Abstract
Fibroblast growth factor (FGF) signaling has been implicated in the pathogenesis of pulmonary fibrosis. Mice lacking FGF2 have increased mortality and impaired epithelial recovery after bleomycin exposure, supporting a protective or reparative function following lung injury. To determine whether FGF2 overexpression reduces bleomycin-induced injury, we developed an inducible genetic system to express FGF2 in type II pneumocytes. Double-transgenic (DTG) mice with doxycycline-inducible overexpression of human FGF2 (SPC-rtTA; TRE-hFGF2) or single-transgenic controls were administered intratracheal bleomycin and fed doxycycline chow, starting at either day 0 or 7. In addition, wild-type mice received intratracheal or intravenous recombinant FGF2, starting at the time of bleomycin treatment. Compared to controls, doxycycline-induced DTG mice had decreased pulmonary fibrosis 21 days after bleomycin, as assessed by gene expression and histology. This beneficial effect was seen when FGF2 overexpression was induced at day 0 or day 7 after bleomycin. FGF2 overexpression did not alter epithelial gene expression, bronchoalveolar lavage cellularity or total protein. In vitro studies using primary mouse and human lung fibroblasts showed that FGF2 strongly inhibited baseline and TGFβ1-induced expression of alpha smooth muscle actin (αSMA), collagen, and connective tissue growth factor. While FGF2 did not suppress phosphorylation of Smad2 or Smad-dependent gene expression, FGF2 inhibited TGFβ1-induced stress fiber formation and serum response factor-dependent gene expression. FGF2 inhibition of stress fiber formation and αSMA requires FGF receptor 1 (FGFR1) and downstream MEK/ERK, but not AKT signaling. In summary, overexpression of FGF2 protects against bleomycin-induced pulmonary fibrosis in vivo and reverses TGFβ1-induced collagen and αSMA expression and stress fiber formation in lung fibroblasts in vitro, without affecting either inflammation or epithelial gene expression. Our results suggest that in the lung, FGF2 is antifibrotic in part through decreased collagen expression and fibroblast to myofibroblast differentiation.
Keywords: fibroblast growth factor, fibroblast, pulmonary fibrosis, bleomycin, collagen, transforming growth factor beta, alpha smooth muscle actin, ERK, AKT
Introduction
Idiopathic pulmonary fibrosis (IPF) is a progressive scarring disease of the lung, with abnormal production of extracellular matrix by activated fibroblasts, or myofibroblasts [1]. IPF affects approximately 89,000 patients annually in the U.S., and the median survival is 3–5 years after diagnosis [1]. Current antifibrotic agents nintedanib (Ofev®) and pirfenidone (Esbriet®) decrease the rate of decline in lung function and improve survival in IPF [2,3]; however, overall outcomes remain poor. The cause of fibroblast activation and matrix deposition in IPF is unknown, although it is suggested to be a result of aberrant healing from an injury of unknown origin [4]. Activated myofibroblasts are a critical component of IPF pathogenesis; they express smooth muscle-specific contractile proteins, and secrete extracellular matrix and pro-fibrotic growth factors. Improved understanding of pathways that regulate myofibroblast differentiation is needed to develop effective treatments for pulmonary fibrosis.
The family of fibroblast growth factors (FGFs) comprises eighteen secreted ligands that bind to four distinct signaling receptors (FGFRs) [5]. FGFs are involved in development and tissue repair in a variety of organ systems, including the skin [6], heart [7–9], and lung [10]. FGFRs signal through the ERK/MAPK, AKT/PI3K, PLCγ, and STAT pathways [11], and activate transcription factors such as ETV4 and ETV5 [12]. FGFs have been implicated in the pathogenesis of pulmonary fibrosis, although the specific ligands and mechanisms involved are not well understood. Non-specific pharmacologic and genetic inhibition of FGFRs decreases bleomycin-induced pulmonary fibrosis [13–16]. FGFR1 signaling has also been shown to be required for fibroblast migration in IPF [17]. FGF2 is increased in bronchoalveolar lavage (BAL) fluid from patients with IPF [18], in IPF lung mast cells [19], and in mouse lung macrophages and mast cells after bleomycin [20].
Although FGF2 has been reported to be pro-fibrotic [21], we recently found that endogenous FGF2 is dispensable for generation of fibrosis, but is a critical component for recovery from lung injury [22]. Whereas other FGFs decrease experimental pulmonary fibrosis in vivo [23–28], no studies have shown an antifibrotic effect of FGF2 in the lung, or have demonstrated an antifibrotic effect of any recombinant FGFs when administered after induction of experimental pulmonary fibrosis. FGF2 has been shown to be antifibrotic in liver [29], and studies in corneal endothelium [30,31], valvular interstitial cells [32], vascular endothelium [33–35], and vascular smooth muscle [36–38] suggest that FGF2 inhibits TGFβ1 signaling. We therefore hypothesized that overexpression of FGF2 would be antifibrotic and protective against bleomycin-induced lung injury and pulmonary fibrosis.
Methods
Engineering of doxycycline-inducible FGF2 expression in mice
TRE-hFGF2-IRES-GFP (or TRE-hFGF2) mice were generated by subcloning a cDNA fragment encoding human FGF2 (nucleotides 106–1042 from NM_002006 with additional poly(A) sequence) into the pTRE2/EGFP plasmid. A human FGF2 plasmid was generously provided by Marja Hurle. FGF2 was excised as a 960-bp EcoR1 fragment encoding both high and low molecular weight isoforms. The TRE-hFGF2 transgene (3.4 kb) was excised for injection into FVB blastocysts. Nine founder lines were obtained, mated to MHC-rtTA mice [39], and MHC-rtTA, TRE-hFGF2 double-transgenic mice were induced with doxycycline (Dox) and screened for GFP fluorescence in the heart. Two lines showed very low background expression (without Dox) and strong GFP expression after Dox induction. Line #2 was used for all experiments.
Animal care and use
Mice were housed in a pathogen-free barrier facility and handled in accordance with standard protocols and animal welfare regulations. All procedures complied with the Guide for the Care and Use of Laboratory Animals (NIH publication No. 85-23, revised 1996), and all protocols were approved by the Animal Studies Committee at Washington University School of Medicine and the University of Chicago. SPC-rtTA mice [40] were mated to TRE-hFGF2 mice to generate SPC-rtTA;TRE-hFGF2 (DTG) mice, single-transgenic control mice, and wild-type mice. All mice were maintained in a C57BL/6J;129X1 mixed background.
Bleomycin-induced lung fibrosis
Bleomycin-induced lung injury and pulmonary fibrosis were induced as previously described [16]. Adult mice between 8 and 10 weeks of age were sedated, orally intubated, and administered a single dose of intratracheal bleomycin (1 unit/kg) (Sigma-Aldrich, St. Louis, MO) in sterile phosphate-buffered saline (PBS) or PBS alone. For some experiments, 5 μg of recombinant human low molecular weight FGF2 (Peprotech, #100-18B) in 50 μl PBS was administered intratracheally at the time of bleomycin or intravenously via the retro-orbital route at days 0, 3, 7, and 10 after bleomycin.
