Abstract
Purpose:
PD-L1 expression in the pre-treatment tumor microenvironment enriches for response to anti-PD-1/PD-L1 therapies. The purpose of this study was to quantitatively compare the performance of five monoclonal anti-PD-L1 antibodies used in recent landmark publications.
Experimental Design:
PD-L1 immunohistochemistry (IHC) was performed on thirty-four formalin-fixed paraffin-embedded archival melanoma samples using the 5H1, SP142, 28-8, 22C3 and SP263 clones. The percentage of total cells (including melanocytes and immune cells) demonstrating cell surface PD-L1 staining, as well as intensity measurements/H-scores, were assessed for each melanoma specimen using a computer-assisted platform. Staining properties were compared between antibodies.
Results:
Strong correlations were observed between the percentage of PD-L1(+) cells across all clones studied (R2=0.81–0.96). When present, discordant results were attributable to geographic heterogeneity of the melanoma tissue section rather than differences in PD-L1 antibody staining characteristics. PD-L1 intensity/H-scores strongly correlated with percentage of PD-L1(+) cells (R2>0.78, all clones).
Conclusions:
The 5H1, SP142, 28-8, 22C3 and SP263 clones all demonstrated similar performance characteristics when used in a standardized IHC assay on melanoma specimens. Reported differences in PD-L1 IHC assays using these antibodies are thus most likely due to assay characteristics beyond the antibody itself. Our findings also argue against the inclusion of an intensity/H-score in chromogenic PD-L1 IHC assays.
Keywords: PD-L1, anti-PD-1, immunohistochemistry, 5H1, SP142, 22C3, 28-8, SP263
Introduction
The PD-1/PD-L1 immune checkpoint is a physiologic mechanism that dampens ongoing immune responses in peripheral tissues. PD-L1 may also be expressed in the tumor microenvironment, facilitating immune evasion. In melanomas, PD-L1 expression is most often observed on malignant melanocytes and immune cells at the host-tumor interface in a focal and geographically heterogeneous pattern.1 Anti-PD-1/PD-L1 therapies block this resistance mechanism,2 unbridling the anti-tumor immune response and frequently leading to tumor regression. Anti-PD-1 monotherapy has demonstrated durable, objective response rates of 30–40% in patients with advanced melanoma.3–5
Numerous clinical studies have demonstrated that PD-L1 expression in pre-treatment tumor specimens can enrich for an anti-tumor response in patients with melanoma and other cancer types.4–8 For example, if the 30–40% of unselected melanoma patients who demonstrate an objective anti-tumor response to anti-PD-1/PD-L1 are stratified by PD-L1 expression, PD-L1(+) and PD-L1(−) patients have average response rates of approximately 50–60% and 10–20%, respectively.9,10 Findings such as these have led to recent FDA approvals for PD-L1 immunohistochemistry (IHC) assays, including the Dako PD-L1 IHC 28-8 PharmDx™ assay as a complementary diagnostic for patients with metastatic melanoma who may receive nivolumab (anti-PD-1). They also raise concern as to whether PD-L1 status should be used as the sole selection criterion for treatment with anti-PD-1 agents, since a proportion of patients with PD-L1(−) tumors also respond to therapy.11
One of the challenges in assessing the potential role of pre-treatment expression of PD-L1 as a predictive biomarker has been the variation in assays used across studies. The vast majority of studies to date have used chromogenic IHC assays for the detection of PD-L1, the results of which are interpreted by a pathologist who reports the percentage of tumor cells and/or immune cells demonstrating expression. However, the studies have used different antibodies, assay conditions, cell types scored and thresholds of positivity.12 It is not yet clear whether some of the observed differences in PD-L1 status as it relates to patient response to anti-PD-1/PD-L1 between studies are a function of the antibody used, different assay conditions, tumor types studied, or how the assay is ultimately scored by the pathologist. Questions also remain as to whether the inclusion of additional parameters--for example the intensity of PD-L1 staining-- provides added information beyond the percentage of cell staining. Given the increasing importance being placed on the immunohistochemical detection of PD-L1 in the melanoma microenvironment, the purpose of this study was to quantitatively compare the staining properties of five different PD-L1 antibodies that have been used in recent clinical trials.
