Summary
Background
While several commercial dermoepidermal scaffolds can promote wound healing of the skin, the achievement of complete skin regeneration still represents a major challenge.
Objectives
To perform biological characterization of self‐assembled extracellular matrices (ECMs) from three different subpopulations of fibroblasts found in human skin: papillary fibroblasts (Pfi), reticular fibroblasts (Rfi) and dermal papilla fibroblasts (DPfi).
Methods
Fibroblast subpopulations were cultured with ascorbic acid to promote cell‐assembled matrix production for 10 days. Subsequently, cells were removed and the remaining matrices characterized. Additionally, in another experiment, keratinocytes were seeded on the top of cell‐depleted ECMs to generate epidermal‐only skin constructs.
Results
We found that the ECM self‐assembled by Pfi exhibited randomly oriented fibres associated with the highest interfibrillar space, reflecting ECM characteristics that are physiologically present within the papillary dermis. Mass spectrometry followed by validation with immunofluorescence analysis showed that thrombospondin 1 is preferentially expressed within the DPfi‐derived matrix. Moreover, we observed that epidermal constructs grown on DPfi or Pfi matrices exhibited normal basement membrane formation, whereas Rfi matrices were unable to support membrane formation.
Conclusions
We argue that inspiration can be taken from these different ECMs, to improve the design of therapeutic biomaterials in skin engineering applications.
Short abstract
What's already known about this topic?
There are several types of skin fibroblasts within the dermis that can be defined by their spatial location: papillary fibroblasts (Pfi), reticular fibroblasts (Rfi) and dermal papilla fibroblasts (DPfi).
Extracellular matrix (ECM) composition is distinct with regard to composition and architecture within the papillary, reticular and hair follicle dermis in vivo.
When skin is injured, dermal replacement substitutes used for tissue repair do not reflect the heterogeneity observed within the skin dermis.
What does this study add?
Self‐assembled ECMs from different subpopulations of skin fibroblasts can be generated in vitro.
Cell‐assembled ECMs made in vitro from Pfi, Rfi and DPfi reflect dermal heterogeneity seen in vivo and are morphologically, functionally and compositionally distinct from one another.
Inspiration should be taken from cell‐assembled ECMs from distinct fibroblast subpopulations, to improve the design of therapeutic biomaterials in skin engineering applications.
What is the translational message?
Cell‐assembled ECMs from DPfi and Pfi, but not Rfi, can support formation of a basement membrane in adjacent keratinocytes in vitro.
Inspiration should be taken from cell‐assembled ECMs from distinct fibroblast subpopulations, to improve the design of therapeutic biomaterials in skin engineering applications.
Linked Comment: https://doi.org/10.1111/bjd.16773.
https://doi.org/10.1111/bjd.16946 available online
Skin is a multilayered structure comprised of an underlying supporting dermis and functional epithelium at the skin surface. This is oversimplified, as within each layer there are multiple cells with different functions that contribute to tissue homeostasis. Within the epidermis, keratinocytes are the most abundant cell type; these differentiate and stratify towards the skin surface, equipping skin with its barrier function.1 Within the skin dermis the most abundant cell type is fibroblasts, the primary role of which is to secrete components of the extracellular matrix (ECM) for structural support.2 Largely unconsidered, there are several types of skin fibroblasts within the dermis, which can be defined by their spatial location,3 and exist as morphologically and functionally heterogeneous subpopulations.4, 5, 6 For example, the fibroblast cells within the dermal layer adjacent to the basement membrane and epidermis known as the papillary dermis are called papillary fibroblasts (Pfi). These are distinct from those residing within the lower reticular dermis, which are termed reticular fibroblasts (Rfi).4, 6 There are also fibroblasts with a fibrogenic tendency, which are thought to be responsible for scar deposition after wounding.5 However these are distributed in a speckled pattern throughout the reticular and papillary dermis, and are therefore harder to define by spatial location.5 If we label cells according to their location, rather than their behaviour, then Pfi reside adjacent to the basement membrane, where they direct growth and differentiation of epidermal keratinocytes.7 Comparatively, the deeper Rfi produce the bulk of the dermal ECM and are responsible for the first wave of dermal repair following a full‐thickness wound.6 In hairy skin there are also fibroblasts associated with the hair follicle, located in the dermal papilla and the connective tissue sheath.3, 8, 9, 10, 11, 12 Dermal papilla fibroblasts (DPfi) have specialized signalling properties required for hair follicle morphogenesis and coordination of hair growth.13 In the hair follicle a specialized basement membrane, termed a glassy membrane, separates the dermal papilla from the surrounding epithelial matrix.14 In addition, the human dermal papilla is rich in interstitial collagens, such as type I and type III, in addition to fibrillar matrix proteins such as fibronectin 1 (FN1), glycoproteins such as thrombospondin 1 (THBS1),14, 15, 16, 17 and proteoglycans such as versican.18 Various studies have also demonstrated that ECMs produced by Pfi and Rfi within the interfollicular dermis are distinct with regard to their composition and architecture.4, 12, 19, 20 Pfi in the upper dermis secrete ECM, which is constituted of thin, poorly organized collagen fibre bundles, whereas thick, well‐organized collagen bundles are characteristic within the lower dermis, which is produced by resident Rfi.4 The papillary dermis also has a higher ratio of collagen type III to type I, higher levels of the dermatan sulfate proteoglycan decorin, yet lower levels of the chondroitin sulfate proteoglycan versican than the reticular dermis.4, 20, 21, 22 Not only do differences between fibroblast subtypes control how they behave and interact with surrounding cell types, but the ECM secreted by fibroblasts in distinct subanatomical locations also has a key role in the regulation of these interactions. When fibroblasts are isolated from the skin and grown in culture they retain distinct transcriptional signatures despite being removed from environmental cues.23 Concomitantly with this, Pfi have been shown to synthesize more decorin than Rfi in culture, reflecting the expression pattern of decorin in the skin in vivo.20
It is important to understand fibroblast behaviour within the skin, especially in the context of wound healing, if we want to successfully modulate this process. After injury in the skin, there are coordinated processes that lead to re‐epithelialization and establishment of a new functional barrier; however, it is the repair and ECM deposition within the skin dermis that results in scar formation. Specifically, fibroblasts synthesize highly aligned and bulky ECM fibres after injury, and a consequence of this is that complete skin regeneration cannot be achieved.24, 25
After injury, or in the case of a chronic skin wound, there are a number of biological products available on the market that promote healing which have been developed by combining primary skin cells with biomaterials.26, 27, 28, 29 These skin substitutes are biologically active through the release of growth factors and cytokines that aid the recruitment and adhesion of host cells.30, 31, 32 There are a small number of bioengineered skin products that replace both the epidermal and dermal layers of the skin; one such example is Apligraf®, which is composed of neonate‐derived fibroblasts cultured in a bovine collagen matrix, over which neonate‐derived keratinocytes are seeded to produce a stratified epidermis. In recent years, a group of biological scaffolds comprised of ECMs that are used solely as a dermal replacement have received increasing interest.33, 34, 35, 36 One of these is Integra®, a dermal regeneration template composed of a layer of bovine tendon collagen type I matrix and shark chondroitine‐6‐sulfate juxtaposed against a silicone layer that acts as a temporary pseudo‐epidermis.37, 38 Despite the success of Integra, there are some limitations. Two operations are necessary, and there are risks of infection under the silicone layer and a risk that the silicone will become detached.39 An alternative to this is MatriDerm®, which is an engineered dermal template specially developed to provide a one‐step grafting procedure. MatriDerm is a scaffold consisting of a native bovine type I, III and V collagen fibre template incorporating elastin hydrolysate that is converted into native host collagen within weeks of application.40 In addition to engineered scaffolds, decellularized skin dermis from cadaveric donors is now becoming more popular as a scaffold for use after injury where there is extensive skin loss.41 The common feature of all these decellularized dermal scaffolds is that the native collagen fibres guide fibroblasts and possibly other cells toward dermal regeneration, whereas the presence of elastin in the collagen matrix diminishes the formation of granulation tissue in the early phase of wound healing.42 As a result, a high‐quality neodermis with randomly organized collagen bundles can be regenerated. These decellularized dermal scaffolds are commonly taken from reticular dermis rather than papillary dermis. Reticular dermis is much thicker than papillary dermis, and therefore provides a greater area for cell infiltration. In addition, its relatively low cellularity in comparison with the papillary dermis means it is easy to decellularize. Lastly, the reticular dermis shows a strong mechanical resistance when compared with papillary dermis, making it easy to handle.43 Despite these advantages, decellularized reticular dermis does not have a basal membrane and this feature can make re‐epithelialization of grafts by host keratinocytes cells more difficult.44
As highlighted above, one problem with dermal replacement scaffolds is they do not reflect the heterogeneity observed within the skin dermis, where different fibroblasts subtypes, including the ones described herein, have divergent functions.1 Furthermore, with decellularized scaffolds there can be issues associated with inflammation, and these foreign body reactions have led scientists to investigate new strategies based on in vitro organogenesis approaches.45 A therapeutic strategy such as this requires cells to first be obtained from their native tissue, then kept in long‐term culture with appropriate growth factors to produce a tissue substitute rich in ECM.46, 47, 48, 49 In this respect, ECM produced by cells via a self‐assembly approach has been shown to act as a key cell adhesion site and a mechanically strong scaffold‐free support for tissue engineering.50, 51, 52 Finally, cell‐derived ECM of mesenchymal cells from anatomically distinct sites, such as the bone marrow and adipose tissue, can be created in vitro and reflect the distinct ECM found in these different sites.53
Based on the above observations, in this study we decided to investigate cell‐derived ECMs as potential scaffolds for use in skin engineering. We had two main objectives: the first was to generate and biologically characterize ECMs produced by fibroblast subpopulations found both within the hair follicle dermis, and the papillary and reticular dermis of human skin. We refer to these fibroblasts using names based on their subanatomical origin. Our second aim was to evaluate how ECMs from fibroblasts in different dermal locations, which are from areas juxtaposed to basement membrane or devoid of basement membrane, can interact with and instruct basement membrane formation in epithelial only skin constructs.