Human lung fibroblast (HLF) culture
Primary HLFs were isolated from explanted non-fibrotic human lungs unsuitable for transplantation and donated for research. Protocols were approved by the University of Chicago IRB. Fibroblasts were generated by mincing lung tissue into sub-millimeter pieces and allowing migration out on collagen-coated culture dishes. Fibroblasts were grown in DMEM with 10% FBS and used until passage 10.
For experiments, fibroblasts were grown to 90% confluence, serum-starved overnight in media containing 0.1% FBS, and treated with recombinant human low-molecular weight FGF2 + heparin sulfate (2 mg/ml), TGFβ1 (R&D, 2 ng/ml), or FGF2 + heparin + TGFβ1. When indicated, cells were treated with 0.1 μM PD173074 (Cayman Chemical) during the culture or pretreated with 20 μM U0126 (Cell Signaling) or 1 μM MK-2206 (Selleckchem) for 1 hour before the addition of FGF2 + heparin sulfate and/or TGFβ1.
Statistical analysis
Significant differences in mean values were calculated using paired Student’s t-tests or one-way ANOVA, and survival fractions were calculated using the Kaplan-Meier method. A p-value of less than 0.05 was considered to be significant. Fold change, when used, was calculated as the ratio of two measures (i.e. B/A).
Additional Materials and Methods are provided in Supplementary materials on-line.
Results
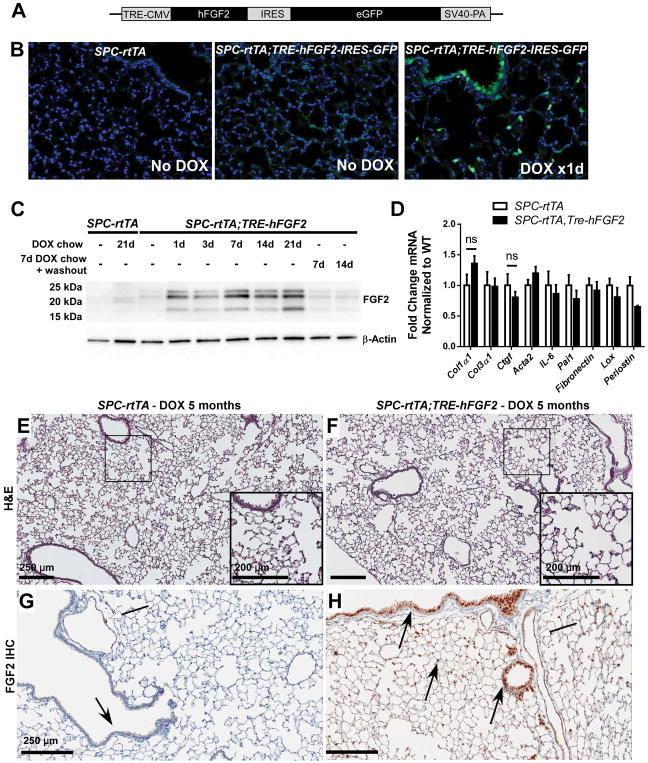
Although transgenic mice with overexpression of FGF2 exist [9], animals with lung-specific or inducible overexpression of FGF2 have not been previously generated. We therefore engineered a mouse line that expresses human FGF2 under the control of a tetracycline-responsive element (TRE-hFGF2), which was used in combination with an alveolar epithelial-driven reverse tetracycline transactivator (SPC-rtTA) to generate mice with doxycycline-inducible, lung-specific overexpression of FGF2. TRE-hFGF2-IRES-GFP (Tre-hFGF2) mice were generated by blastocyst injection with a cDNA fragment encoding human FGF2 within the pTRE2/EGFP plasmid (Figure 1A). TRE-hFGF2 and SPC-rtTA mice were mated to generate SPC-rtTA;TRE-hFGF2 double-transgenic (DTG) mice and littermate SPC-rtTA and TRE-hFGF2 single-transgenic (STG) controls. When fed doxycycline chow, expression of GFP was seen within one day in type II pneumocytes and distal airway epithelium (Figure 1B). All isoforms of human FGF2 were detected within 24 hours via whole-lung Western blot (Figure 1C and supplementary material, Figure S1A) and quantitative RT-PCR (not shown). Significantly elevated levels of FGF2 were not seen in plasma (supplementary material, Figure S1B) or other organs (not shown) of doxycycline-fed DTG mice compared to controls. FGF2 overexpression is reversible following replacement of doxycycline chow with normal chow (Figure 1C). Control and DTG mice were fed doxycycline chow for 3 weeks with no significant alteration in baseline gene expression (Figure 1D), and for up to 5 months with no observable alteration to lung histology (Figure 1E–F). IHC for FGF2 in DTG mice demonstrated increased FGF2 throughout the lung, particularly in epithelium (Figure 1G–H).
Figure 1.
Doxycycline-inducible and reversible overexpression of human FGF2 in mouse lung. (A) Diagram of the TRE-hFGF2-IRES-GFP construct used to generate transgenic TRE-hFGF2 mice. (B) SPC-rtTA and double-transgenic SPC-rtTA;TRE-hFGF2-IRES-GFP mice were fed either normal chow (No DOX) or doxycycline chow (DOX), and lungs were collected for frozen sections. GFP was directly visualized with DAPI as a nuclear stain. (C) SPC-rtTA and double-transgenic SPC-rtTA;TRE-hFGF2-IRES-GFP (or SPC-rtTA;TRE-hFGF2) mice were fed normal chow or DOX chow for specified times and whole lung protein lysates were immunoblotted for FGF2. Where indicated, mice were fed DOX chow for 7 days followed by doxycycline washout with normal chow for 7 or 14 days. (D) Quantitative RT-PCR analysis of whole-lung RNA from DTG and STG control mice fed DOX chow for 21 days. ΔCt values were normalized to Gapdh and expressed as fold change from STG controls. “ns” = not significant. (E–H) SPC-rtTA and SPC-rtTA;TRE-hFGF2 mice were fed DOX chow for 5 months, and lungs were collected for H&E staining (E, F), and FGF2 immunohistochemistry (G, H). White-headed arrows indicate FGF2 staining in vascular smooth muscle; black-headed arrows indicate FGF2 staining in airway and alveolar epithelial cells, which is increased in SPC-rtTA;TRE-hFGF2 mice. Higher magnification of marked areas in E and F is shown in insets, scale bar = 250 μM (200 μM in insets).