Methods
Clinicopathologic characteristics
Following institutional review board approval, 34 formalin-fixed paraffin-embedded melanoma specimens from 34 patients were obtained from the surgical pathology archives. Seven were primary tumors, with 1 representing a local recurrence, and 27 were metastases.
Immunohistochemistry (IHC) for PD-L1
Positive and negative controls for PD-L1 IHC were created using 624-mel lines that were transfected with full length human PD-L1 and untransfected 624-mel, respectively.1 Tonsil tissue as well as 10 melanomas with previously-characterized levels of PD-L1 expression13,14 were used for assay development to assess for anticipated patterns of staining in certain cell types as well as assess for potential background staining. Additionally, Horizon systems PD-L1 IHC Reference Standard with engineered protein expressing cell lines (Horizon Discovery, Cambridge UK) were used when finalizing antibody concentrations and antigen retrieval conditions to ensure that we had comparable and dynamic ranges of staining for all antibodies studied, Supplementary Figure S1.
The five different primary antibodies were compared using a manual IHC assay for PD-L1 expression. Slides were deparaffinized, and antigen retrieval was performed for 10 min at 120oC (Decloaking chamber, Biocare Medical) in a citrate buffer, pH 6.0 (Dako S1699) for clones SP142, SP263, 22C3 and 28-8. A Tris-EDTA buffer, pH 9.0 was used for clone 5H1. Following blocking for endogenous peroxidases, protein, and biotin (Fisher Scientific H325–500, Serotec Block ACE and Vector Avidin/Biotin Blocking Kit, respectively), the primary antibody was applied and allowed to incubate at 4oC for 22 hours. The primary antibodies were tested at concentrations of: SP142 (Spring Bioscience, Pleasanton, CA) at 0.096 ug/mL; 5H1 (gift from Dr. Lieping Chen) at 1.8 μg/mL; SP263 (Ventana, Tucson, AZ) at 1.6 μg/mL; 22C3 (Dako, Carpinteria, CA) at 3.0 μg/mL and 28-8 at 0.1 μg/mL (Abcam, Cambridge, MA). After washing, a biotinylated anti-rabbit IgG (BD Biosciences, San Jose, CA) for clones SP142, SP263, and 28-8 or an anti-mouse IgG1 (BD Biosciences) secondary antibody for the other clones was applied at 1.0 μg/mL and allowed to incubate for 30 minutes at room temperature. Signal was developed using an ABC kit (Vector Elite PK-6100, Burlingame, CA) for 15 minutes, followed by amplification with the TSA plus biotin kit (Perkin Elmer, Waltham, MA) at a dilution of 1:50, incubated for 5 minutes. Samples were visualized with Streptavidin-HRP diluted to 1:300 in TBST (Dako P0397, Carpinteria, CA), followed by DAB chromogen (Sigma, St. Louis, MO) and a hematoxylin counterstain. In addition, isotype controls were performed for each clone on each specimen.
