Abstract
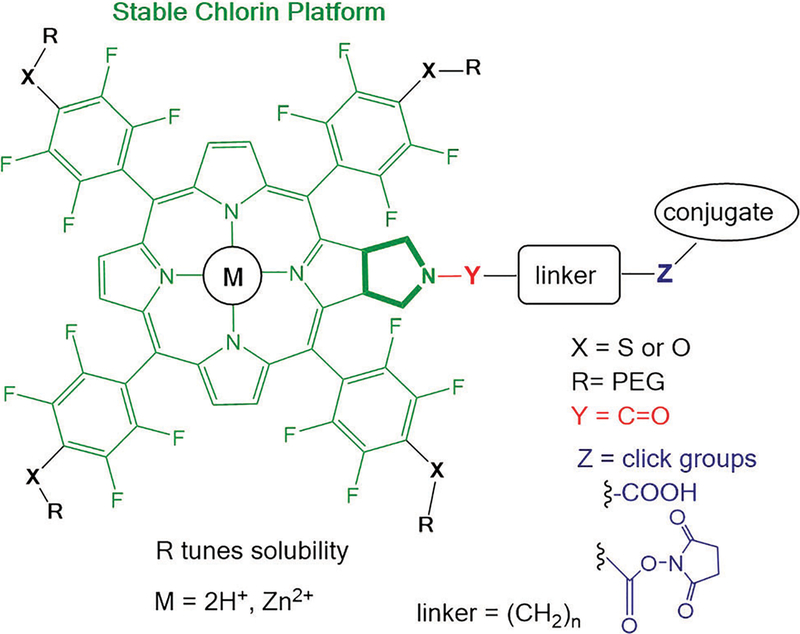
A multifunctional chlorin platform appended with four short poly ethylene glycols and a carboxylate-linker allows rapid conjugation to biotargeting motifs such as proteins and oligonucleotides. The stability and photophysical properties of the chlorin enable develop ment of diagnostics, imaging, molecular tracking, and theranostics.
Porphyrinoid compounds have diverse materials, biological, and diagnostic applications. Chlorins, porphyrins missing one double bond, have excellent photophysical properties for these applications that include a strong red absorption around 650 nm, a balance between the singlet state and intersystem crossing to the triplet manifold, and many are stable to photo-bleaching.1,2 For example chlorin conjugates are used as photo-dynamic therapy (PDT) agents, as fluorescent dyes for biochemical trackers, diagnostics, imaging, as antiviral and antibacterial agents, and for sequence specific photochemical cleaving of DNA.3–11 The optimal properties of biomedical dyes include: chemical purity, amphipathic solubility, strong red light absorption in the window with minimal absorbance from biomolecules and water (ca. 650–850 nm), and photophysical properties that can be tuned for specific applications. Therapeutic, diagnostic imaging, and biochemical applications of phorphyrinoids such as chlorins often require biomolecular recognition motifs that selectively target diseased tissues or other biological structures for tracking. Biotargeting motifs can include sugars, DNA, RNA (e.g. aptamers) and proteins (e.g. antibodies).12–14
However, de novo porphyrinoid synthesis is difficult because of low yields and tedious purification. Considering the large chemical diversity of biotargeting motifs, it is desirable to create modular, multifunctional chlorin platforms that can be readily adapted to target the many forms of cancer, target specific bacteria, or as specific sensors in other photonic bioanalytic applications. The optimization of chlorins is often an iterative process requiring new synthetic schemes with each generation. Modular systems that allow facile optimization of each of the components (dye, linker, targeting, photonic properties, solubility, binding affinity/specificity, and pharmacokinetics) can enable rapid development of new technologies.
To obviate the design of new synthetic strategies and enable rapid development and deployment of chlorins in the aforementioned applications, we report herein a modular chlorin platform that uses successive click reactions to rapidly and efficiently append diverse biotargeting motifs and diverse polyethylene glycols (PEG) onto the dye. Thus, the well-known properties of PEGs to modulate small molecule solubility, pharmacokinetics, targeting, and reactivity are also readily exploited.15
When the chlorin is formed using ylide chemistry that results in pyrrolidine N–H or N–R-groups on the b positions of a pyrrole, an additional issue is that targeting motifs with chiral centers appended to the meso positions results in the formation of diastereomers at the bridgehead carbons1 (ESI†). Herein we provide a synthetic route to a chlorin platform (Scheme 1) for the modular construction of targeted diagnostics, therapeutics, and trackers that exploits well-established linking protocols. This allows researchers to concentrate on the role of the biotargeting motif and tuning the pharmacokinetics rather than the de novo synthesis of new dye systems.