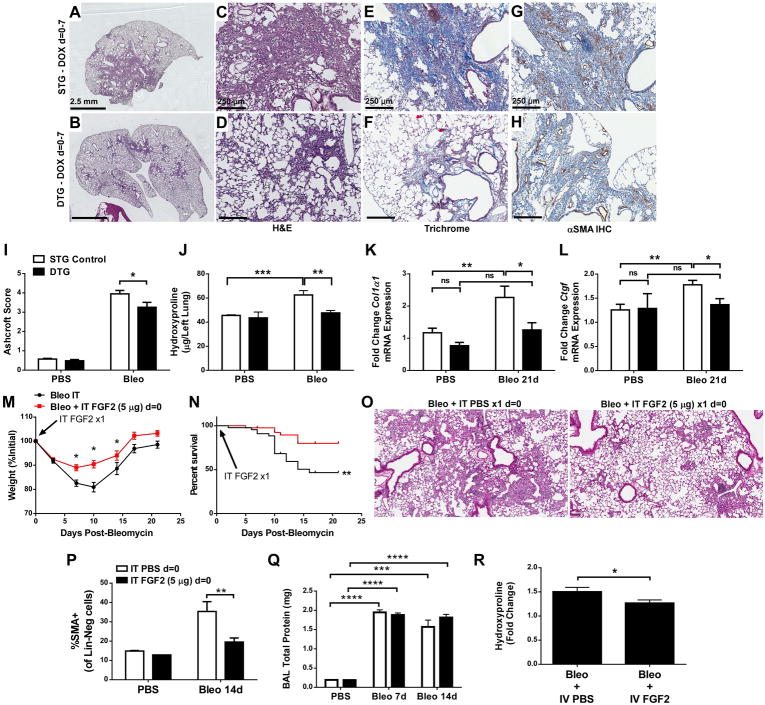
To determine whether overexpression of FGF2 in lung affects the response to bleomycin, STG and DTG mice were treated with intratracheal bleomycin and fed doxycycline chow for the first 7 days after bleomycin treatment. Lungs were collected at 21 days, and lung sections were stained with H&E and Masson’s Trichrome. Bleomycin-treated DTG mice had less fibrosis than STG controls (Figure 2A–F, I) and fewer αSMA+ fibroblasts (Figure 2G, H). Lung collagen was decreased in bleomycin-treated DTG mice compared to STG controls, as assessed by quantification of hydroxyproline (Figure 2J). When doxycycline chow was provided for the entire 21 days following bleomycin, a similar decrease in fibrosis and collagen accumulation in DTG mice compared to STG controls was observed, and no alterations in Tgfβ1 or Tgfβ-r1 expression were seen in DTG mice (Supplementary material, Figure S2). In addition, whole-lung expression of Col1a1, Ctgf (Figure 2K, L), Col3a1, and Pai1 (Supplementary material, Figure S3A, B) in DTG mice was not increased post-bleomycin compared to PBS-treated DTG mice, and was significantly decreased compared to bleomycin-treated STG controls. FGF2 overexpression did not alter BAL total protein or cell count (Supplementary material, Figure S3C, D) and there were no differences in BAL neutrophils, macrophages, lymphocytes, or eosinophils (not shown) following bleomycin treatment. FGF2 overexpression also did not alter whole-lung expression of surfactant protein C (Sftpc) or club cell secretory protein (Scgb1a1) (Supplementary material, Figure S3E, F), and DTG mice had decreased E-cadherin (Cdh1) expression at 7 days, but not at 14 or 21 days, post-bleomycin (Supplementary material, Figure S3G).
Figure 2.
FGF2 immediately post-bleomycin decreases pulmonary fibrosis and collagen accumulation. Single-transgenic (STG) control mice and double-transgenic SPC-rtTA;TRE-hFGF2 (DTG) mice were treated with a single intratracheal dose of bleomycin (1 U/kg) and fed DOX chow starting on day = 0 for 7 days, followed by normal chow. (A–D) Lungs were collected for H&E staining at 21 days post-bleomycin, and low-magnification (1.25 ×, scale bar = 2.5 mm) and 10 × (scale bar = 250 μm) images are shown. (E, F) Masson’s Trichrome staining, 10 × magnification. (G,H) Immunohistochemistry for αSMA, with DAB and hematoxylin as counter-stain, 10 × magnification. (I) Quantification of histologic fibrosis by Ashcroft scoring. (J–L) At 21 days post-bleomycin lungs were collected for quantitative hydroxyproline analysis (J), or for whole-lung RNA isolation. qRT-PCR was performed for Col1α1 (K), and Ctgf (L). ΔCt values were normalized to Gapdh and expressed as fold change from PBS-treated STG controls. Wild-type mice were treated with a single intratracheal dose of recombinant FGF2 at the time of intratracheal bleomycin administration. Weight (M) and mortality (N) was recorded, and lungs were collected for H&E staining 21 days post-bleomycin (O). At 14 days post-bleomycin, lungs were dissociated and stained for αSMA, and % of CD45–CD31–EpCAM–(Lin-Neg) cells that were αSMA+ was measured via flow cytometry (P). Whole lung BAL was measured 7- and 14-days post-bleomycin and total protein from cell-free supernatants was measured (Q). Wild-type mice treated with bleomycin were administered intravenous recombinant FGF2 at d = 0, 3, 7, and 10 post-bleomycin, and quantitative hydroxyproline analysis was performed (R). “ns” = not significant. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.005, **** indicates p < 0.001 using an unpaired 2-way t-test. Analysis of weight and survival was performed using one-way ANOVA and Kaplan-Meier analysis, respectively.
Wild-type mice were then administered intratracheal recombinant FGF2 (rFGF2, 5 μg in PBS) or PBS alone at the time of bleomycin. rFGF2 reduced bleomycin-induced weight loss, mortality, and histologic fibrosis (Figure 2M–O). rFGF2 also significantly decreased αSMA+ fibroblasts 14 days post-bleomycin (Figure 2P) but did not alter BAL cellularity (not shown) or BAL total protein (Figure 2Q). A single dose of intratracheal rFGF2 did not alter total lung collagen 14 days after bleomycin treatment (not shown); however, repeated dosing of intravenous rFGF2 reduced whole lung collagen post-bleomycin (Figure 2R).