Slide imaging and sampling
In a previous study, we showed that > 90% of melanocytic lesions that are PD-L1 positive demonstrate focal, geographic membranous expression in the range of 0–30%.1 Accordingly, we chose three melanoma cases each showing 0 - ≤ 5%, 10–15%, and 25–30% PD-L1 expression as scored by a pathologist. These slides were used to determine the optimal high power field sampling frequency, such that the fraction sampled was representative of PD-L1 expression on the entire slide. Slides were imaged using the Vectra Automated Quantitative Pathology Imaging System (PerkinElmer). Specifically, low power fields (40x, LPF) images were acquired using an automated, scanning protocol that utilized a trainable tissue finder to select the tissue. Within the acquired low power image, multiple high power fields (200x, HPF) were then imaged at different sampling ratios. A grid system was used to generate fractional coverage maps of 4% or 1:25, 6% or 1:16, 11% or 1:9 of the entire tissue region on the slide, Supplementary Figure 2. Slides that were sampled at these ratios were also rotated by 180 degrees and resampled at the same fraction and re-scored. For the cases where no sampling was performed and the entire slide was imaged, HPF selection was performed manually to cover the entire tumor, i.e., 1:1 coverage ratio. Sampling frequency was optimized by comparing the percent cells scored as positive for PD-L1 expression at the three sampling ratios (1:25, 1:16, or 1:9), both normally and inverted, and comparing the measured values of PD-L1 expression to those obtained by 20x imaging of the entire tumor, Supplementary Table 1. All acquired HPF fields were then individually reviewed and were advanced to automated cell scoring if they contained >10% viable tumor. Fields were excluded if they contained large areas of necrosis or tissue artifact, such as tears or folds. To be conservative, we used 1:9 sampling for all subsequent cases across the five PD-L1 antibodies tested, except in the cases where that approach resulted in fewer than ten 20x images being obtained for the case. For smaller samples, a minimum of 10 fields were studied, resulting in a higher sampling ratio.
Computer-assisted scoring of PD-L1 expression
Pathologists tend to overestimate the proportion of cells within a tumor staining positive at low levels and at high levels.15 To aid with maintaining scoring accuracy across a wide range of expression values, computer-assisted cell scoring was performed using inForm 2.1.1 software (PerkinElmer). A spectral library for DAB and hematoxylin was created using the Nuance 3.0.2 software (PerkinElmer). Viable tumor and associated peritumoral stroma were separated from blank space or areas of necrosis via a trainable tissue classifier algorithm. Cells were segmented by the software into nuclear, cytoplasm, and membrane components by using the hematoxylin stain to establish the nuclear center. Thresholds for scoring cells as positive for PD-L1 expression were defined by two pathologists (JMT and RAA) for each antibody. The software was then used to quantify the total number and percentage of cells in the specimen expressing PD-L1. To control for the possible contribution of melanin to the score, each matched isotype control was also scored, and the final % of cells demonstrating PD-L1 expression was determined by subtracting the score of the isotype control from the PD-L1-stained slide. Additionally, subset analysis was performed on cases with either higher or lower melanin content to determine whether this could potentially impact the correlative studies, Supplementary Figure 3A.
PD-L1 staining intensity was also scored. Specifically, after image processing, the software generated a mean-membrane intensity score for each cell. These scores were divided into 50-bins by 0.01 units, such that the 20th bin encapsulated all cells with mean-membrane staining intensities of 0.19–0.20, and the 50th bin includes all cells with intensities of 0.49 or higher. A 50-bin H-score was calculated for each patient specimen by taking the sum of all of the products of the bin number and the fraction of cells within the sample that occupied that bin; for example, if 5% of cells occupied bin 20, the 50-bin H-score would equal one, i.e. 5% x 20, for that bin, and the totals generated from each of the 50-bins are summed to produce an H-score that ranges from 0–50. Raw 50-bin H-scores were calculated for these samples and their respective isotype control slides. The H-score from the isotype control was subtracted from the raw H-score of the PD-L1-stained slide to correct for the presence of background melanin and generate a final H-score for each specimen.
Pathologist qualitative assessment of PD-L1 staining
The pathologists (RAA and JMT) independently reviewed the cases to determine if the antibody clones studied differentially labeled PD-L1 on macrophages, lymphocytes, or melanocytes. Additionally, the pathologists independently reviewed the staining properties of the antibodies to determine if there were noticeable differences in membranous vs. cytoplasmic PD-L1 labeling by antibody clone, and specifically to see if certain clones demonstrated a more accentuated membrane-staining pattern.16 Potential cytoplasmic staining was assessed by reviewing the PD-L1-stained slide in conjunction with its matched isotype control.
Statistical analysis
Correlations in antibody labeling for percent cells staining and intensity of cells staining were performed using Pearson’s correlation coefficient. GraphPad software was used for correlation plots.