Scheme 1.
Core chlorin platform in green is modified by successive click chemistries to append PEG groups that modulate solubility, and conjugate biomolecules for targeting and tracking. The chelated metal ions modulate the photophysics.
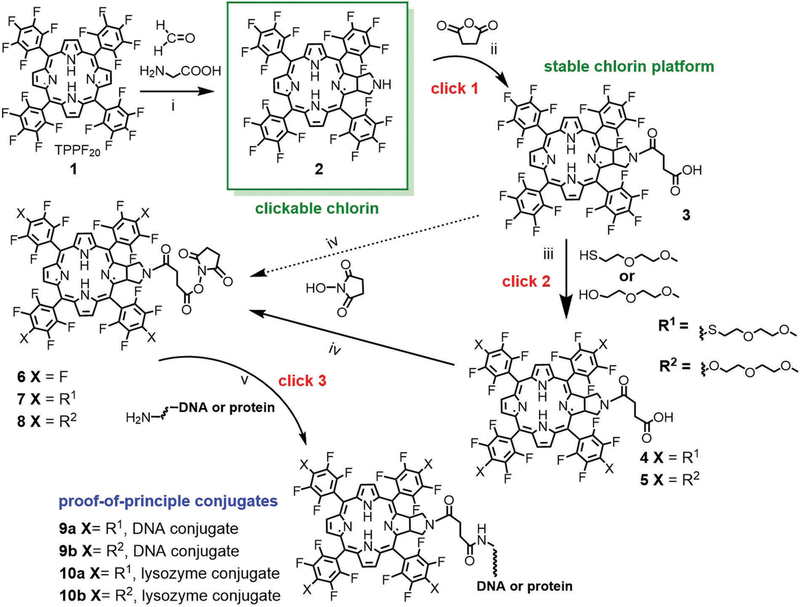
As a demonstration of the sequential click reactions used to make the photonic dye platforms for bioanalytics and theranostics, we report the synthesis of the core meso perfluorophenyl chlorin, the chlorin appended with four 2-(2-methoxyethoxy)ethanol groups (O-PEG) and the chlorin platform appended with four 2-(2-methoxyethoxy)ethanethiol (S-PEG) groups at the 4’ positions of the meso perfluorophenyls (Scheme 2). These chlorins feature a carboxylate linker tethered to the nitrogen of a fused pyrrolidine ring used to make and stabilize the chlorin. The carboxylate is readily activated by the N-hydroxysuccinimide group to enable clicking onto amines of biomolecules or biomolecular recognition motifs.
Scheme 2.
(i) Glycine/paraformaldehyde (1 : 1), added in four aliquots in chlorobenzene, 145 °C, 8 h, 36% yield. (ii) 5 eq. succinic anhydride, CH2Cl2, NEt3, 25 °C, overnight, 95% yield. (iii) For HS-PEG, 4.5 eq. 2-(2-methoxyethoxy)ethane thiol and stoichiometric K2CO3 in acetone, 25 °C, 6 h, 85%; for HO-PEG 10 eq. 2-(2-methoxyethoxy)ethanol and stoichiometric K2CO3 in dry acetone, reflux, overnight, 85%. (iv) 1.2 eq. N-hydroxysuccinimide in dioxane or tetrahydrofuran with 1.2 eq. dicyclohexylcarbodiimide at 25 °C, overnight, quantitative yield. (v) Bio-conjugation depends on the specific targeting molecule but is generally accomplished in buffer or DMSO at 25 °C for 2–10 h. Chlorins were reacted in a 5 μM solution of a 14 nt DNA with an amino terminated tether (10 : 1 ratio; 5’-NH2-(CH2)6-NH-TTCTTCTCCTTTCT-3’), pH 7.4 phosphate buffer overnight, 37 °C. For the lysozyme (hydrolases) conjugate, a 100 μM solution of 8 was allowed to react with lysozyme in DMSO over night at room temperature and purified by gel filtration on a 5 mL column of Sephadex G-25 with water as eluent.
