing bacteria in a coastal marine surface sediment by RNA‐stable isotope probing in anoxic slurries and intact cores. FEMS Microbiol Ecol 84: 373–386. [DOI] [PubMed] [Google Scholar]
- Vaughan, D.G. , Comiso, J.C. , Allison, I. , Carrasco, J. , Kaser, G. , Kwok, R. , et al (2013) Observations: Cryosphere In Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge. [Google Scholar]
- Waite, D.W. , Vanwonterghem, I. , Rinke, C. , Parks, D.H. , Zhang, Y. , Takai, K. , et al (2017) Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol 8: 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F.‐Q. , Lin, X.‐Z. , Chen, G.‐J. , and Du, Z.‐J. (2015) Colwellia arctica sp. nov., isolated from Arctic marine sediment. Antonie Van Leeuwenhoek 107: 723–729. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Garrity, G.M. , Tiedje, J.M. , and Cole, J.R. (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmund, K. , Mußmann, M. , and Loy, A. (2017) The life sulfuric: Microbial ecology of sulfur cycling in marine sediments. Environ Microbiol Rep 9: 323–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Experimental overview.
A. Setup of anoxic sediment incubations.
B. Timeline for substrate additions and sampling. LD, low substrate dose (50 μg ml‐1 spirulina and 50 μM acetate). HD, high substrate dose (1 mg ml‐1 spirulina and 1 mM acetate). SR‐inhibitor, the sulfate reduction inhibitor molybdate.
Fig. S2. Volatile fatty acids concentrations in acetate incubations. Concentrations of volatile fatty acids in (A) LD and (B) HD incubations with acetate. 13C‐inhibited, sediment incubations with 13C‐substrate and molybdate. Note that acetate was periodically added (black arrows) right before the measurement. LD, low dose (50 μM) of acetate. HD, high dose (1 mM) of acetate. Note that the scales are different for each treatment and each VFA.
Fig. S3. Principal coordinates analysis of bacterial beta‐diversity in anoxic sediment incubations. The analyses were based on Bray‐Curtis dissimilarities of relative 16S rRNA gene (DNA) and transcript (RNA) abundances of bacterial phylotypes. Sample colour indicates type and concentration of added substrate. Sample shape indicates the day of sampling.
Fig. S4. Bacterial community dynamics in spirulina and acetate amended anoxic sediment incubations. Only phyla/classes with a relative 16S rRNA gene (DNA) and/or transcript (RNA) abundance of ≥ 1% at day 0 are indicated. 13C., sediment incubations with 13C‐substrate. 13Ci., sediment incubations with 13C‐substrate and molybdate. 12C., sediment incubations with 12C‐substrate. None., no‐substrate control. LD, low substrate dose (50 μg ml−1 spirulina or 50 μm acetate). HD, high substrate dose (1 mg ml−1 spirulina or 1 mM acetate).
Fig. S5. Phylogeny of abundant phylotypes. Only phylotypes with ≥ 1% relative 16S rRNA gene or transcript abundance in at least one incubation sample are shown. The tree was calculated using FastTree (Price et al., 2009) and an alignment of close relatives of phylotypes selected from the SILVA database SSU_Ref_NR99_128 (Quast et al., 2013). Short amplicon sequences were subsequently aligned with SINA (Pruesse et al., 2012) and added to the reference tree without changing its topology using the EPA algorithm (Berger et al., 2011) in RAxML (Stamatakis, 2014). Numbers in parentheses indicate average relative 16S rRNA gene/transcript abundances at day 0. Blue and red squares indicate significant enrichment (P ≤ 0.01) in spirulina and acetate incubations respectively, compared to the no substrate control. Numbers within the squares give the number of samples (of 24 spirulina and 22 acetate incubation samples in total) in which the phylotype was significantly enriched. Yellow squares indicate sulfate‐reduction‐associated phylotypes, numbers in the square indicate numbers of samples (of 15 in total) in which the phylotype was significantly enriched in uninhibited incubations with substrate compared to molybdate‐inhibited controls.
Fig. S6. Raman spectra of responsive phylotypes and their relative sequence abundance.
A. Overlays of single cell Raman spectra are displayed for the CARD‐FISH‐ probe‐labelled phylotypes in 13C‐spirulina (1 mg ml−1) or 13C‐acetate (1 mM) incubations. Clostridiales phylotype 1452 (Firmicutes), Marinifilum phylotype 4400 (Bacteroidetes), Psychrilyobacter phylotype 4749 (Fusobacteria), Psychromonas phylotype 7435 (Gammaproteobacteria), Arcobacter phylotype 10615 (Epsilonbacteraeota) and Desulfobacteraceae phylotype 2011 (Deltaproteobacteria). Random, Raman spectra from randomly selected cells in the 13C‐spirulina (1 mg ml−1) incubations. Negative 13C‐spirulina, Raman spectra from randomly selected cells from the 12C‐spirulina incubations. Negative 13C‐ acetate, Raman spectra from cells of Desulfobacteraceae phylotype 2011 (Deltaproteobacteria) from incubations with 12C‐acetate at day 0. Insets show enlarged Raman spectrum regions containing 13C‐phenylalanine (wavelength of 960–970 cm−1) and 12C‐phenylalanine (wavelength of 1000–1005 cm−1) peaks.
B. Relative 16S rRNA gene and transcript abundance of CARD‐FISH targeted cell populations in the respective substrate supplemented sediment incubation.
Table S1. Relative abundances of phylotypes with ≥ 1% of all bacterial 16S rRNA genes or transcripts at day 0.
Table S2. Read number, coverage and alpha‐diversity of bacterial 16S rRNA gene and transcript libraries.
Table S3. Data for calculation of in situ Gibbs energy of acetoclastic sulfate reduction.
Table S4. Thermodynamic properties used for the calculation of standard state Gibbs energy change of reaction [DG0 (T, p)]. Using the SUPCRT92 software (Johnson et al., 1992). References: (a) Schock, (1995) and (b) Schock and Helgeson (1988).
Table S5. 16S rRNA‐targeted FISH probes.