Materials and methods
Human tissue samples and cell culture
Scalp skin tissues were taken from healthy donors, using Imperial College Research Ethics Committee‐approved consent forms, and used for the isolation of epidermal keratinocytes and dermal subpopulations. Specifically, Pfi and Rfi were obtained from the upper and lower regions of interfollicular dermal tissue, respectively. To isolate cells, the adipose tissue was first cut off the scalp tissue using a scalpel. The tissue was then laid flat within a Petri dish containing phosphate‐buffered saline (PBS) then, using a stereo‐dissection microscope to view the tissue, a scalpel was used to transect the dermis 100–200 μm below the epidermis. The piece of tissue containing epidermis and papillary dermis was used to isolate Pfi, whereas the deeper dermis was used for isolation of Rfi. These pieces of superficial (papillary) and deep (reticular) tissue were minced, then placed in culture in different 35‐mm dishes in Dulbecco's Modified Eagle's Medium (DMEM) containing GlutaMAX (Gibco, Invitrogen, Carlsbad, CA, U.S.A.) supplemented with 20% fetal bovine serum (FBS) (Gibco, Invitrogen) and 1% antibiotics–antimycotics (Gibco, Invitrogen). Tissue was maintained at 37 °C in a humidified atmosphere of 5% CO2/95% air for 10 days, during which time Pfi and Rfi migrated from their respective dermal explants. After migration of fibroblasts outward from the explants, cells were amplified in culture in DMEM GlutaMAX, 10% FBS and 1% antibiotics at 37 °C in a humidified atmosphere of 5% CO2/95% air, and passaged until passage 3 (P3), at which point they were used to establish cell‐derived ECMs.
Intact dermal papillae were also isolated from the same piece of scalp skin using a microdissection approach to obtain DPfi cultures from the same donors as the Pfi and Rfi.54 Follicles were transected just above the level of the dermal papilla to isolate end bulbs, which were inverted using 27‐gauge needles to remove the matrix and expose the dermal papilla. Papillae were then separated from the follicle by cutting through their stalk. For culture, eight papillae were transferred to 35‐mm dishes containing 20% FBS in DMEM GlutaMAX, with 1% antibiotics–antimycotics. Cells migrated from the papillae and when the dish was approaching confluence, DPfi were passaged at a 1 : 2 ratio using 0·5% trypsin–ethylenediaminetetraacetic acid (EDTA; Gibco, Invitrogen) for detachment. After the initial 2 weeks of culture, cells were grown in DMEM GlutaMAX, 10% FBS and 1% antibiotics. After passaging, DPfi were cultured in the same manner as Pfi and Rfi. Matched sets of cells, from three different male donors, were used for ECM generation and subsequent analysis.
To assess proliferation characteristics of fibroblast subtypes, cells were seeded at 6000 cells per cm2. After 24 and 168 h, 100 μl alamarBlue® reagent (Invitrogen) was added directly to cells in 1 mL culture medium. The cells were incubated for 3 h at 37 °C, protected from direct sunlight. After 3 h, 100 μL aliquots were taken in duplicate and their absorbance was measured at 570 nm, using 600 nm as a reference wavelength. The fluorescence intensity is proportional to cell number.
For keratinocyte isolation, skin samples were washed briefly in PBS containing 2% antibiotics–antimycotics, then incubated in Dispase (Stem Cell Technologies, Vancouver, Canada) overnight at 4 °C. The epidermis and dermis were then separated from each other with fine forceps. Next, the epidermal layer was minced with scissors and incubated in 0·5% trypsin–EDTA at 37 °C for 30 min. Digested tissue was filtered through a 70‐μm cell strainer, then cells were pelleted and resuspended in EpiLife medium with 1% antibiotics and EpiLife Defined Growth Supplement (Gibco, Invitrogen). Cells were grown at 37 °C in a humidified atmosphere of 5% CO2/95% air and used for epithelial‐only skin construct generation at P2.
Self‐assembly approach for matrix generation
To generate ECMs from dermal fibroblast subpopulations we followed previously described protocols.55 Sterile 13‐mm glass coverslips in a 24‐well plate were coated with 0·2% sterile gelatin (gelatin type B; Sigma Aldrich, St Louis, MO, U.S.A.) for 60 min at 37°C. They were washed three times with PBS, cross‐linked with 1% sterile glutaraldehyde (Sigma Aldrich) for 30 min at room temperature (RT) and again washed three times with PBS. Crosslinking was quenched with 1 mol L−1 sterile glycine in PBS for 20 min at RT, followed by three more washes in PBS. Coverslips were then incubated in growth medium (DMEM GlutaMAX, 10% FBS, 1% antibiotics) for 30 min at 37°C. Finally, coverslips were washed three more times with PBS, then used immediately. The three different subpopulations of fibroblasts (DPfi, Pfi, Rfi) were plated onto coverslips in 24‐well plates. A total of 65 000 cells (34 000 cells cm−2) were seeded into each well in DMEM GlutaMAX, 10% FBS and 1% antibiotics, and cultured overnight at 37°C, 5% CO2/95% air to achieve a confluent lawn of fibroblasts. The next day, growth medium supplemented with 50 μg mL−1 ascorbic acid (Sigma Aldrich) was added to each culture to promote self‐assembly of ECM from each cell type. Medium was replaced with fresh medium every 2 days for a total of 10 days. After 10 days of culture in ascorbic acid‐supplemented medium, cells were removed. Medium was aspirated and cells were washed once with PBS before prewarmed extraction buffer (20 mmol L−1 NH4OH, 0·5% Triton X‐100 in PBS) was added and left for 4 min to allow cell lysis. Half the buffer was carefully removed and PBS was added. The same step was repeated until no intact cells were visible. The DNA residue was digested with 10 μg mL−1 DNase I (Roche, Indianapolis, IN, U.S.A.) in PBS for 30 min at 37°C in 5% CO2, followed by two washes with PBS. Matrices were either used immediately or stored at 4 °C in PBS with 1% antibiotics for up to 4 weeks.