The chlorins are formed by a 1,3-dipolar cycloaddition16 on a pyrrole of 5,10,15,20-tetrakis(2,3,4,5,6-pentafluorophenyl)-porphyrin (TPPF20) 1 using an azomethine ylide to afford amine 2 (Scheme 2) almost exclusively depending on reaction conditions. The resulting fused pyrrolidine ring presents a free nitrogen base that is used to add the tether, in this case using succinic anhydride to yield compound 3, but other tethers are amenable. Compound 2 is somewhat labile, but the tertiary amine is stable for over a year in air in a vial. This approach to adding the linker on the pyrrolidine was pursued to avoid the formation of diastereomers that result from using chiral substituents such as sugars on the para phenyl position.1 To make the chlorin amphipathic and facilitate reactions in aqueous solvents, short S-PEG and O-PEG are readily added to 3 to yield chlorin 4 and 5. Chlorins 3, 4 and 5 exhibit a terminal carboxylic acid that is readily reacted with N-hydroxysuccinimide to give the core chlorin platforms 6, 7 and 8. Note that the 4’-fluoro groups can be substituted nearly quantitatively with a large menu of primary thiols, but amines and alcohols also react well.17,18 Thus other functional groups are readily appended to the four aryl groups of chlorin 3, e.g. to modulate solubility, charge, steric bulk or add secondary targeting motifs. Compounds 6, 7 and 8 readily react with amine groups on an array of biomolecules using well-establish conjugation protocols similar to commercial dye-labelling systems. NMR spectra, mass spectrometry and UV-visible spectroscopy were consistent with compounds 2 to 8 (ESI†). The NMR spectrum for chlorin 2 is consistent with that of the N-methyl pyrrolidine derivative previously reported.1,16,19 The NMR data for both PEG derivatives are somewhat complicated by the reduced symmetry of the chlorin resulting from the orientation of the pyrrolidine pointing toward one face of the macrocycle and increased molecular size, resulting in broadened multiplets for both the macrocycle and the PEG units.
The fluorescence and UV-visible spectra are minimally perturbed from known N-methyl derivatives (Table 1).1 The UV-visible spectrum of 2 in CH2Cl2 has a lowest energy Q band at Amax = 652 nm. The Soret bands for the S-PEG (4 and 7) and O-PEG (5 and 8) derivatives are blue shifted by about 5 nm, indicating small electronic differences arising from the substitution of the four –F groups with four –S-PEG and –O-PEG groups. The ca. 35% fluorescence quantum yield and optical density at multiple wavelengths enable using the platform as a fluorescent tracker and for bioimaging.2 The florescence anisotropy spectra has a maximum of 0.35 ± 0.05, making the compound suitable for biophysical applications based on anisotropy changes. As with the N-CH3 derivatives, the chlorin platform 3 and the conjugates are stable to photo-bleaching.
Table 1.
UV-visible and fluorescence
| Compounda | UV-visible Amax(nm) | Fluorescenceb Amax (nm) (τ ns) |
|---|---|---|
| 2 | 412, 506, 597, 652 | 656, 720 (6.6) |
| N-Me-chlorin | 411, 506, 596, 652 | 656, 720 (6.6) |
| 3 | 405, 503, 597, 651 | — |
| 4 | 412, 506, 597, 651 | 657, 725 (6.6) |
| 5 | 410, 506, 597, 652 | 657, 725 (6.7) |
| 7 | 408, 504, 598, 652 | 657, 725 (6.7) |
| 8 | 405, 502, 596, 652 | 655, 725 (6.6) |
In CH2Cl2 in 1 cm cuvette.
ca. 1 mM.
0.1 mM, Aex at Soret band Amax, the FWHM of the 725 nm peak is ca. 50 nm, lifetime, τ ± 0.1 ns by time correlated single photon counting. Fluorescence quantum yields are ca. 35% for these compounds (see ESI).