Results
Computer-assisted scoring of PD-L1 expression
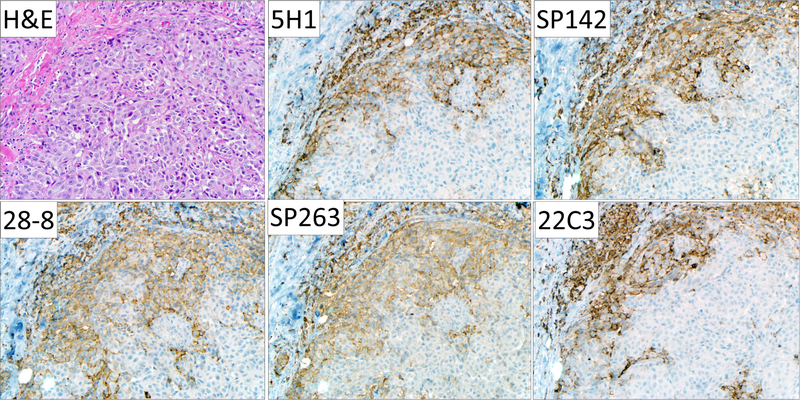
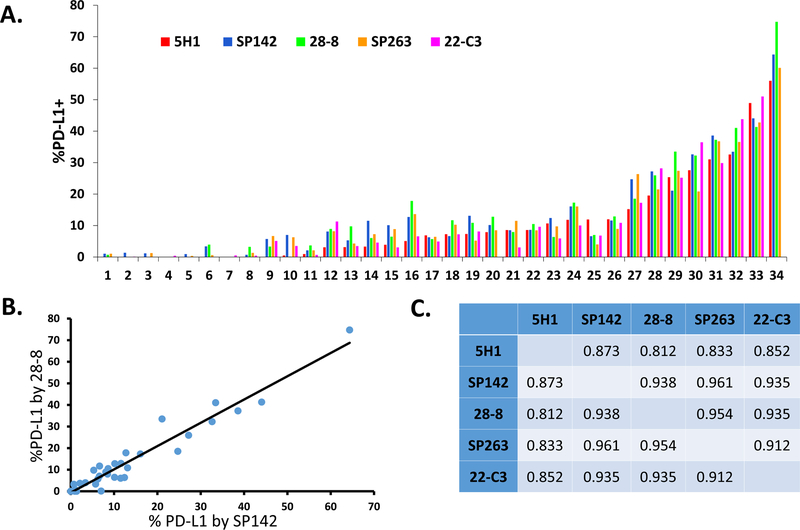
PD-L1 IHC was performed on 34 different melanoma specimens using five different PD-L1 antibodies: 5H1, SP142, 28-8, SP263 and 22C3 and assessed for membranous staining, Figure 1. After PD-L1 IHC was performed, each sample was scanned and PD-L1 expression was scored using a computer-assisted method and with pathologist-derived thresholds set for each antibody, Figure 2A. We plotted the percent PD-L1 expression obtained for each patient sample for each of the antibodies against each other and calculated pairwise correlation coefficients, Figure 2B-2C. The clones that produced the highest concordance were SP142 and SP263 (R2 =0.961) and SP263 and 28-8 (R2 =0.954). The least concordant antibody pairing was 28-8 and 5H1 (R2 = 0.812). Review of slides by pathologists (JMT and RAA) showed that scoring differences were attributable to heterogeneous PD-L1 display in geographically distinct regions that varied even between near adjacent tissue sections.
Figure 1. Representative photomicrographs of PD-L1 immunohistochemistry performed on a cutaneous melanoma metastasis using clones 5H1, SP142, 28-8, SP263, and 22C3.
All five antibodies showed similar regional patterns of PD-L1 expression by tumor cells, macrophages, and lymphocytes. A degree of geographic heterogeneity between the different tissue sections is evident. Qualitative differences in staining with regard to cytoplasmic or membranous staining or non-specific background staining were not observed. Original magnification, 200x, all fields.