Immunofluorescence staining and analysis of cell‐assembled extracellular matrices
We performed immunofluorescence of specific proteins in the self‐assembled matrices using antibodies against individual ECM components. Matrices were fixed using 4% paraformaldehyde (PFA) for 20 min, followed by wash and blocking steps with 5% goat serum (Vector Laboratories, Burlingame, CA, U.S.A.) in PBS for 30 min. Primary antibodies were diluted in PBS and placed on matrices overnight at 4°C (Table 1). Secondary antibodies were used for 1 h at RT (Table 1). Finally, the matrices were washed three times in PBS and mounted on glass slides with Vectashield mounting medium (Vector Laboratories). After staining, ECMs were imaged using a Zeiss LSM‐510 inverted confocal microscope (Carl Zeiss, Jena, Germany). For each FN1‐stained sample, three random Z‐stack images of matrices were acquired; these were used for the evaluation of alignment and fibre measurements. The thickness of the ECMs was determined by taking Z‐stack images on the confocal microscope and subtracting the distance between the top and the bottom of assembled FN1 matrix. The images were subsequently processed with the ImageJ (National Institutes of Health, Bethesda, MD, U.S.A.) and Fiji (http://fiji.sc) programs. Each Z‐stack set of images was converted to a single image by using the maximum projection function. The ‘Dimensionality’ plug‐in was applied to calculate the orientation distribution of fibres, whereas the diameter of fibres and interfibrillar spaces were assessed with the ‘BoneJ’ plug‐in.56 All graphs were generated using GraphPad Prism version 6·01 (GraphPad Inc., La Jolla, CA, U.S.A.). Data are shown as the mean; error bars (±) are the SD of the mean. A one‐way anova followed by Bonferroni's correction was performed for experiments, with P < 0·05 considered significant.
Table 1.
Primary and secondary antibodies used in this study
| Antigen | Source | Catalogue number | Species (raised in) | Dilution used |
|---|---|---|---|---|
| Fibronectin | Sigma‐Aldricha | F3648‐5mL | Rabbit | 1 : 500 |
| Collagen 1 | Abcamb | ab90395 | Mouse | 1 : 300 |
| Tenascin C | Abcamb | ab6393 | Mouse | 1 : 300 |
| Collagen 6 | Abcamb | ab6588 | Rabbit | 1 : 500 |
| Thrombospondin | Abcamb | ab1823 | Mouse | 1 : 50 |
| Collagen 7 | Abcamb | ab93350 | Rabbit | 1 : 500 |
| Collagen 4 | Abcamb | ab6583 | Mouse | 1 : 50 |
| Antimouse–Alexa Fluor 488 | Molecular Probesc | A‐11001 | Goat | 1 : 200 |
| Antirabbit–Alexa Fluor 546 | Molecular Probesc | A‐11010 | Goat | 1 : 200 |
aSt Louis, MO, U.S.A.; bCambridge, U.K.; cEugene, OR, U.S.A.
Generation of epidermal‐only skin constructs
For this experiment we used fibroblasts and keratinocytes isolated as described above. To start, we placed several cell inserts (Millicell‐24 Cell Culture Insert Plate, polycarbonate, 0·4 μm; Millipore, Burlington, MA, U.S.A.) into a 60‐mm cell culture dish, and coated them using the same method for coverslip coating as described earlier. We seeded each fibroblast subpopulation at a density of 30 000 cells per insert in DMEM GlutaMAX, 10% FBS, 1% antibiotics and allowed the fibroblasts to produce ECMs for 10 days in ascorbic acid‐supplemented medium on the inside of each insert. After removing fibroblasts from these matrices, we seeded 250 000 keratinocytes inside each insert on the top of the cell‐depleted ECMs in Cnt Prime Medium (CellnTec, Bern, Switzerland). After 2 days, we removed the Cnt Prime Medium from the inserts and added 3D Barrier Medium (CellnTec) both inside and outside the inserts. The next day all 3D Barrier Medium was removed and replaced with fresh 3D Barrier Medium on the outside of the inserts only. This enabled establishment of an air–liquid interface, which is required for epithelial stratification. Finally, we kept the cells for 14 days in culture at the air–liquid interface, prior to analysis as described earlier.
Skin construct viability, immunofluorescence staining and transmission electron microscopy
We used an alamarBlue assay to assess cell viability of epidermal skin constructs after 14 days of culture. Resazurin, the active ingredient of alamarBlue, undergoes reduction on exposing cells to resorufin, a compound that is red in colour and highly fluorescent. Viable cells continuously convert resazurin to resorufin, increasing the overall fluorescence and colour of the medium surrounding cells. alamarBlue dye, at a concentration of 10% in PBS, was added to each skin construct and incubated for 4 h at 37 °C in 5% CO2/95% air. Subsequently, 100 μL medium was transferred to a fresh 96‐well plate and absorbance was read at 570 nm. Data are expressed as absorbance value units.
Alternatively, after the growth period of 14 days at the air–liquid interface, samples were snap‐frozen in OCT mounting medium and stored at –80 °C until processing. Frozen samples were sectioned into 7‐μm slides using a cryostat. For staining, these were fixed with either 4% PFA in PBS for 10 min at RT, or 100% methanol for 7 min at –20 °C. After fixation, the samples were rinsed three times with PBS and pretreated for 30 min with PBS containing 5% goat serum, followed by incubation overnight at 4°C with primary antibodies (Table 1). The sections were thoroughly rinsed with PBS and then incubated with secondary antibodies for 1 h at RT. Coverslips were mounted using Vectashield containing DAPI (Vector Laboratories), which subsequently labels nuclei. Confocal microscopy (Zeiss LSM‐510 inverted) was used to visualize and capture immunostained cells. As a positive control, human skin was stained using the same antibodies.
For transmission electron microscopy (TEM), 14‐day‐old constructs were fixed in 2·5% glutaraldehyde in 0·1 M cacodylate buffer at pH 7·2–7·4, washed in 0·1 mol L−1 cacodylate buffer at pH 7·2, postfixed in 1% OsO4 in 0·1 mol L−1 cacodylate buffer at pH 7·2, dehydrated in graded ethanol and embedded in Araldite (Serva, Heidelberg, Germany). Ultrathin sections were counterstained with uranyl acetate and lead citrate, and examined under a Zeiss EM109 transmission electron microscope to evaluate basement membrane formation within the constructs.
Extracellular matrix samples directly analysed by mass spectrometry
The protocol we used for mass spectrometry (MS) was described previously by Pflieger et al.57 After cell elimination, ECM proteins were directly proteolysed in each well using 2·5 μg trypsin (Gibco, Invitrogen) in 250 μL 30 mmol L−1 Tris (pH 8·0) at 37°C overnight. Predigested matrix proteins were scraped off from the wells using a cell scraper and collected into 1·5 mL microfuge tubes. The samples were then reduced by the addition of dithiothreitol (final concentration 10 mmol L−1) and incubated for 30 min at 56 °C. After reduction, iodoacetamide (final concentration 55 mmol L−1) was added to prevent the disulfide bonds re‐forming and thus keep the protein unfolded, and samples were incubated for 30 min at RT in the dark. Finally, the samples were further digested by trypsin at 37 °C overnight before terminating the digest with the addition of 10% trifluoroacetic acid (final concentration 0·5%).