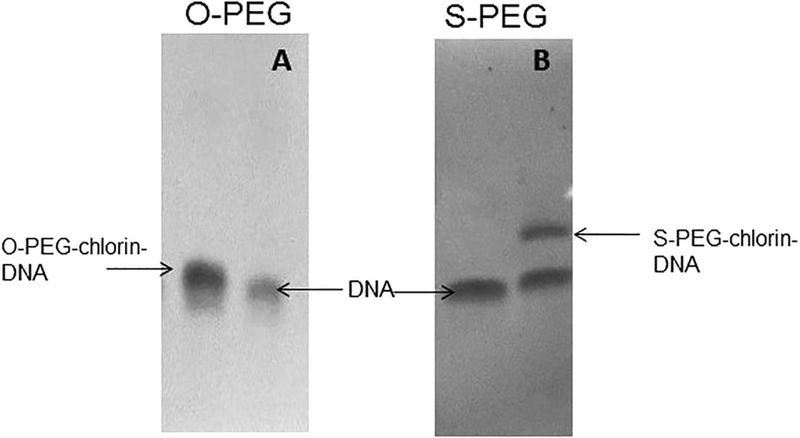
As proof-of-concept, chlorins 7 and 8 with the activator were coupled to a primary amine tethered on the 5’ end of a 14 nt single strand DNA (ssDNA) and to the exposed lysine amino groups on the lysozyme enzyme, yielding 9a, 9b, 10a, and 10b, respectively. Chlorin conjugation to the biomolecules was performed under standard coupling reaction conditions.20 Chlorins 7 and 8 were coupled to the 5’ end of 14 nt DNA via 6-carbon amine linker (5’-NH2-(CH2)6-NH-TTCTTCTCCTTTCT-3’). The chlorins were added to 5 μM DNA in phosphate buffer (0.1 M, pH 7.4) with 10 : 1 ratio. The reaction was carried out overnight at 37 °C. Some aggregation was observed under these conditions if greater DNA concentrations are used. PAGE assays confirmed the coupling of chlorin to the DNA (Fig. 1).
Fig. 1.
20% denaturing polyacrylamide gel for (A) the O-PEG–chlorin– DNA conjugate, (B) the S-PEG–chlorin–DNA conjugate. Gels are stained with Nuclistain to visualize DNA and conjugates: the conjugation reaction mixture and the free 14 nt DNA.
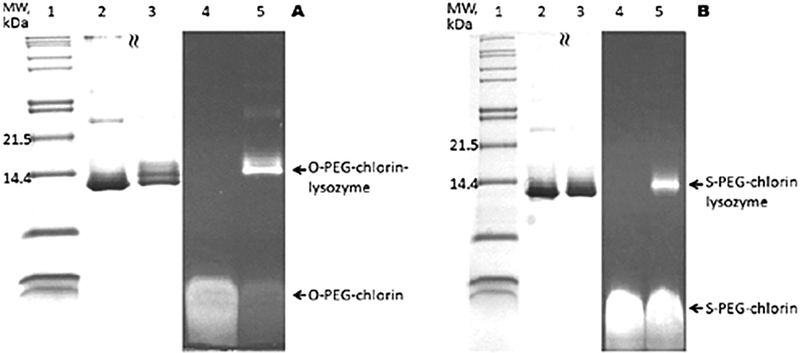
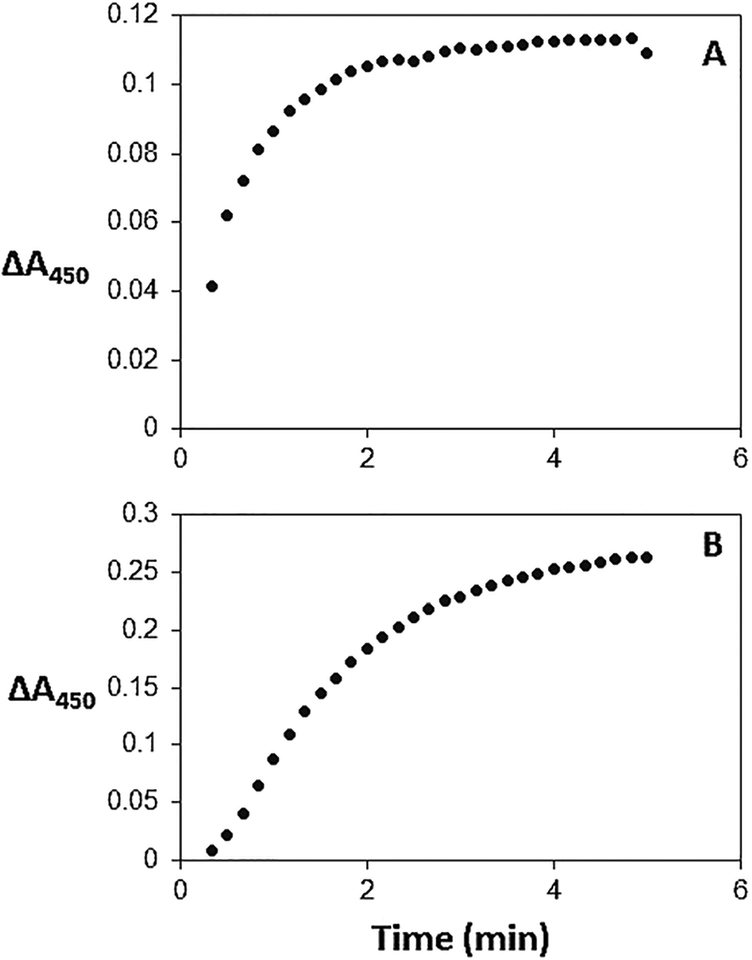
Coupling of lysozyme to chlorin 7 and 8 in DMSO is accomplished at room temperature overnight in a ca. 1 : 1 reaction or a ca. 1 : 10 reaction (ESI†) followed by gel filtration. SDS-PAGE of the purified conjugate imaged by the chlorin fluorescence clearly indicates that either the amino group on the N-terminal amine or one of the six lysine residues is conjugated to the chlorin. Small amounts of a double and triple labelled conjugate (Fig. 2) are observed. These conclusions are corroborated by staining the gel with Coomassie blue to visualize the protein at the same location in the gel as the fluorescent band. Electrospray ionization mass spectrometry indicates the lysozyme is appended with one chlorin. Importantly, activity of the enzyme is maintained after conjugation (Fig. 3).