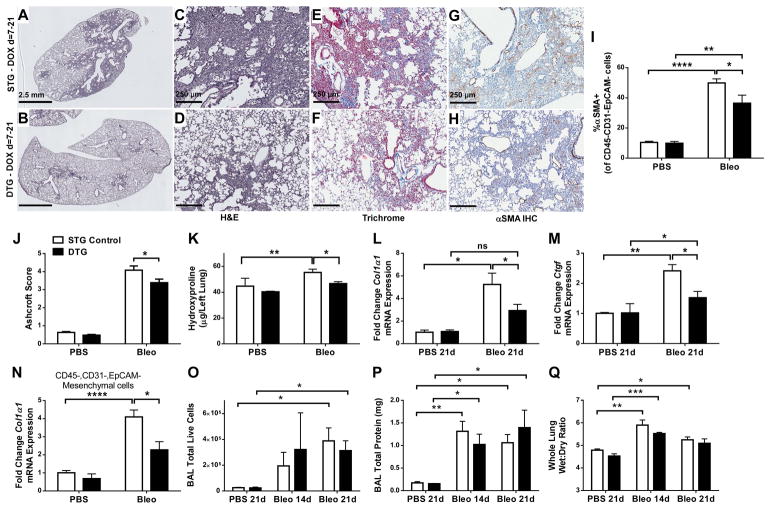
To model therapeutic applications, STG and DTG mice were provided doxycycline chow starting 7 days after bleomycin treatment. Bleomycin did not alter overexpression of human FGF2 in DTG mice (Supplementary material, Figure S4A, B) and IHC for FGF2 in lungs from bleomycin-treated mice revealed diffuse staining for FGF2 in fibrotic areas from STG controls, and strong epithelial FGF2 staining in DTG mice (Supplementary material, Figure S4C, D). Bleomycin-treated DTG mice had decreased histologic pulmonary fibrosis compared to controls, as assessed both quantitatively and qualitatively (Figure 3A–F, J). Fewer αSMA+ mesenchymal cells were seen in bleomycin-treated DTG mouse lungs compared to STG controls, using both IHC and flow cytometry (Figure 3G, H, I). Quantitative hydroxyproline assays indicated decreased lung collagen in bleomycin-treated DTG mice compared to bleomycin-treated STG controls (Figure 3K). Whole-lung expression of Col1a1, Ctgf (Figure 3L,M), and Pai1, but not Col3a1 (Supplementary material, Figure S5A, B), was significantly decreased in bleomycin-treated DTG mice compared to controls, and DTG mice failed to demonstrate a significant increase in Pai1 following bleomycin treatment compared to PBS controls. In addition, Col1a1 expression in freshly sorted lung mesenchymal cells 14 days post-bleomycin was decreased in DTG mice compared to STG controls (Figure 3N). No significant alterations to BAL total protein, BAL cell count, or whole-lung wet:dry ratio were seen in DTG mice post-bleomycin (Figure 3O–Q). Expression of whole-lung Sftpc and Scgb1a1 (Supplementary material, Figure S5C, D) was decreased equally in STG and DTG mice following bleomycin. Also, STG controls, but not DTG mice, demonstrated a decrease in Cdh1 expression 21 days post-bleomycin; however, there was no statistically significant difference between bleomycin-treated STG and DTG mice (Supplementary material, Figure S5E).
Figure 3.
FGF2 overexpression starting 7 days post-bleomycin decreases pulmonary fibrosis and collagen accumulation. Single-transgenic (STG) control mice and double-transgenic SPC-rtTA;TRE-hFGF2 (DTG) mice were treated with a single intratracheal dose of bleomycin (1 U/kg) and fed DOX chow starting on day = 7 until the time of collection. (A–D) Lungs were collected for H&E staining at 21 days post-bleomycin, and 1.2 × (scale bar = 2.5 mm) and 10 × (scale bar = 250 μm) magnified images are shown. (E,F) Masson’s Trichrome staining, 10 × images. (G,H) Immunohistochemistry for αSMA, with DAB and hematoxylin as a counter-stain, 10 × images. (I) At 14 days post-bleomycin, lungs were dissociated and stained for αSMA, and % of CD45−,CD31−,EpCAM- (Lin-Neg) cells that were αSMA+ was measured via flow cytometry. (J) Quantification of histologic fibrosis by Ashcroft scoring. (K–M) At 21 days post-bleomycin, lungs were collected for hydroxyproline quantification (K), or for whole-lung RNA isolation. qRT-PCR was performed for Col1α1 (L), and Ctgf (M). (N) Lin-neg cells from dissociated lungs 14 days post-bleomycin were collected via FACS and qRT-PCR from total RNA was performed for Col1α1. ΔCt values were normalized to Gapdh and expressed as fold change from PBS-treated STG controls. (O–P) At days 14 and 21, whole lung BAL was performed, total live cells were counted (O), and total protein was measured from cell-free supernatants (P). (Q) On days 14 and 21 post-bleomycin whole lungs were collected and weighed before and after dessication to generate wet:dry ratios. “ns” = not significant. * indicates p < 0.05, ** indicates p < 0.01 using an unpaired 2-way t-test, *** indicates p < 0.005 using an unpaired 2-way t-test.
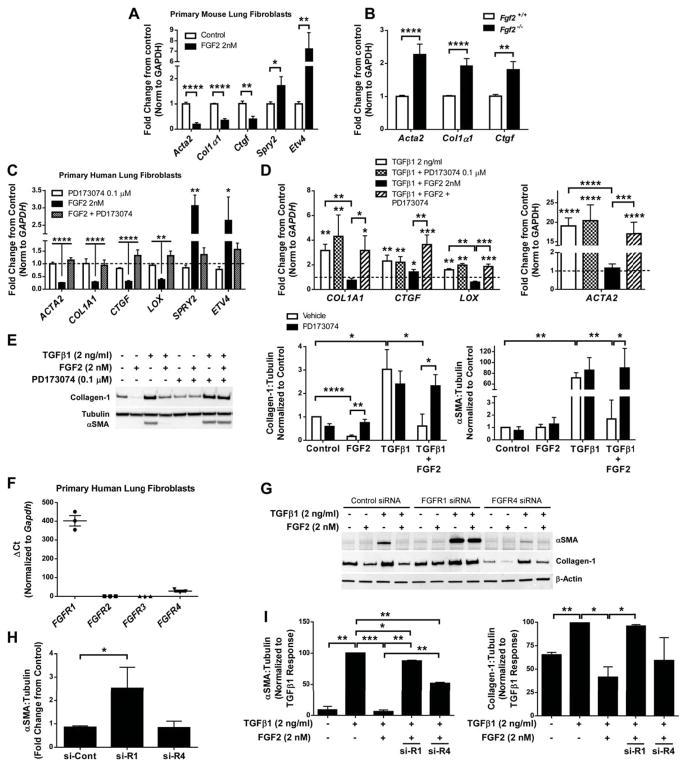
Since FGF2 decreased bleomycin-induced collagen and αSMA expression in whole lung and mesenchymal cells, but did not alter BAL cell counts, BAL total protein, or epithelial gene expression, we hypothesized that FGF2 was antifibrotic, in part, through a direct effect on lung fibroblasts. We found that primary mouse lung fibroblasts treated with recombinant FGF2 (2 nM) had decreased expression of Acta2, Col1a1, and Ctgf, and increased expression of FGF-dependent genes Spry2 and Etv4 (Figure 4A). Expression of Acta2, Col1α1, and Ctgf was increased in cultured primary lung fibroblasts from Fgf2−/− mice compared to Fgf2+/+ controls (Figure 4B), suggesting that endogenous FGF2 produced by fibroblasts can suppress expression of αSMA, collagen, and CTGF.