Samples were analysed by liquid chromatography MS using a nanoACQUITY UPLC™ system (Waters MS Technologies, Manchester, U.K.). One microlitre (1–3 μg protein digest) of sample was injected onto each trapping column (C18, 180 μm × 20 mm; Waters) using partial loop injection, for 1 min at a flow rate of 15 μL min−1 with 0·1% (v/v) formic acid. Samples were resolved on an analytical column (nanoACQUITY UPLC™ M‐class HSS T3 75 μm × 150 mm 1·8 μm column; Waters) using a gradient of 97% A [0·1% (v/v) formic acid], 3% B [99·9% acetonitrile 0·1% (v/v) formic acid] to 60% A, 40% B over 36 min at a flow rate of 300 nL min−1. The nanoACQUITY UPLC was coupled to a Synapt ™ G2 mass spectrometer (Waters) and data were acquired using a MSE program with 1‐s scan times and a collision energy ramp of 15–40 eV for elevated energy scans. The mass spectrometer was calibrated before use and throughout the analytical run at 1‐min intervals using the NanoLockSpray™ (Waters) source with Glu‐fibrinopeptide.
To analyse quantitatively the MS results, peptide identification was performed using the ProteinLynx Global SERVER™ (PLGS) v3·0·3 (Waters). The data were processed using a low‐energy threshold of 250. A fixed carbamidomethyl modification for cysteine was specified. The search thresholds used were as follows: minimum fragment ion matches per peptide 3; minimum fragment ion matches per protein 7; minimum peptides per protein 1; a false‐positive value of 4. The maximum protein mass identified was 50 kDa. Data were searched against the most recent UniProtKB/Swiss‐Prot human database entry.58 For normalization, each sample had a known trypsin concentration, which was used as an internal standard. All protein hits were identified with a confidence of more than 95%. The PLGS protein score was based on the probability that the observed match between the experimental data and the PLGS database was not a random event, thus minimizing potential random hits. Furthermore, if two or more proteins shared an identical peptide, but the peptide in question was regulated differently, then the peptide was not included in the analysis. Peptide intensity was determined as the sum of the peptide ions for all unique peptides for each protein. A total peptide score was taken as the integrated total of all normalized peptide intensities, for all identified ECM proteins.59 Analysis was performed on matrices derived using cells from two different patients.
Results
Fibroblast subpopulations produced self‐assembled extracellular matrices with different morphology and architectural structure
We first generated cell‐assembled matrices from each of the three fibroblast subpopulations: DPfi, Pfi and Rfi. These were seeded to achieve full confluency immediately and switched to matrix assembly media at 24 h, to ensure differential proliferation profiles had no effect on matrix production (Fig. 1a). Using confocal microscopy images of FN1 coupled with image analysis algorithms (Fig. 1b–d), we found that the architecture of the self‐assembled ECMs varied between each of the three fibroblast subpopulations used. Specifically, Pfi generated matrices with significantly thicker fibres than DPfi and Rfi, which were associated with the highest amount of interfibrillar space between each fibre (Fig. 1e, f). The Pfi‐derived ECM fibres were also significantly anisotropic compared with both Rfi‐ and DPfi‐derived matrices (Fig. 1g). This bears resemblance to the papillary dermis, which is disorganized in comparison with the reticular dermis.4 Lastly, when we assessed matrix thickness, the DPfi deposited significantly more matrix than the Pfi (Fig. 1h).
Figure 1.
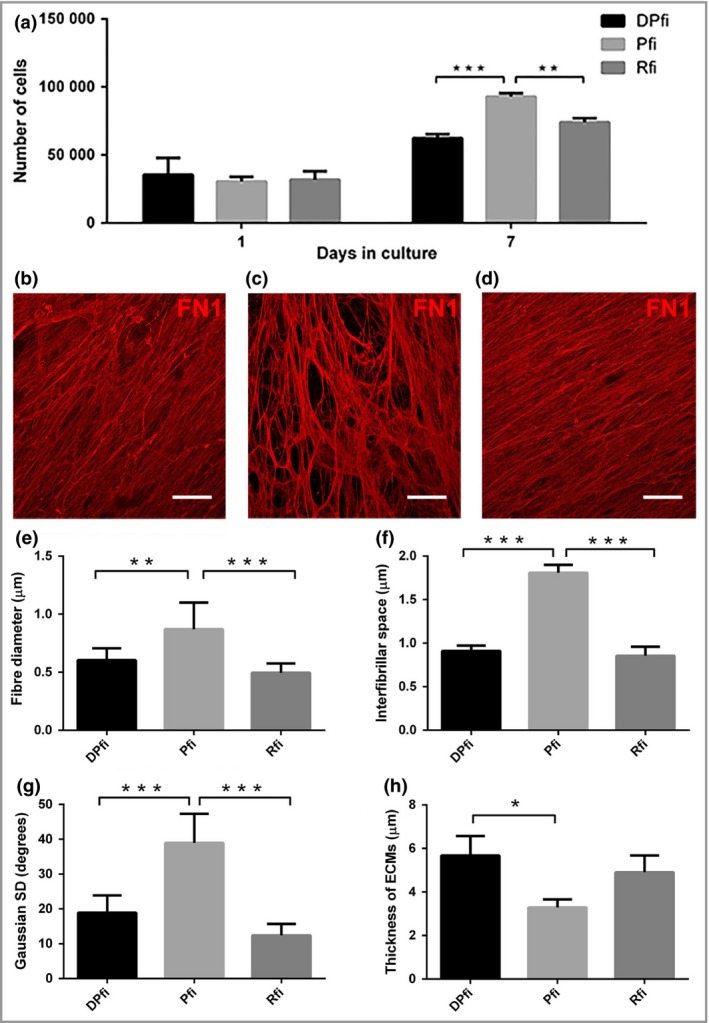
Morphological characterization of cell‐assembled extracellular matrices (ECMs). (a) Cell numbers, 1 and 7 days after seeding cells at equivalent densities, highlight fibroblast subtype‐specific differences. Representative immunofluorescence images of fibronectin 1 (FN1)‐stained ECMs generated by (b) dermal papilla fibroblasts (DPfi), (c) papillary fibroblasts (Pfi) or (d) reticular fibroblasts (Rfi) after 10 days in culture. Corresponding graphs determined from analysis of nine representative images show quantification of fibre diameter for each (e) cell‐assembled ECM, (f) interfibrillar space, (g) Gaussian SD of fibre orientation and (h) total ECM thickness. Data are mean ± SD (n = 3). *P < 0·05, **P < 0·01, ***P < 0·001. Scale bars in (a–c) are 50 μm.
Fibroblast subpopulations differentially support basement membrane formation
After determining that ECMs could be successfully generated from each fibroblast subtype, we decided to employ a protocol to generate epithelial‐only skin constructs, replacing the collagen coating that is usually used with ECM assembled from each cell type. Using alamarBlue to assess keratinocyte viability, we found no significant statistical differences between epithelial‐only constructs grown on different self‐assembled ECMs compared with the control (Fig. 2a). Next, we specifically wanted to assess basement membrane formation, as our fibroblast subpopulations were derived from subanatomical locations subjacent to, and devoid of, basement membrane. We used antibodies against collagen IV (COL4) and collagen VII (COL7) to assess basement membrane characteristics. COL4, which is deposited by both fibroblasts and keratinocytes,60 was observed only in keratinocytes grown on Pfi ECM and DPfi ECM (Fig. 2), but not in keratinocytes supported by Rfi ECM or control constructs (without matrix). COL7, the production of which in keratinocytes is stimulated by fibroblasts,61 was also found in keratinocytes in both DPfi and Pfi matrix‐supported constructs. However, COL7 was not expressed in the control group or in the Rfi ECM supported constructs (Fig. 2c). In addition to immunofluorescence, we used TEM to assess basement membrane formation in our epithelial‐only skin constructs (Fig. 2d). Basement membranes were detected in the control group, despite not identifying COL4 or COL7 by immunofluorescence. We also detected basement membrane in constructs supported by Pfi and DPfi matrices but not Rfi‐derived matrices. The skin constructs grown on Pfi matrices had a very clear and continuous basal lamina structure compared with the control. In contrast, the skin constructs supported by DPfi‐derived ECM had a very thick basement membrane; however, this was not continuous across the entire construct.