Figure 2. Comparison of anti-PD-L1 antibody clones in melanoma showing percentage of total PD-L1 (+) cells scored by automated image analysis.
(A) Distribution of PD-L1 expression across antibodies for all melanoma specimens. (B) Representative plot showing percentage of total cells in 34 archival melanoma specimens demonstrating membranous PD-L1 staining with clones SP142 and 28-8. Each dot represents a single specimen. A strong correlation was observed, R2=0.938. (C) Pearson’s correlation coefficients for comparisons of percentage of total cells staining for PD-L1 between all studied clones.
Automated scoring was reviewed by a pathologist for each case, and one quarter (43/170) of cases required additional, individualized tuning of the scoring algorithm. This was not specific to any one antibody tested and in almost every instance was required due to incorrect cell segmentation. To ensure this did not introduce a selection bias into our results, we performed a secondary, subset analysis of concordances where the cases requiring additional tuning were excluded, Supplementary Table S2. We found the range of concordances between all the antibodies to be very similar, and the antibodies showing the greatest concordance were the same. We also performed a subset analysis on cases with higher and lower melanin content, and demonstrated that melanin content did not appear to influence the consistently high concordances seen across antibodies, Supplementary Figures 3B and 3C.
Comparison of PD-L1 staining intensity vs percentage of cells staining
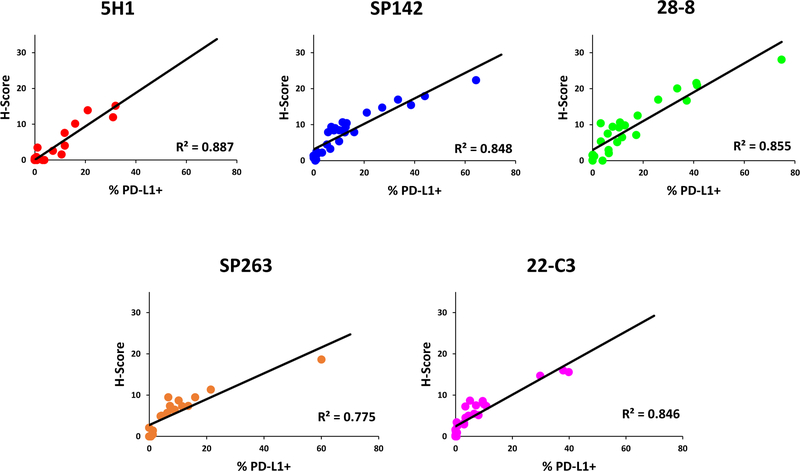
To date, most PD-L1 IHC tests are scored by percentage of cells displaying PD-L1 expression. A metric that includes the intensity of staining, for example an “H-score” that incorporates both the proportion of cells staining and the staining intensity, has not been included. To address the question of whether staining intensity and the percentage of cells staining were independent variables, we compared intensity scoring by a 50-bin H-score to % PD-L1 positivity. For this analysis, we excluded the cases that required individualized cutoffs of positivity by a pathologist. There were strong correlations for all clones between the H-score and percentage of PD-L1(+) cells (R2 = 0.775–0.887), with 5H1 and SP142 showing the closest relationship between intensity and percentage of total cells staining (0.887 and 0.849 respectively), Figure 3.
Figure 3. Comparison of percentage of total PD-L1(+) cells and intensity/H-score using automated image analysis.
Strong correlations were observed between the percentage of cells demonstrating membranous PD-L1 expression and the intensity/H-score across all clones studied, arguing against the inclusion of intensity measurements as an independently scored parameter in PD-L1 IHC assays.
Pathologist qualitative assessment of PD-L1 staining
The slides were independently reviewed by two board-certified pathologists (RAA and JMT) and qualitative differences were not observed in the cell types stained (i.e., immune cells vs. tumor cells) or in the quality of the staining (e.g., cytoplasmic vs. membranous staining patterns or generalizable differences in nonspecific background staining).