Figure 4.
Recombinant FGF2 inhibits expression of pro-fibrotic genes in primary cultured lung fibroblasts in an FGFR1-dependent manner. (A) Wild-type mouse lung fibroblasts were serum-starved overnight and treated with recombinant FGF2 (2 nM). 48 hours after treatment total RNA was collected and analyzed via qRT-PCR for expression of Acta2, Col1α1, Ctgf, Spry2, and Etv4. (B) Total RNA was collected from primary lung fibrobasts isolated from Fgf2 +/+ and Fgf2−/− mice, and qRT-PCR was performed for Acta2, Col1α1, and Ctgf. ΔCt values were normalized to Gapdh and expressed as fold change from untreated controls. (C) Primary HLFs were serum-starved overnight and treated with FGF2 (2 nM) and/or PD173074 (0.1 μM) for 48 hours prior to total RNA collection. qRT-PCR was performed for ACTA2, COL1A1, CTGF, LOX, SPRY2, and ETV4. (D) Serum-starved primary HLFs were treated with TGFβ1 (2 ng/ml) or FGF2+TGFβ1 +/− PD173074 (0.1 μM) for 48 hours. qRT-PCR was performed for COL1A1, LOX, and ACTA2. ΔCt values for C and D were normalized to GAPDH and expressed as fold change from untreated controls (represented as dashed line). (E) Total protein was collected from primary HLFs treated with FGF2, TGFβ1, or FGF2+TGFβ1 +/− PD173074 for 48 hours. Collagen-1 and αSMA were measured via western blot, normalized to tubulin, and expressed as fold change from untreated controls. (F) RNA from untreated early passage HLFs was analyzed via qRT-PCR for FGFR1, FGFR2, FGFR3, and FGFR4. ΔCt values were normalized to GAPDH. (G) HLFs were transfected with control siRNA, FGFR1 siRNA, or FGFR4 siRNA, and treated with FGF2, TGFβ1, or TGFβ1+FGF2. Total protein was analyzed via western blot for αSMA, collagen-1, and beta-actin. Densitometric data was quantified and is expressed as fold change from control siRNA (H) and % of TGFβ1 response (I). “ns” = not significant. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.005, **** indicates p < 0.001 using an unpaired 2-way t-test.
We also found that treatment of primary human lung fibroblasts (HLFs) with FGF2 reduced expression of ACTA2, COL1A1, CTGF, and LOX, while increasing FGF-dependent genes SPRY2 and ETV4 (Figure 4C). The effect of FGF2 was inhibited by the FGFR-specific tyrosine kinase inhibitor PD173074 (0.1 μM) (Figure 4C, Supplementary material, Figure S6A). PD173074 on its own did not alter expression of the above genes (Figure 4C). HLFs were then serum-starved and treated with FGF2 (2 nM, or 34 ng/ml), TGFβ1 (2 ng/ml), or TGFβ1 + FGF2 for 48 hours. TGFβ1 increased mRNA expression of COL1A1, CTGF, LOX, and ACTA2, and this was inhibited by co-treatment with FGF2 (Figure 4D). FGF2-mediated inhibition of TGFβ1-induced COL1A1, CTGF, LOX, and ACTA2 expression was inhibited by PD173074 (Figure 4D). FGF2 inhibited TGFβ1-induced Collagen-1 and αSMA protein expression, and this was blocked by PD173074 (Figure 4E). Neither PD173074 nor heparin (co-factor for FGF2/FGFR binding) altered TGFβ1-induced expression of Collagen-1 or αSMA (Figure 4E; Supplementary material, Figure S6B). Lastly, we found that while FGF2 led to an increase in cell migration and proliferation in primary HLFs, it did not reverse the inhibition of migration induced by TGFβ1 and did not increase proliferation when combined with TGFβ1 (Supplementary material, Figure S7).
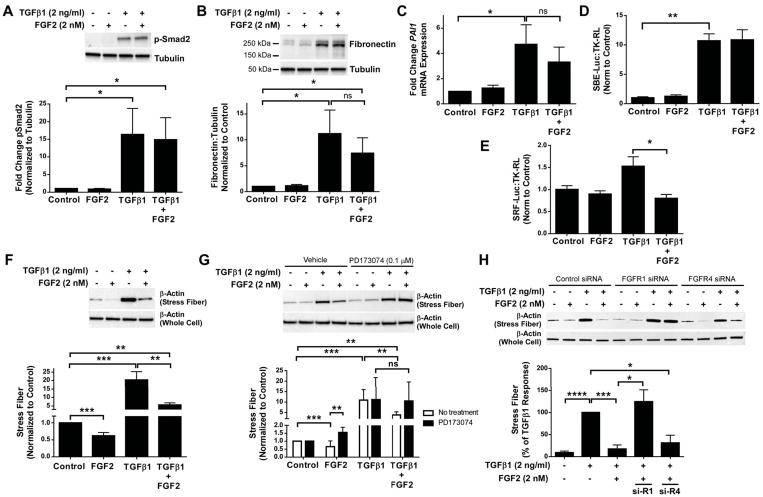
To determine which FGFR is required for FGF2-mediated suppression of collagen and αSMA, we first measured relative expression of FGFRs in HLFs. We found FGFR1 mRNA abundance to be 15-fold higher than FGFR4 (Figure 4F). We did not detect any FGFR2 or FGFR3 in HLFs using either qRT-PCR (Figure 4F) or western blot (not shown). We then used siRNA for selective suppression of either FGFR1 or FGFR4 and found FGFR1 probe #10 and FGFR4 probe #5 to be the most efficient and selective (Supplementary material, Figure S8 and Table S1). siRNA of FGFR1 increased baseline αSMA expression, and completely blocked the inhibitory effect of FGF2 on collagen and αSMA (Supplementary material, Figure S8, Figure 4G–I). siRNA of FGFR4 had no effect on baseline αSMA or collagen expression, only partially blocked FGF2-mediated inhibition of αSMA, and had no effect on FGF2-mediated inhibition of collagen (Figure 4G–I). siRNA of FGFR4 did appear to decrease TGFβ1-induced expression of αSMA independent of FGF2.
To investigate further the mechanism by which FGF2 inhibits TGFβ1-induced αSMA and collagen, we first determined whether FGF2 inhibits TGFβ1-induced Smad signaling. At one hour, FGF2 did not inhibit phosphorylation of Smad2 induced by TGFβ1 in both mouse (not shown) and HLFs (Figure 5A). At 48 hours, FGF2 did not inhibit TGFβ1-induced expression of Smad-dependent genes such as fibronectin (Figure 5B, Supplementary material, Figure S6C) or PAI1 (Figure 5C). Finally, FGF2 did not inhibit TGFβ1-induced Smad-binding element reporter (SBE-luciferase) activity (Figure 5D).