Figure 2.

Epithelial‐only skin constructs grown on cell‐assembled extracellular matrices (ECMs). (a) Viability assay of cultured keratinocytes (KCs) on different fibroblast‐derived matrices (mean ± SD). Immunofluorescence images of (b) collagen IV (COL4) and (c) collage VII (COL7) in entire skin and epithelial‐only constructs cultured on coating matrix, or dermal papilla fibroblast (DPfi), papillary fibroblast (Pfi) and reticular fibroblast (Rfi)‐derived ECM. (d) Ultrastructural analysis of the basement membrane (arrows) in the epithelial‐only skin constructs (n = 3). Scale bars for (b, c) are 50 μm. Scale bars for (d) are 400 nm. RFU, relative fluorescence unit; CTR, control; DAPI, 4’,6‐diamidino‐2‐phenylindole.
Mass spectrometry revealed different matrix compositions
Given the different fibre morphologies in our self‐assembled ECMS from different fibroblast subpopulations, and a divergent ability to support establishment of a basement membrane, we wanted to determine whether there were also differences in matrix composition. To do this, we performed MS analysis of digested matrices devoid of cells.
Our analysis identified a number of different proteins with the maximum protein mass of 50 kDa. In total, 12 ECM proteins were identified that passed the 95% accuracy threshold within the DPfi, Pfi and Rfi cell‐assembled ECMs (Table 2). Based on the total peptide ion count, DPfi produced significantly more ECM compared with either the Rfi or Pfi over a 10‐day time frame. All identified proteins were interstitial ECM rather than basement membrane ECM. Specifically, FN1, which was used for image analysis, was produced equally by all three different fibroblast subpopulations. Surprisingly, collagen I (COL1), which is generally thought to be expressed throughout the dermis, was not found in either of the Pfi ECM samples, even if we considered peptides that did not pass the accuracy cut‐off threshold.
Table 2.
Mass spectrometry data of extracellular matrices (ECM) for each fibroblast subtype
| Protein symbol | Protein ID | DPfi ECM | Pfi ECM | Rfi ECM | ECM type | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PLGS scorea | Coverage (%)b | Peptide intensity sumc | PLGS scorea | Coverage (%)b | Peptide intensity sum | PLGS scorea | Coverage (%)b | Peptide intensity sumc | |||
| FN1 | P02751 | 13937 | 49 | 9138740 | 4245 | 44 | 4349942 | 4511 | 47 | 6198948 | Adhesive glycoprotein |
| COL1A1 | P02452 | 241 | 25 | 319566 | – | – | – | – | – | – | Fibrous |
| COL1A2 | P08123 | 1018 | 15 | 398538 | – | – | – | 89 | 14 | 26986 | Fibrous |
| COL6A1 | P12109 | 5787 | 42 | 1399723 | 208 | 18 | 79824 | 259 | 26 | 142226 | Fibrous |
| COL6A2 | P12110 | 2448 | 28 | 721444 | – | – | – | – | – | – | Fibrous |
| COL6A3 | P12111 | 2561 | 34 | 3888636 | 226 | 23 | 263054 | 268 | 25 | 683339 | Fibrous |
| TNC | P24821 | 1931 | 30 | 1579325 | 549 | 27 | 647935 | 681 | 30 | 885657 | Adhesive glycoprotein |
| THBS1 | P07996 | 1014 | 31 | 575557 | – | – | – | 129 | 21 | 106958 | Adhesive glycoprotein |
| FBLN2 | P98095 | 431 | 23 | 171414 | 129 | 17 | 61035 | 238 | 24 | 217243 | Ca2+ binding glycoprotein |
| VTN | P04004 | 403 | 16 | 44674 | 95 | 18 | 12991 | – | – | – | Adhesive glycoprotein |
| FBN1 | P35555 | – | – | – | – | – | – | 118 | 17 | 136846 | Adhesive glycoprotein |
| EMIL1 | Q9Y6C2 | – | – | – | – | – | – | 95 | 9 | 21431 | Adhesive glycoprotein |
| Total peptide intensityd | 18237614 | 5414781 | 8392648 | ||||||||
Listed are the protein symbol and accession number (ID) from Swiss‐Prot for all the ECM proteins detected. aA statistical measure of peptide assignment accuracy. bPercentage of protein sequence covered by identified peptides for each protein. cSum of the peptide ion intensities for each protein, normalized against a trypsin internal control. dSum of the peptide ions for all peptides identified, for each ECM protein. Values are the average of matrices from n = 2. DPfi, dermal papilla fibroblasts; Pfi, papillary fibroblasts; Rfi, reticular fibroblasts; PLGS, ProteinLynx Global SERVER; FN1, fibronectin 1; COL1A1, α1 chain collagen I; COL1A2, α2 chain collagen II; COL6A1, α1 chain collagen VI; COL6A2, α2 chain collagen VI; COL6A3, α3 chain collagen VI; TNC, tenascin; THBS1, thrombospondin 1; FBLN2, fibulin 2; VTN, vitronectin; FBN1, fibrillin 1; EMIL1, emilin 1.
Interstitial ECM is usually classified into fibrous ECMs (collagens and elastin), proteoglycans or ‘other’. Intriguingly, our analysis identified collagens and proteins that fell into the ‘other’ category, but no proteoglycans (Table 2). Of the proteins in the ‘other’ category, nearly all were adhesive glycoproteins; these are linker proteins that connect cells with fibrous ECM. Of the collagens, the MS revealed that while α1 chain collagen VI (COL6A1) and α3 chain COL6 (COL6A3) were present in all ECMs derived from all cell types, α2 chain COL1 (COL1A2) was only in the DPfi and Rfi matrices, whereas α2 chain COLVI (COL6A2) and α1 chain COL1 (COL1A1) were only in the DPfi ECM. Of the glycoproteins, tenascin (TNC) and fibulin 2 were present in all ECMs, whereas THBS1 was present at high levels in the DPfi ECM, low levels in the Rfi ECM and was absent from the Pfi ECM. Vitronectin was present in the both the DPfi and Pfi ECM, whereas fibrillin 1 (FBN1) and emilin 1 were only found in the Rfi ECM (Fig. 3a). As noted above, we did not find any proteoglycans when the accuracy threshold was set to 95%. However, we did identify one proteoglycan – biglycan (BGN) – in the DPfi matrix at low levels when the accuracy threshold was lowered to 50%. We cannot be sure if other proteoglycans were expressed within our matrices as no other proteoglycans were detected with our technique. It is highly possible that the selected ECM extraction method selectively obtained only fibrous ECM and adhesive glycoproteins anchored to it.
Figure 3.