Discussion
PD-1 and its ligand PD-L1 are important immunotherapeutic targets, and antibody blockade of this checkpoint has resulted in remarkable antitumor efficacy and durability of responses in patients with melanoma and many other tumor types. Immunohistochemical detection of PD-L1 expression in pre-treatment tumor specimens has been identified as one approach to pre-selecting patients who are more likely to respond to these agents. These findings have largely been generated in the context of clinical trials using four different therapeutic agents (Nivolumab/Opdivo/BMS-936558, Pembrolizumab/Keytruda/MK-3475, Durvalumab/MEDI-4736, and Atezolizumab/MPDL3280A/RG7446) along with the parallel development of four distinct IHC assays for PD-L1. In some instances, FDA approval has been granted for these assays to serve as so-called Companion and Complementary diagnostics for anti-PD-1 therapy.17 Such a “one assay-one drug” paradigm presents many challenges to the health care team, including questions about interchangeability amongst assay results. Comparisons between assays and across trials have been challenging due to different antibodies employed, proprietary staining platforms, and different scoring systems.
In the current study, we compared the performance of five different anti-PD-L1 antibodies on melanoma specimens, including the four antibodies that have been prominently co-developed in trials of PD-1/PD-L1 inhibitors. Importantly, we standardized the other assay reagents, staining methodology, and scoring approaches. We found that all five of the studied antibodies showed consistent qualitative and quantitative performance in highlighting PD-L1 expression in melanoma specimens. Similar findings were recently reported by Gaule, et al, using a combination of quantitative immunofluorescence and chromogenic PD-L1 IHC on non-small cell lung carcinoma (NSCLC) specimens and cell lines in a tissue microarray format.18 When present, differences in staining in both our melanoma cohort and the NSCLC specimens studied by Gaule, et al. were attributable to the previously recognized focal, geographic nature of PD-L1 expression.1,6,19 The antibodies tested in this study would likely demonstrate an even higher concordance if they were repeated on the same tissue section, rather than on adjacent, geographically-distinct sections. We also looked at whether the potential inclusion of an intensity measurement could have added value beyond simply the percentage of total cells staining. We found that intensity of PD-L1 expression was highly correlated with the percentage of total cells staining, arguing against reporting this feature as a separate parameter.
Comparative review by pathologists of slides stained with each of the clones revealed that there was no difference in the antibodies’ ability to highlight PD-L1 expression by immune cells when compared to tumor cells. It is important to emphasize that we did not perform the commercially marketable assays, but used the antibodies employed in those assays in a custom, laboratory-developed test. The concurrent Phase I of the Blueprint PD-L1 Assay Comparison Project compared the marketed assay systems, including the entire packaged assay reagents and proprietary staining platforms, on slides from NSCLC specimens. They showed that the three assays developed by Bristol-Myers Squibb, Merck, and AstraZeneca employing the 28-8, 22C3, and SP263 clones, respectively, were analytically similar for tumor cell staining, while the Genentech assay using SP142 was less sensitive for tumor cell expression of PD-L1.20 A separate study conducted by Rimm, et al on NSCLC also indicated that the assay using SP142 (in this instance a laboratory-derived test (LDT) mimicking the IUO assay) was an outlier in its ability to highlight tumor cell PD-L1 display, when compared to the FDA-approved assays using 28-8 and 22C3 on the Dako Link 48, and an LDT using the E1L3N clone.21 Taken together with our results, these findings suggest that the Genentech SP142 assay likely differs in some other component —primary antibody concentration, antigen retrieval, amplification, or signal development — rather than the intrinsic ability of the antibody to label PD-L1 expression on tumor cells.