Fig. 2.
(A) SDS-PAGE for conjugation of O-PEG–chlorin to lysozyme in DMSO. (lane 1) Molecular weight marker. (2) Free lysozyme. (3) Crude product reaction (1 : 1). (4) Free SPEG–chlorin. (5) Crude product reaction (1 : 1). (B) SDS-PAGE for conjugation of S-PEG–chlorin to lysozyme in DMSO. (lane 1) Molecular weight marker. (2) Free lysozyme. (3) Crude product reaction (1 : 1). (4): Free SPEG–chlorin. (5) Crude product reaction (1 : 1). Lanes 1–3 visualization of protein after staining with Coomassie blue. Lanes 4 and 5 visualization of chlorin fluorescence after excitation at 365 nm.
Fig. 3.
Enzymatic activity assay of (A) lysozyme in DMSO; (B) O-PEG– chlorin–lysozyme conjugate in DMSO using Micrococcus lysodeikticus in 66 mM potassium phosphate pH 6.2 as substrate (see ESI†).
Though these are proof-of-concept conjugates, to investigate the possible uptake of DNA–chlorin conjugates by MDA-MB-231 breast cancer cells, we incubated 1 nM to 10 nM of 9b with the cultured cells for 24 hours. Afterwards, we observed a decrease in cell density, a change in cell morphology, and cell clumping with concentrations of 9b above 5 nM. Fluorescence microscopy studies indicate that the conjugates are aggregated in/on the cells and slowly disaggregate (ESI†). Neither the DNA nor chlorins elicit cell morphology changes or death at these concentrations.
The efficient synthetic route for amphipathic chlorins with widely used bioconjugation linkers yields a platform for diverse photonic applications in biochemistry, biology, imaging and therapeutics. S-PEGs have the advantage of shorter reaction time, ambient conditions, and require fewer equivalents (4–5 eq.). O-PEGs of various lengths are widely available commercially and cheaper but require heating, 10–15 eq. and longer reaction times. PEGs and other water soluble moieties can be used to modulate solubility and pharmacokinetics;21 conversely secondary targeting groups such as sugars are readily affixed to the 4’ position using the same chemistry.13,18 The N-hydroxysuccinamide ester is widely used to form stable covalent amide bonds efficiently, but the linking chemistry at the end of the tether can also be readily changed to accommodate other common conjugation chemistries, e.g. esters, disulfides, thioureas and different click groups. Since chlorins strongly bind most metal ions, these ions can be readily incorporated into 3, 4, 5 to modulate the photophysical properties: e.g. Zn2+ or Pt2+ to enhance intersystem crossing to the triplet manifold for PDT, Ni2+ to enhance internal conversion for photothermal applications, 64Cu2+ for positron emission tomography imaging. This molecular design is amenable to attaching chlorins to surfaces such as nanoparticles for other photonic applications.22
Supplementary Material
Acknowledgments
Supported by the U.S. National Science Foundation (NSF) CHE-1213962, IGERT-0965983, to C. M. D., the U.S. National Institutes of Health MBRS GM R25–60665 at Hunter College and CTSC TL1-TR000459 at Weill Cornell Medical College supported J. G. Hunter College science infrastructure is supported by the NSF, the National Institute on Minority Health and Health Disparities 8G12 MD007599, and the City University of New York. We thank Robert Collison, Kanchana Wijerathne, Daniel Hart, and Christopher Farley for helping with the purification and photophysical characterization of the chlorins.