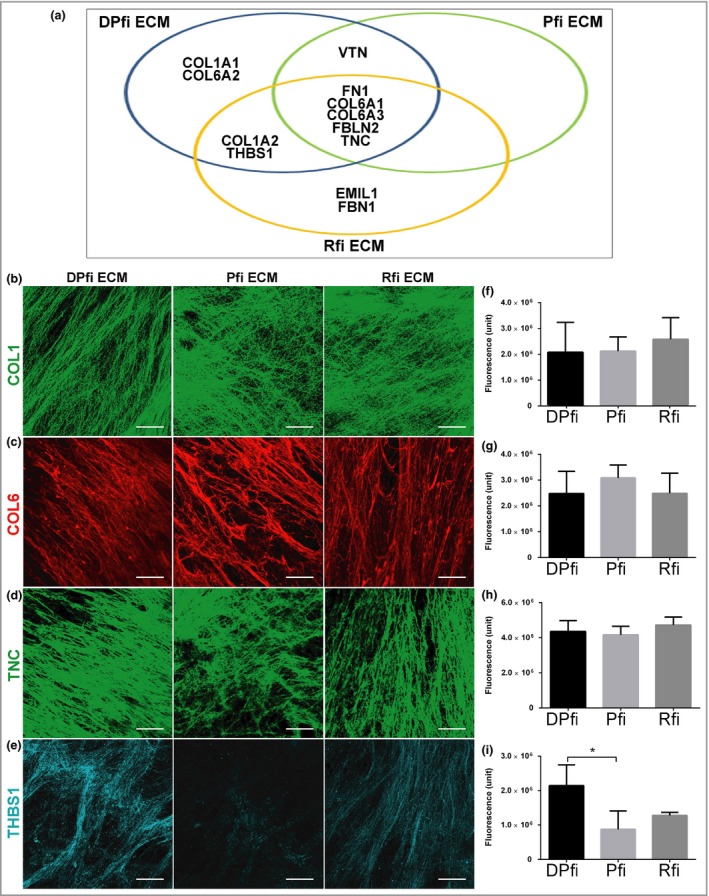
Extracellular matrix (ECM) composition. (a) Venn diagram showing distribution of 12 ECM components in matrices assembled from dermal papilla fibroblast (DPfi), papillary fibroblast (Pfi) and reticular fibroblast (Rfi). Representative immunofluorescence images of (b) collagen I (COL1), (c) collagen VI (COL6), (d) tenascin (TNC) and (e) thrombospondin 1 (THBS1) in ECMs generated by DPfi, Pfi or Rfi cells after 10 days in ascorbic acid‐supplemented medium. (f–i) Quantification of immunofluorescence is plotted in graphs as the mean ± SD (n = 3). *P < 0·05. Scale bars in (b–e) are 50 μm. COL1A1, α1 chain collagen I; COL6A2, α2 chain collagen VI; COL1A2, α2 chain collagen II; VTN, vitronectin; FN1, fibronectin 1; COL6A1, α1 chain collagen VI; COL6A3, α3 chain collagen VI; FBLN2, fibulin 2.
In order to verify that proteins detected in our MS analysis were expressed within our cell‐derived ECMS, we stained each cell‐assembled ECM with antibodies against COL1 (all chains), COL6 (all chains), TNC and THBS1. Despite not finding COL1A1 or COL1A2 in our Pfi ECM by MS we found COL1 in all samples (Fig. 3b). The COL6 antibody we used also detects all α chains, and it appeared to be slightly more, but not significantly, abundant in the Pfi ECM, which correlates with observations from an earlier study (Fig. 3c).55 Likewise, TNC was present at similar levels across all cell‐derived ECMs (Fig. 3d). THBS1 was predominantly expressed in DPfi‐derived matrix (Fig. 3e), which corroborates our MS results. We also used image analysis to quantify expression levels, and confirm these observations (Fig. 3f–i). These results highlight that cells in vitro deposit matrices similar to the ECM they produce in vivo. For example, THBS1 expression is higher within the dermal papilla than the interfollicular skin dermis.16
Discussion
In the last two decades the role of ECM in cell biology has become evident. Indeed, ECM is known to regulate cell behaviour, and it plays an essential role in organ development, function and repair.62, 63 On this basis, ECM as a molecular scaffold is fundamental for tissue homeostasis, and alterations in a specific ECM component can lead to disruption of this process.64 Physical properties such as topography and porosity of ECM can influence various anchorage‐related biological functions, such as cell division and migration.65
Given the importance of ECM in cell biology, ECM scaffolds are becoming commonplace for tissue repair.45 However, within the skin at least, dermal scaffolds used to promote regeneration are insufficient. They are composed of only a simple ECM, or decellularized reticular dermis, and they do not take into account the complexity of heterogeneity within whole‐skin dermis.1 These limitations highlight the need for bioinspired scaffolds for skin repair.
ECM scaffolds derived from cultured cells offer several advantages over decellularized tissues; there are lower risks of pathogen transmission and undesirable inflammatory and immunological reactions.34 In this study we showed that ECMs derived from three distinct fibroblast subtypes located within the skin dermis were morphologically, functionally and compositionally distinct from one another. In particular, in our study PFI were observed to generate matrices with significantly thicker fibers than DPfi and Rfi. This is different to how dermal ECM is organized in vivo, where the papillary dermis contains thinner fibers than the reticular dermis.4 However, with regard to organization we observed parallels in our cell‐assembled ECMs to ECM in different subanatomical locations from which the fibroblast subtypes were initially isolated. Specifically, the Pfi‐assembled matrix had significantly anisotropic fibres, which is analogous to the organization of the papillary dermis. The composition of cell‐assembled ECMs in vitro is also similar to the ECM in the spatial locations from where the cells were initially derived, indicating that fibroblasts in vitro have a memory of their subanatomical origin (Fig. 4). In particular, THBS1 was preferentially deposited by DPfi, whereas the in vivo dermal papilla is rich in THBS1. In addition, it is worth mentioning the absence of elastin and proteoglycans, which are usually key components of dermal ECMs.66 While we did detect BGN in our DPfi matrices, this was only at low levels when the accuracy threshold was lowered to 50%. However, previous studies of cell‐assembled ECM (by G.T. and J.T.C.) from dermal fibroblasts have identified proteoglycans, including versican and decorin, using antibodies for detection.55 We therefore believe the lack of proteoglycans is an artefact of matrix processing for MS. With regard to the lack of elastin in our matrices, we have an alternative explanation. In development, elastin is deposited relatively late, onto a scaffold of FBN1 microfibrils, which forms at an earlier time point.67 It may be the case that elastin was just not deposited in the 10‐day time frame over which ECMs were generated. In support of this, matrisome analysis of cell‐assembled matrices derived from neonatal dermal fibroblasts indicates that they also lack elastin.53 Despite this, fibrillins were observed in both our Rfi‐assembled matrices and the neonatal dermal fibroblast matrices, which act as a guide for elastin fibre deposition.
Figure 4.
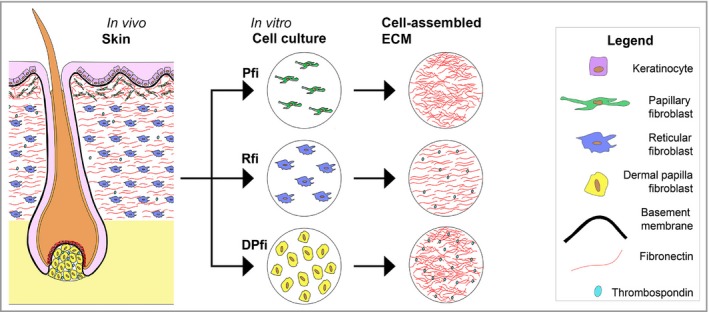
Summary of the results. Human skin in vivo has distinct features established by the fibroblast subtypes within the dermis that create and deposit extracellular matrix (ECM). For example, the dermal papilla is rich in thrombospondin, whereas the reticular fibroblast (Rfi)‐deposited matrix contains aligned ECM fibres. Fibroblasts isolated from the skin maintain distinct identities reflecting their origin, and subsequently deposit ECM in vitro, which reflects the location from which they were initially isolated. Pfi, papillary fibroblasts; DPfi, dermal papilla fibroblasts.