Figure 5.
Recombinant FGF2 inhibits TGFβ1-induced stress fiber formation and activation of the SRF pathway. Primary HLFs were serum-starved and treated with FGF2 (2 nM), TGFβ1 (2 ng/ml), or FGF2 + TGFβ1 for 1 hour or 48 hours. (A) Total protein was collected at 1 hour and immunoblotting was performed for phosphorylated Smad2, normalized to tubulin, and expressed as fold change from untreated control. (B) Total protein was collected at 48 hours and immunoblotting was performed for fibronectin and tubulin. Densitometry was performed for fibronectin, normalized to tubulin, and expressed as fold change from untreated control. (C) Total RNA was collected at 48 hours, and qRT-PCR was performed for PAI1. ΔCt values were normalized to GAPDH and expressed as fold change from untreated controls. (D, E) Primary HLFs were transfected with SBE-luciferase and TK-renilla luciferase (D) or SRF-luciferase and TK-renilla luciferase (E), serum-starved overnight, and treated with FGF2 (2 nM), TGFβ1 (2 ng/ml), or FGF2 + TGFβ1 for 24 hours. Firefly luciferase activity was measured and normalized to renilla luciferase, and expressed as fold change from untreated controls. (F, G) Primary HLFs were treated with FGF2, TGFβ1, or FGF2 + TGFβ1 ± PD173074 for 48 hours prior to collection of purified stress fibers and total cell lysates. (H) HLFs were transfected with control siRNA, FGFR1 siRNA, or FGFR4 siRNA, and treated with FGF2, TGFβ1, or TGFβ1 + FGF2 for 48 hours prior to collection of purified stress fibers and total cell lysates. Western blot for β-actin was performed on stress fiber preparations and whole cell lysates. Densitometry for stress fiber β-actin was normalized to total cell lysate β-actin, and expressed as fold change from untreated control. “ns” = not significant. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.005 using an unpaired 2-way t-test.
We then tested whether FGF2 altered TGFβ1-induced stress fiber formation and serum response factor (SRF) activity, which are critical mediators of TGFβ1-induced αSMA expression [41,42]. Using a SRF-luciferase reporter, we found that FGF2 decreased TGFβ1-induced SRF activity (Figure 5E). FGF2 also decreased baseline and TGFβ1-induced stress fiber formation (Figure 5F), an effect not seen with heparin alone, and inhibition of stress fiber formation by FGF2 was blocked by PD173074 (Figure 5G, Supplementary material, Figure S6B). Furthermore, inhibition of stress fiber formation by FGF2 was inhibited by siRNA of FGFR1, but not FGFR4 (Figure 5H). In total, these data suggest that FGF2 signals through FGFR1 to reduce TGFβ1-induced gene expression in part through inhibition of stress fiber formation and SRF-dependent gene transcription.
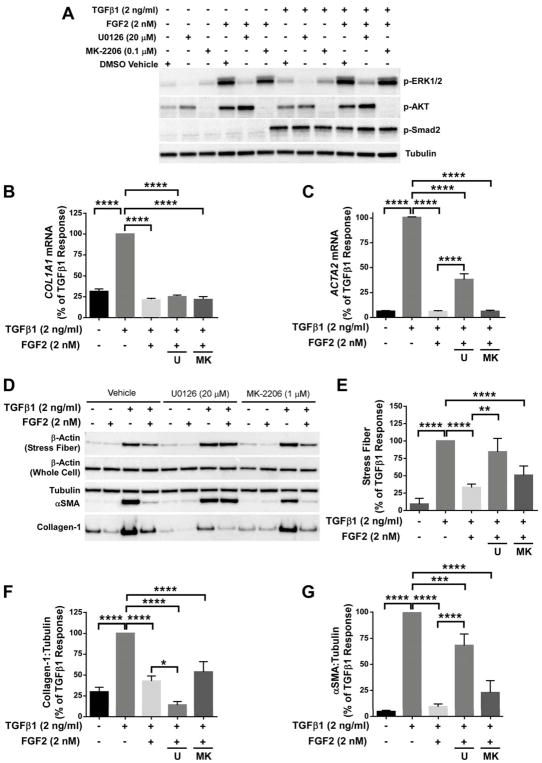
FGF2 signals through multiple known pathways downstream of FGFR1, including MEK/ERK and AKT [11]. We tested whether the inhibition of collagen and αSMA by FGF2 was mediated by MEK/ERK or AKT, using U0126 and MK-2206, respectively. These compounds result in selective inhibition of ERK1/2 and AKT phosphorylation without altering TGFβ1-induced Smad2 phosphorylation (Figure 6A). When added to HLFs in combination with TGFβ1 and FGF2, U0126 blocked FGF2-mediated inhibition of ACTA2, but not COL1A1 mRNA (Figure 6B, C). Inhibition of MEK/ERK, but not AKT, blocked inhibition of stress fiber formation and αSMA expression by FGF2, while inhibition of collagen-1 by FGF2 was not blocked by either U0126 or MK-2206 (Figure 6D–G). These results suggest that multiple pathways are involved in the antifibrotic effect of FGF2, as FGF2 inhibited stress fiber formation and αSMA expression through MEK/ERK signaling, while inhibition of collagen did not require MEK/ERK or AKT.
Figure 6.
FGF2 inhibition of TGFβ1 induced αSMA requires MEK/ERK, but not AKT. Primary HLFs were serum-starved and treated with FGF2 (2 nM), TGFβ1 (2 ng/ml), or FGF2 + TGFβ1 for 1 hour or 48 hours in the presence or absence of U0126 (20 μM) or MK-2206 (0.1 μM). (A) Total protein was collected at 1 hour and immunoblotting was performed for phosphorylated ERK1/2, AKT, and Smad2, and normalized to tubulin. (B, C) Total RNA was collected at 48 hours, and qRT-PCR was performed for COL1A1 (B) or ACTA2 (C). ΔCt values were normalized to GAPDH and expressed as % of TGFβ1 response. (D) Purified stress fibers and total protein were collected at 48 hours, and immunoblotting was performed for beta-actin, αSMA, collagen-1, and tubulin. (E–G) Densitometry was performed and expressed as % of TGFβ1 response. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.005, **** indicates p < 0.001 using an unpaired 2-way t-test.
Discussion
Overall, this report demonstrates a protective effect of FGF2 against bleomycin-induced pulmonary fibrosis and suggests that the antifibrotic effect occurs in part through suppression of fibroblast collagen production and differentiation of lung fibroblasts to αSMA+ myofibroblasts. TRE-hFGF2 mice represent a useful reagent to study tissue-restricted effects of FGF2 overexpression in vivo, when combined with cell type-specific rtTA expression. Similar to previous reports showing that generalized overexpression or administration of FGF2 does not cause pulmonary or cardiac abnormalities [9,43], we found that overexpression of FGF2 for up to 5 months does not lead to histologic changes. This suggests that treatment with FGF2 is well-tolerated without causing spontaneous lung abnormalities.