Footnotes
Electronic supplementary information (ESI) available: Synthesis, spectroscopy.
Notes and references
- 1.Singh S, Aggarwal A, Thompson S, Tomé JPC, Zhu X, Samaroo D, Vinodu M, Gao R and Drain CM, Bioconjugate Chem, 2010, 21, 2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal A, Thompson S, Singh S, Newton B, Moore A, Gao R, Gu X, Mukherjee S and Drain CM, Photochem. Photobiol, 2014, 90, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Da˛browski JM, Pucelik B, Regiel-Futyra A, Brindell M, Mazuryk O, Kyzioł A, Stochel G, Macyk W and Arnaut LG, Coord. Chem. Rev, 2016, 325, 67–101. [Google Scholar]
- 4.Bonnett R, Charlesworth P, Djelal BD, Foley S, McGarvey DJ and Truscott TG, J. Chem. Soc., Perkin Trans 2, 1999, 325–328. [Google Scholar]
- 5.Detty MR, Gibson SL and Wagner SJ, J. Med. Chem, 2004, 47, 3897–3915. [DOI] [PubMed] [Google Scholar]
- 6.Vrouenraets MB, Visser GW, Snow GB and van Dongen GA, Anticancer Res, 2003, 505–522. [PubMed] [Google Scholar]
- 7.Yano S, Hirohara S, Obata M, Hagiya Y, Ogura S.-i., Ikeda A, Kataoka H, Tanaka M and Joh T, J. Photochem. Photobiol., C, 2011, 12, 46–67. [Google Scholar]
- 8.Zhang Y and Lovell JF, Theranostics, 2012, 2, 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesquita MQ, Menezes JCJMDS, Neves MGPMS, Tomé AC, Cavaleiro JAS,Cunha Â, Almeida A, Hackbarth S, Röder B and Faustino MAF, Bioorg. Med. Chem. Lett, 2014, 24, 808–812. [DOI] [PubMed] [Google Scholar]
- 10.Pereira PMR, Korsak B, Sarmento B, Schneider RJ, Fernandes R and Tomé JPC, Org. Biomol. Chem, 2015, 13, 2518–2529. [DOI] [PubMed] [Google Scholar]
- 11.Boutorine AS, Brault D, Takasugi M, Delgado O and Hélène C, J. Am. Chem. Soc, 1996, 118, 9469–9476. [Google Scholar]
- 12.Ferreira CSM, Cheung MC, Missailidis S, Bisland S and Gariépy J, Nucleic Acids Res, 2009, 37, 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh S, Aggarwal A, Bhupathiraju NVSDK, Arianna G, Tiwari K and Drain CM, Chem. Rev, 2015, 115, 10261–10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogura S.-i., Fujita Y, Kamachi T and Okura I, J. Porphyrins Phthalocyanines, 2001, 5, 486–489. [Google Scholar]
- 15.Banerjee SS, Aher N, Patil R and Khandare J, J. Drug Delivery, 2012, 2012, 103973, 17 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva AMG, Tomé AC, Neves MGPMS, Silva AMS and Cavaleiro JAS, J. Org. Chem, 2005, 70, 2306–2314. [DOI] [PubMed] [Google Scholar]
- 17.Drain CM and Singh S, in The Handbook of Porphyrin Science with Applications to Chemistry, Physics, Materials Science, Engineering, Biology and Medicine, ed. Kadish K, Smith KM and Guilard R, World Scientific Publisher, Singapore, 2010, ch. 15, vol. 3, pp. 485–537. [Google Scholar]
- 18.Bhupathiraju NVSDK, Rizvi W, Batteas JD and Drain CM, Org. Biomol. Chem, 2016, 14, 389–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho CMB, Neves MGPMS, Tomé AC, Paz FAA, Silva AMS and Cavaleiro JAS, Org. Lett, 2011, 13, 130–133. [DOI] [PubMed] [Google Scholar]
- 20.Brinkley M, Bioconjugate Chem, 1992, 3, 2–13. [DOI] [PubMed] [Google Scholar]
- 21.Caliceti P and Veronese FM, Adv. Drug Delivery Rev, 2003, 55, 1261–1277. [DOI] [PubMed] [Google Scholar]
- 22.Schuckman AE, Ewers BW, Yu LH, Tomé JPC, Pérez LM, Drain CM, Kushmerick JG and Batteas JD, J. Phys. Chem. C, 2015, 119, 13569–13579. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.