PD-L1 is a transmembrane molecule, and it is reasonable to speculate that the location of this epitope recognized by the different antibodies may give rise to variations in PD-L1 staining patterns. As many of the antibodies we studied are proprietary, the information available about the recognized epitopes is limited. It is known that 5H1, 22C3, and 28-8, target the extracellular domain of the protein, while SP142 and SP263 target the intracellular domain. PD-L1 splice variants may demonstrate cleavage of the intracellular domain of PD-L1, but the frequency or significance of this phenomenon is not well-understood.16 In our cohort, we did not observe any cases that were positive by the three extracellular antibodies and negative with SP142 or SP263. When pathologists reviewed the staining properties, differences in the general qualitative staining pattern between those antibodies that label the extracellular vs. intracellular domains of PD-L1, including distributions of staining between the membranous and cytoplasmic compartments as has been previously reported for other tumor types, were not observed.17 However, it is worth noting that the strongest correlation on computer-assisted scoring was observed between the two antibodies targeting the cytoplasmic domain. It is possible that subtle differences in the better delineation of cell membranes by certain antibodies may be masked in melanoma specimens by the presence of melanin in the cytoplasm.
The overall size of our cohort is one limitation of our study, though the high concordances observed suggests that this does not impact significantly on the results. Notwithstanding, there may be subtle differences between the antibodies that may only become apparent when larger cohorts or different tumor types are studied. True appraisal of potential interchangability of antibodies between clinical-grade assays would require extensive assessment of analytic and clinical concordance, including rigorous testing of all assay components (not just antibody performance as was emphasized here), threshold determination, interpretation guidelines, and concordance with clinical response for comprehensive diagnostic-therapeutic validation.
The utility of PD-L1 as a predictive biomarker is further limited by the fact that some patients with PD-L1(−) tumors respond to anti-PD-1/PD-L1 therapy. Some of this discordance is likely attributable to the spatial and temporal heterogeneity of PD-L1 expression as well as the assay methods used to detect it.22 The role that PD-L1 will play as a predictive biomarker is still being evaluated in a number of clinical trials across multiple tumor types. It is possible that in the future, PD-L1 status in patients with melanoma may be used to help triage patients between immunotherapy regimens in an effort to increase the risk-to-benefit ratio. In this scenario, patients whose melanomas are PD-L1(+) might receive first line anti-PD-1/PD-L1 monotherapy, while those whose melanomas are PD-L1(−) would receive combined anti-PD-1 and anti-CTLA-4.23 Before PD-L1 expression is used routinely to guide clinical care decisions, it would be beneficial to understand the relationship of reported results from different assays and even potentially to harmonize PD-L1 IHC assays. Our results suggest that for melanoma specimens, it is differences in the assay protocols, and not in the antibodies themselves, which contribute to the current observed variation in PD-L1 assessments between assays. Our results also further highlight the heterogeneity of PD-L1 expression as a limiting factor of using PD-L1 expression as a solitary biomarker. These findings inform PD-L1 assay comparison efforts and future assay development and standardization.
Supplementary Material
Statement of Translational Relevance:
Drugs blocking PD-1 and PD-L1 have received FDA approval for the treatment of patients with melanoma and other advanced cancers. PD-L1 immunohistochemistry (IHC) companion and complementary diagnostics have been co-developed for each agent to help identify patients who are more likely to respond to therapy. Differences exist amongst these diagnostic assays, and their comparative performance is not yet clear. Our results indicate that the staining characteristics of the 5H1, SP142, 28-8, SP263, and 22C3 clones used in these PD-L1 IHC assays are actually very similar when other assay conditions are held constant. The few differences in PD-L1 staining we observed between antibodies could be attributed to the spatial heterogeneity of PD-L1 expression in melanoma. These findings are important for the comparative understanding of currently marketed assays and potential future reconciliation of these diagnostic tests.
Acknowledgements
The authors would like to thank Dr. David Rimm at Yale University as well as Hao Wang and Brandon Luber at Johns Hopkins University for helpful discussions. This work was supported by the Bloomberg~Kimmel Institute for Cancer Immunotherapy (JMT, EJL, RAA), Dermatology Foundation (JMT), WW. Smith Foundation (JMT), Moving for Melanoma Delaware (JMT, EJL), NIH R01 CA142779 (JMT), NIH T32 CA193145 (TRC), and the Melanoma Research Alliance (JMT, TRC). JT and RAA were also supported by a Stand Up To Cancer—Cancer Research Institute Cancer Immunology Translational Cancer Research Grant (Grant Number: SU2C-AACR-DT1012). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C.