It is well known that the skin dermis and its interaction and crosstalk with epithelial cells are important for the production of the basement membrane components.17 In our study of epithelial‐only constructs, COL4 and COL7 were only found in keratinocytes of Pfi and DPfi ECM‐supported constructs, but not keratinocytes on Rfi ECM or control constructs. This observation is supported by previous studies, where authors have observed that decellularized fibroblast ECM stimulated the production and deposition of COL7 by keratinocytes.61 Refocusing on constructs supported by Rfi matrices, here we found no evidence of basement membrane establishment with either antibody staining or TEM. This is despite basement membrane formation occurring within our control samples and suggests that there may be an inhibitor of membrane formation within the Rfi matrices. While we removed cells from our matrices, growth factors may have remained attached to the ECMs onto which keratinocytes were seeded. We therefore cannot conclude that this observation is due specifically to the different ECMs, and it is highly likely that the growth factor constituents produced by the different fibroblast subtypes play an important role in directing the establishment of a basement membrane.68 Other studies have shown that when Pfi and Rfi are grown in co‐cultures with keratinocytes, they form cysts.4 In Rfi–keratinocyte cysts COL7 is absent; however, Pfi–keratinocyte cysts express all basement membrane components evaluated.4 Based on our observation and the aforementioned study, we postulate that basement membrane formation cannot be supported by ECM produced by Rfi. Again, this reflects the dermal locations of the cells in vivo, where supporting establishment of a basement membrane is not a usual Rfi cell function. In contrast, DPfi support establishment of a very thick basement membrane, reflecting their location subjacent to a thickened basement membrane, termed a glassy membrane in vivo.14 It is postulated that the expression of a high number of basement membrane ECMs in the dermal papilla, as opposed to the interfollicular dermis, means they have a key role in directing follicular physiology.15
In conclusion, the differences we observed in ECM composition, fibre morphology and behaviour between the different cell‐derived ECMs reflects differences that are physiologically present between the papillary, reticular and hair follicle dermis within the skin. We believe that inspiration can be taken from these physiologically different cell‐derived ECMs to improve the design of reliable biomimetic materials with improved therapeutic potential for skin tissue engineering. Self‐assembled scaffolds may be useful for clinical application because they mimic natural ECM structurally and compositionally, yet they do not elicit an immunogenic response. The self‐assembly process can also be used to establish ECMs of different thicknesses and strengths.49 Decellularized papillary dermis is not usually used as a tissue engineered product as it is very thin compared with the reticular dermis. Moreover, dermal papillae, which are < 100 μm in diameter, have never, to our knowledge, been decellularized and used in tissue engineering; they are simply too small. Using a self‐assembly approach, matrices derived from both DPfi or Pfi cultured in vitro could be superior to ex vivo Rfi‐derived dermis, or engineered dermal templates, as an effective product for skin engineering.
Supporting information
Powerpoint S1. Journal Club Slide Set.
Acknowledgments
We thank both the Facility for Imaging by Light Microscopy (FILM) and the CISBIO Mass Spectrometry Core Facility at Imperial College London for assistance with confocal imaging and mass spectrometry analysis.
Funding sources This work was financially supported, in part, by the Cassa dei Risparmi di Cesena Foundation, La Fenice ONLUS Association and a Marco Polo Grant from the University of Bologna to M.G., and by a Royal Society Grant and Medical Research Council (MR/M01858X/1) funding to C.A.H.
Conflicts of interest None to declare.
https://doi.org/10.1111/bjd.16946 available online
References
- 1. Sriram G, Bigliardi PL, Bigliardi‐Qi M. Fibroblast heterogeneity and its implications for engineering organotypic skin models in vitro. Eur J Cell Biol 2015; 94:483–512. [DOI] [PubMed] [Google Scholar]
- 2. Tracy LE, Minasian RA, Caterson EJ. Extracellular matrix and dermal fibroblast function in the healing wound. Adv Wound Care (New Rochelle) 2016; 5:119–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harper RA, Grove G. Human skin fibroblasts derived from papillary and reticular dermis: differences in growth potential in vitro. Science 1979; 204:526–7. [DOI] [PubMed] [Google Scholar]
- 4. Sorrell JM, Caplan AI. Fibroblast heterogeneity: more than skin deep. J Cell Sci 2004; 117:667–75. [DOI] [PubMed] [Google Scholar]
- 5. Rinkevich Y, Walmsley GG, Hu MS et al Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 2015; 348:aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Driskell RR, Lichtenberger BM, Hoste E et al Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 2013; 504:277–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Werner S, Smola H. Paracrine regulation of keratinocyte proliferation and differentiation. Trends Cell Biol 2001; 11:143–6. [DOI] [PubMed] [Google Scholar]
- 8. Chuong CM, Cotsarelis G, Stenn K. Defining hair follicles in the age of stem cell bioengineering. J Invest Dermatol 2007; 127:2098–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Honardoust D, Ding J, Varkey M et al Deep dermal fibroblasts refractory to migration and decorin‐induced apoptosis contribute to hypertrophic scarring. J Burn Care Res 2012; 33:668–77. [DOI] [PubMed] [Google Scholar]
- 10. Jahoda CA, Reynolds AJ. Hair follicle dermal sheath cells: unsung participants in wound healing. Lancet 2001; 358:1445–8. [DOI] [PubMed] [Google Scholar]
- 11. Ohyama M, Zheng Y, Paus R et al The mesenchymal component of hair follicle neogenesis: background, methods and molecular characterization. Exp Dermatol 2010; 19:89–99. [DOI] [PubMed] [Google Scholar]
- 12. Schafer IA, Pandy M, Ferguson R et al Comparative observation of fibroblasts derived from the papillary and reticular dermis of infants and adults: growth kinetics, packing density at confluence and surface morphology. Mech Ageing Dev 1985; 31:275–93. [DOI] [PubMed] [Google Scholar]
- 13. Sennett R, Rendl M. Mesenchymal–epithelial interactions during hair follicle morphogenesis and cycling. Semin Cell Dev Biol 2012; 23:917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jahoda CA, Horne KA, Mauger A et al Cellular and extracellular involvement in the regeneration of the rat lower vibrissa follicle. Development 1992; 114:887–97. [DOI] [PubMed] [Google Scholar]
- 15. Messenger AG, Elliott K, Westgate GE et al Distribution of extracellular matrix molecules in human hair follicles. Ann N Y Acad Sci 1991; 642:253–62. [DOI] [PubMed] [Google Scholar]
- 16. Pablos JL, Everett ET, Leroy EC et al Thrombospondin 1 is expressed by mesenchymal cells in mouse post‐natal skin and hair follicle development. Histochem J 1998; 30:461–5. [DOI] [PubMed] [Google Scholar]
- 17. Werner S, Krieg T, Smola H. Keratinocyte‐fibroblast interactions in wound healing. J Invest Dermatol 2007; 127:998–1008. [DOI] [PubMed] [Google Scholar]
- 18. Soma T, Tajima M, Kishimoto J. Hair cycle‐specific expression of versican in human hair follicles. J Dermatol Sci 2005; 39:147–54. [DOI] [PubMed] [Google Scholar]
- 19. Azzarone B, Macieira‐Coelho A. Heterogeneity of the kinetics of proliferation within human skin fibroblastic cell populations. J Cell Sci 1982; 57:177–87. [DOI] [PubMed] [Google Scholar]
- 20. Schonherr E, Beavan LA, Hausser H et al Differences in decorin expression by papillary and reticular fibroblasts in vivo and in vitro . Biochem J 1993; 290:893–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sorrell JM, Carrino DA, Baber MA et al A monoclonal antibody which recognizes a glycosaminoglycan epitope in both dermatan sulfate and chondroitin sulfate proteoglycans of human skin. Histochem J 1999; 31:549–58. [DOI] [PubMed] [Google Scholar]
- 22. Zimmermann DR, Dours‐Zimmermann MT, Schubert M et al Versican is expressed in the proliferating zone in the epidermis and in association with the elastic network of the dermis. J Cell Biol 1994; 124:817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Janson DG, Saintigny G, van Adrichem A et al Different gene expression patterns in human papillary and reticular fibroblasts. J Invest Dermatol 2012; 132:2565–72. [DOI] [PubMed] [Google Scholar]