This is the first report of an antifibrotic effect of FGF2 in the lung in vivo. Studies in eye [30,31,38], synovium [44], breast glands [45], and blood vessels [32,33,35–38] support our findings that FGF2 inhibits TGFβ1-induced αSMA expression. Other FGFs, including FGF1, FGF7, FGF9, and FGF10, can decrease experimental pulmonary fibrosis in vivo, and FGF1 and FGF9 decrease myofibroblast differentiation in vitro [17,24–28,46,47]. While pre-treatment with recombinant FGF7 decreases bleomycin-induced pulmonary fibrosis in rodents [47–49], no study to date has demonstrated an antifibrotic effect of recombinant FGFs at the time of or subsequent to bleomycin treatment. Recombinant FGF2 improves recovery from myocardial ischemia, hindlimb ischemia, gastric ulcers, bone fractures, and skin wounds [8,9,50–53], and reduces liver fibrosis [29,54]. In the lung, recombinant FGF2 protects from experimental emphysema, allergen-induced airway hyperreactivity, and influenza [50,55–60], but has not previously been tested in models of pulmonary fibrosis. In this report, we show not only that overexpression of FGF2 reduces pulmonary fibrosis, but also that administration of intratracheal recombinant FGF2 reduces bleomycin-induced mortality and pulmonary fibrosis. More studies are still needed to determine the effect of FGF2 on lung function and physiology after bleomycin injury, as well as the optimal dose, route, and timing of FGF2 administration to maximize its antifibrotic effect.
Differentiation of lung fibroblasts to myofibroblasts is a key event in the pathogenesis of pulmonary fibrosis and fibrotic diseases of other organs [61]. Stress fiber formation is required for TGFβ1-induced αSMA expression through MRTF nuclear translocation and the SRF transcription factor [41,42,62,63]. Both in vivo and in vitro, we show that FGF2 treatment reduces the number of αSMA+ fibroblasts and αSMA expression in mouse and human fibroblasts, as well as other myofibroblast-associated genes such as CTGF. This inhibition was Smad-independent, and could be blocked with an FGFR-specific tyrosine kinase inhibitor, selective siRNA knockdown of FGFR1, or with a MEK/ERK inhibitor. Inhibition of myofibroblast differentiation by FGF9 was shown to require FGFR3 [27,28]; however we were unable to detect FGFR3 in our primary HLFs. We show that FGF2 blocks TGFβ1 induction of stress fibers and SRF reporter activity, suggesting that the mechanism involved in FGF2 inhibition of αSMA expression is downstream of MEK/ERK, and upstream of stress fiber formation and actin polymerization.
Multiple mechanisms of inhibition of TGFβ1 by FGFs in different organs and cell types have been proposed, including ERK [32,36,38,64], focal adhesion kinase [65], Nkx2.5/Csx [66], let-7 miRNA [34,37], downregulation of TGFβ1 and TGFβR1 [34], decreased Smad2 phosphorylation [67], miR-20a [35], alteration in Rho activity [68,69], and PI3-kinase [31], suggesting that the effect of FGF2 may not be universal in all cell types. In contrast to some of these studies, we found that FGF2 did not inhibit Smad2 phosphorylation or Smad-dependent gene expression. We found that, in primary human lung fibroblasts, FGF2 inhibited TGFβ1-induced stress fiber formation and αSMA expression through an ERK, but not AKT, signaling pathway. These data are supported by recent findings in skin fibroblasts that demonstrate ERK-dependent inhibition of αSMA by FGF2 [70]. The pathway involved in the inhibition of collagen expression by FGF2 remains unclear, as FGF2 inhibition of collagen mRNA does not appear to require intact MEK/ERK or AKT signaling, suggesting that multiple pathways are involved in the antifibrotic effect of FGF2. Overall, these data indicate that an FGF2-FGFR1 signaling axis via MAPK pathways suppresses stress fiber formation and αSMA expression in lung fibroblasts. FGF2 overexpression had no effect on BAL protein and cell count or whole lung epithelial gene expression following bleomycin treatment. FGF2 binds with high affinity to principally the “c”, or mesenchymal, isoforms of FGFRs [11], but can also interact with the “b”, or epithelial, isoform of FGFR1 and FGFR2 in vitro [71]. FGF2 overexpression may have a dominant effect on mesenchymal cells expressing “c” isoforms relative to “b” isoform-expressing epithelium in vivo. This does not exclude an effect of FGF2 on epithelium or other cell types in the lung, and the effect of FGF2 on fibroblasts in vivo may be indirect. For example, while we observed a decrease in bleomycin-induced Pai1 expression due to FGF2 overexpression in vivo, this was not seen in isolated fibroblasts, suggesting that other cell types are influenced by FGF2. Additional studies are needed to determine the effect of FGF2 overexpression on other cell types in experimental pulmonary fibrosis.
The current study does not address how endogenous FGF2 alters gene expression in lung fibroblasts in vivo or in fibroblasts from patients with pulmonary fibrosis. In the first place, the amounts of FGF2 used far exceed endogenous FGF2 described in vivo after lung injury and in IPF [18–20,22]. Additionally, endogenous FGF2 [22], FGF7, and FGF10 [72] are not essential for pulmonary fibrosis in mice, probably due to compensation by other FGF ligands. Interestingly, we found that deletion of FGF2 and siRNA knockdown of FGFR1 leads to increased expression of collagen and αSMA in the absence of TGFβ1 stimulation, suggesting that FGF2 may play a role in the homeostatic regulation of these genes. We have not observed increased fibrosis in Fgf2−/− mice, probably due to compensation by other FGFs in vivo [22], and we have also found that FGF2 expression is excluded from fibroblastic foci in IPF [22]. This suggests that fibroblasts in IPF may have alterations in FGF signaling, and additional studies are needed to explore this further.
As outcomes in pulmonary fibrosis and IPF remain poor despite the emergence of antifibrotic agents, improved understanding of factors that influence myofibroblast differentiation and gene expression are needed. Our data suggest that FGF2 signaling is important in regulating myofibroblast differentiation and collagen production, but further investigation is still needed to better understand FGF2-dependent pathways that mediate this effect. Administration of FGF2 or activation of FGF2-dependent signaling may prove to be useful for the treatment of pulmonary fibrosis.