Footnotes
COI for RAA: Consultant (compensated): Adaptive Biotech; Research Funding from Merck and BMS. For EJL: Consultant (compensated): Bristol-Myers Squibb, EMD Serono, Merck, Novartis; Research funding: AstraZeneca, Genetech, Merck. For JMT: Consultant (compensated) Bristol-Myers Squibb, Merck, AstraZeneca; Research Funding from BMS.
References
- 1.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 2012;4:127ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumeh P, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topalian ST, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving Nivolumab. J Clin Oncol 2014;32:1020–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 2015;372:320–30. [DOI] [PubMed] [Google Scholar]
- 5.Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti- CTLA-4 treatment (CheckMate 037): a randomized, controlled, open-label, phase 3 trial. Lancet Oncol 2015;16:375–84. [DOI] [PubMed] [Google Scholar]
- 6.Topalian ST, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 2014;20:5064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipson EJ, Forde PM, Hammers HJ, Emens LA, Taube JM, Topalian SL. Antagonists of PD-1 and PD-L1 in cancer treatment. Semin Oncol 2015;42:587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunshine J and Taube JM. PD-1/PD-L1 inhibitors. Curr Opin Pharmacol 2015;23:32–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taube JM. Unleashing the immune system: PD-1 and PD-Ls in the pre-treatment tumor microenvironment and correlation with response to PD-1/PD-L1 blockade. Oncoimmunology 2014;3:e96341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen AR, Siu LL. PD-L1 Testing in Cancer: Challenges in Companion Diagnostic Development. JAMA Oncol 2016;2:15–6. [DOI] [PubMed] [Google Scholar]
- 13.Taube JM, Young GD, McMiller TL, Chen S, Salas JT, Pritchard TS, et al. Differential Expression of Immune-Regulatory Genes Associated with PD-L1 Display in Melanoma: Implications for PD-1 Pathway Blockade. Clin Cancer Res 2015;21:3969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodić N, Anders RA, Eshleman JR, Lin MT, Xu H, Kim JH, et al. Cancer Immunol Res 2015;3:110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarrella ER, Coulter M, Welsh AW, Carvajal DE, Schalper KA, Harigopal M, et al. Automated measurement of estrogen receptor in breast cancer: a comparison of fluorescent and chromogenic methods of measurement. Lab Invest 2016;96:1016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahoney KM, Sun H, Liao X, Hua P, Callea M, Greenfield EA, et al. PD-L1 antibodies to its cytoplasmic domain most clearly delineate cell membranes in immunohistochemical staining of tumor cells. Cancer Immunol Res 2015;3:1308–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garber K Predictive biomarkers for checkpoints, first tests approved. Nat Biotechnol 2015;33:1217–8. [DOI] [PubMed] [Google Scholar]
- 18.Gaule P, Smithy JW, Toki M, Rehman J, Patell-Socha F, Cougot D, et al. A Quantitative Comparison of Antibodies to Programmed Cell Death 1 Ligand 1. JAMA Oncol. 2016. August 18 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Topalian ST, Taube JM, Anders RA, and Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16(5): 275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsh FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kalangara K, et al. PD-L1 assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. Session: The Blueprint Project: Comparing PD-L1 IHC Diagnostics for Immune Checkpoint Inhibitors. J Thorac Oncol 2017;12:208–222. [DOI] [PubMed] [Google Scholar]
- 21.Rimm D, Han G, Taube JM, Yi ES, Bridge JA, Flieder DB, et al. ORAL01.01: A Prospective, Multi-Institutional Assessment of Four Assays for PD-L1 Expression in NSCLC by Immunohistochemistry: Topic: Pathology. J Thorac Oncol 2016;11:S249. [Google Scholar]
- 22.McLaughlin J, Han G, Schalper KA, Carvajal-Hausdorf D, Pelekanou V, Rehman J, et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small cell lung cancer. JAMA Oncol 2016;2:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. New Engl J Med 2015;373:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.