- 24. Lee NM, Erisken C, Iskratsch T et al Polymer fiber‐based models of connective tissue repair and healing. Biomaterials 2016; 112:303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xue Y, Cohen JM, Tsiaras WG. Skin autotomization and regeneration – the African spiny mouse. JAMA Dermatol 2015; 151:1199. [DOI] [PubMed] [Google Scholar]
- 26. Catalano E, Cochis A, Varoni E et al Tissue‐engineered skin substitutes: an overview. J Artif Organs 2013; 16:397–403. [DOI] [PubMed] [Google Scholar]
- 27. MacNeil S, Shepherd J, Smith L. Production of tissue‐engineered skin and oral mucosa for clinical and experimental use. Methods Mol Biol 2011; 695:129–53. [DOI] [PubMed] [Google Scholar]
- 28. Patel M, Fisher JP. Biomaterial scaffolds in pediatric tissue engineering. Pediatr Res 2008; 63:497–501. [DOI] [PubMed] [Google Scholar]
- 29. Shevchenko RV, James SL, James SE. A review of tissue‐engineered skin bioconstructs available for skin reconstruction. J R Soc Interface 2010; 7:229–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma PX. Biomimetic materials for tissue engineering. Adv Drug Deliv Rev 2008; 60:184–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Metcalfe AD, Ferguson MW. Tissue engineering of replacement skin: the crossroads of biomaterials, wound healing, embryonic development, stem cells and regeneration. J R Soc Interface 2007; 4:413–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Supp DM, Boyce ST. Engineered skin substitutes: practices and potentials. Clin Dermatol 2005; 23:403–12. [DOI] [PubMed] [Google Scholar]
- 33. Bourget JM, Gauvin R, Larouche D et al Human fibroblast‐derived ECM as a scaffold for vascular tissue engineering. Biomaterials 2012; 33:9205–13. [DOI] [PubMed] [Google Scholar]
- 34. Lu H, Hoshiba T, Kawazoe N et al Cultured cell‐derived extracellular matrix scaffolds for tissue engineering. Biomaterials 2011; 32:9658–66. [DOI] [PubMed] [Google Scholar]
- 35. Pei M, He F, Kish VL. Expansion on extracellular matrix deposited by human bone marrow stromal cells facilitates stem cell proliferation and tissue‐specific lineage potential. Tissue Eng Part A 2011; 17:3067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sadr N, Pippenger BE, Scherberich A et al Enhancing the biological performance of synthetic polymeric materials by decoration with engineered, decellularized extracellular matrix. Biomaterials 2012; 33:5085–93. [DOI] [PubMed] [Google Scholar]
- 37. Nguyen DQ, Dickson WA. A review of the use of a dermal skin substitute in burns care. J Wound Care 2006; 15:373–6. [DOI] [PubMed] [Google Scholar]
- 38. Frame JD, Still J, Lakhel‐LeCoadou A et al Use of dermal regeneration template in contracture release procedures: a multicenter evaluation. Plast Reconstr Surg 2004; 113:1330–8. [DOI] [PubMed] [Google Scholar]
- 39. Dantzer E, Braye FM. Reconstructive surgery using an artificial dermis (Integra): results with 39 grafts. Br J Plast Surg 2001; 54:659–64. [DOI] [PubMed] [Google Scholar]
- 40. Halim AS, Khoo TL, Mohd Yussof SJ. Biologic and synthetic skin substitutes: an overview. Indian J Plast Surg 2010; 43:S23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bondioli E, Fini M, Veronesi F et al Development and evaluation of a decellularized membrane from human dermis. J Tissue Eng Regen Med 2014; 8:325–36. [DOI] [PubMed] [Google Scholar]
- 42. Bello YM, Falabella AF, Eaglstein WH. Tissue‐engineered skin. Current status in wound healing. Am J Clin Dermatol 2001; 2:305–13. [DOI] [PubMed] [Google Scholar]
- 43. Ferrando PM, Balmativola D, Cambieri I et al Glycerolized reticular dermis as a new human acellular dermal matrix: an exploratory study. PLOS ONE 2016; 11:e0149124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brown B, Lindberg K, Reing J et al The basement membrane component of biologic scaffolds derived from extracellular matrix. Tissue Eng 2006; 12:519–26. [DOI] [PubMed] [Google Scholar]
- 45. Kumar P, Satyam A, Cigognini D et al Low oxygen tension and macromolecular crowding accelerate extracellular matrix deposition in human corneal fibroblast culture. J Tissue Eng Regen Med 2016. Sep 4 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 46. Guillame‐Gentil O, Semenov O, Roca AS et al Engineering the extracellular environment: Strategies for building 2D and 3D cellular structures. Adv Mater 2010; 22:5443–62. [DOI] [PubMed] [Google Scholar]
- 47. Kelm JM, Fussenegger M. Scaffold‐free cell delivery for use in regenerative medicine. Adv Drug Deliv Rev 2010; 62:753–64. [DOI] [PubMed] [Google Scholar]
- 48. Nichol JW, Khademhosseini A. Modular tissue engineering: engineering biological tissues from the bottom up. Soft Matter 2009; 5:1312–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peck M, Gebhart D, Dusserre N et al The evolution of vascular tissue engineering and current state of the art. Cells Tissues Organs 2012; 195:144–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xing Q, Yates K, Tahtinen M et al Decellularization of fibroblast cell sheets for natural extracellular matrix scaffold preparation. Tissue Eng Part C Methods 2015; 21:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. L'Heureux N, Paquet S, Labbe R et al A completely biological tissue‐engineered human blood vessel. FASEB J 1998; 12:47–56. [DOI] [PubMed] [Google Scholar]
- 52. Labbe B, Marceau‐Fortier G, Fradette J. Cell sheet technology for tissue engineering: the self‐assembly approach using adipose‐derived stromal cells. Methods Mol Biol 2011; 702:429–41. [DOI] [PubMed] [Google Scholar]
- 53. Ragelle H, Naba A, Larson BL et al Comprehensive proteomic characterization of stem cell‐derived extracellular matrices. Biomaterials 2017; 128:147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Topouzi H, Logan NJ, Williams G et al Methods for the isolation and 3D culture of dermal papilla cells from human hair follicles. Exp Dermatol 2017; 26:491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Theocharidis G, Drymoussi Z, Kao AP et al Type VI collagen regulates dermal matrix assembly and fibroblast motility. J Invest Dermatol 2016; 136:74–83. [DOI] [PubMed] [Google Scholar]
- 56. Doube M, Klosowski MM, Arganda‐Carreras I et al BoneJ: free and extensible bone image analysis in ImageJ. Bone 2010; 47:1076–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pflieger D, Chabane S, Gaillard O et al Comparative proteomic analysis of extracellular matrix proteins secreted by two types of skin fibroblasts. Proteomics 2006; 6:5868–79. [DOI] [PubMed] [Google Scholar]
- 58. Boutet E, Lieberherr D, Tognolli M et al UniProtKB/Swiss‐Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol Biol 2016; 1374:23–54. [DOI] [PubMed] [Google Scholar]
- 59. Zhang G, Ueberheide BM, Waldemarson S et al Protein quantitation using mass spectrometry. Methods Mol Biol 2010; 673:211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marinkovich MP, Verrando P, Keene DR et al Basement membrane proteins kalinin and nicein are structurally and immunologically identical. Lab Invest 1993; 69:295–9. [PubMed] [Google Scholar]
- 61. Benny P, Badowski C, Lane EB et al Making more matrix: enhancing the deposition of dermal‐epidermal junction components in vitro and accelerating organotypic skin culture development, using macromolecular crowding. Tissue Eng Part A 2015; 21:183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hynes RO. The extracellular matrix: not just pretty fibrils. Science 2009; 326:1216–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Page‐McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol 2007; 8:221–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lu P, Takai K, Weaver VM et al Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 2011; 3:a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gattazzo F, Urciuolo A, Bonaldo P. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim Biophys Acta 2014; 1840:2506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mikesh LM, Aramadhaka LR, Moskaluk C et al Proteomic anatomy of human skin. J Proteomics 2013; 84:190–200. [DOI] [PubMed] [Google Scholar]
- 67. Daamen WF, Veerkamp JH, van Hest JC et al Elastin as a biomaterial for tissue engineering. Biomaterials 2007; 28:4378–98. [DOI] [PubMed] [Google Scholar]
- 68. Konig A, Bruckner‐Tuderman L. Transforming growth factor‐beta promotes deposition of collagen VII in a modified organotypic skin model. Lab Invest 1994; 70:203–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Powerpoint S1. Journal Club Slide Set.