Supplementary Material
siRNA sequences used for knockdown of FGFR1 and FGFR4
Figure S1. Human FGF2 overexpression in DTG mice in response to doxycycline
Figure S2. Continuous overexpression of FGF2 for 21 days following bleomycin reduces pulmonary fibrosis and collagen accumulation
Figure S3. FGF2 overexpression decreases collagen 3 and Pai-1 expression, but does not alter bleomycin-induced lung injury or epithelial gene expression
Figure S4. Doxycycline-induced overexpression of FGF2 is unaltered by bleomycin treatment
Figure S5. FGF2 overexpression beginning 7 days post-bleomycin decreases Pai-1 expression, but does not alter epithelial gene expression
Figure S6. The FGFR-specific tyrosine kinase inhibitor PD173074 blocks the effect of FGF2, but not TGFβ1 in primary human lung fibroblasts
Figure S7. Recombinant FGF2 stimulates fibroblast migration and proliferation, but does not alter effect of TGFβ1 in primary human lung fibroblasts
Figure S8. Efficient and selective knockdown of FGFR1 and FGFR4 using siRNA
Acknowledgments
This work was funded by NIH grant HL111190, Washington University Physician Scientist Training Program, AHA grant 14FTF19840029, American Lung Association Dalsemer Research Grant, and NIH/NHLBI T32HL007317. LMFEB was supported by The Culture Affairs and Mission Sector, Ministry of Higher Education and Scientific Research, Egypt. We thank M. Hurley for providing the hFGF2 cDNA.
Footnotes
Conflict of Interest Statement:
None of the authors have any relevant conflicts of interest or declarations pertaining to this manuscript.
Original Publication Statement:
No portion of this manuscript has been published elsewhere. A portion of this work has previously been presented in abstract form at the American Thoracic Society International Conference.
Statement of Author Contributions
HYK and LMFEB contributed equally to this manuscript. HYK performed and analyzed all in vitro fibroblast experiments, and contributed to manuscript revisions. LMFEB performed and analyzed BAL analysis, whole lung qRT-PCR, western blots, ELISA, and IHC staining and contributed to manuscript revisions. SJD performed BAL analysis and hydroxyproline assays. SLH, SNC, and DMO designed and developed TRE-hFGF2 mice. NMS, MLS, and HSH contributed to manuscript revision. ND assisted with in vitro experimental design and manuscript revision. DMO contributed to experimental design and manuscript revision. RDG designed and supervised all experiments, analyzed data, and wrote the manuscript.
References
* Cited only in on-line supplementary material
- 1.King TE, Jr, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378:1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 2.Richeldi L, du Bois RM, Raghu G, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 3.King TE, Jr, Bradford WZ, Castro-Bernardini S, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell TS, Tager AM, Borok Z, et al. Future Directions in Idiopathic Pulmonary Fibrosis Research. An NHLBI Workshop Report. American journal of respiratory and critical care medicine. 2014;189:214–222. doi: 10.1164/rccm.201306-1141WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149:121–130. doi: 10.1093/jb/mvq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortega S, Ittmann M, Tsang SH, et al. Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virag JA, Rolle ML, Reece J, et al. Fibroblast growth factor-2 regulates myocardial infarct repair: effects on cell proliferation, scar contraction, and ventricular function. The American journal of pathology. 2007;171:1431–1440. doi: 10.2353/ajpath.2007.070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao S, Porter D, Scott A, et al. The cardioprotective effect of the low molecular weight isoform of fibroblast growth factor-2: the role of JNK signaling. Journal of Molecular and Cellular Cardiology. 2007;42:106–120. doi: 10.1016/j.yjmcc.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.House SL, Bolte C, Zhou M, et al. Cardiac-specific overexpression of fibroblast growth factor-2 protects against myocardial dysfunction and infarction in a murine model of low-flow ischemia. Circulation. 2003;108:3140–3148. doi: 10.1161/01.CIR.0000105723.91637.1C. [DOI] [PubMed] [Google Scholar]
- 10.Hokuto I, Perl AK, Whitsett JA. FGF signaling is required for pulmonary homeostasis following hyperoxia. American journal of physiology Lung cellular and molecular physiology. 2004;286:L580–587. doi: 10.1152/ajplung.00278.2003. [DOI] [PubMed] [Google Scholar]
- 11.Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley interdisciplinary reviews Developmental biology. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao J, McGlinn E, Huang P, et al. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev Cell. 2009;16:600–606. doi: 10.1016/j.devcel.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wollin L, Maillet I, Quesniaux V, et al. Antifibrotic and anti-inflammatory activity of the tyrosine kinase inhibitor nintedanib in experimental models of lung fibrosis. J Pharmacol Exp Ther. 2014;349:209–220. doi: 10.1124/jpet.113.208223. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhary NI, Roth GJ, Hilberg F, et al. Inhibition of PDGF, VEGF and FGF signalling attenuates fibrosis. The European respiratory journal. 2007;29:976–985. doi: 10.1183/09031936.00152106. [DOI] [PubMed] [Google Scholar]
- 15.Yu ZH, Wang DD, Zhou ZY, et al. Mutant soluble ectodomain of fibroblast growth factor receptor-2 IIIc attenuates bleomycin-induced pulmonary fibrosis in mice. Biol Pharm Bull. 2012;35:731–736. doi: 10.1248/bpb.35.731. [DOI] [PubMed] [Google Scholar]
- 16.Guzy RD, Li L, Smith C, et al. Pulmonary fibrosis requires cell-autonomous mesenchymal fibroblast growth factor (FGF) signaling. The Journal of biological chemistry. 2017;292:10364–10378. doi: 10.1074/jbc.M117.791764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKenzie B, Korfei M, Henneke I, et al. Increased FGF1-FGFRc expression in idiopathic pulmonary fibrosis. Respiratory research. 2015;16:83. doi: 10.1186/s12931-015-0242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue Y, King TE, Jr, Tinkle SS, et al. Human mast cell basic fibroblast growth factor in pulmonary fibrotic disorders. The American journal of pathology. 1996;149:2037–2054. [PMC free article] [PubMed] [Google Scholar]
- 19.Qu Z, Liebler JM, Powers MR, et al. Mast cells are a major source of basic fibroblast growth factor in chronic inflammation and cutaneous hemangioma. The American journal of pathology. 1995;147:564–573. [PMC free article] [PubMed] [Google Scholar]
- 20.Liebler JM, Picou MA, Qu Z, et al. Altered immunohistochemical localization of basic fibroblast growth factor after bleomycin-induced lung injury. Growth Factors. 1997;14:25–38. doi: 10.3109/08977199709021508. [DOI] [PubMed] [Google Scholar]
- 21.Xiao L, Du Y, Shen Y, et al. TGF-beta 1 induced fibroblast proliferation is mediated by the FGF-2/ERK pathway. Front Biosci. 2012;17:2667–2674. doi: 10.2741/4077. [DOI] [PubMed] [Google Scholar]
- 22.Guzy RD, Stoilov I, Elton TJ, et al. Fibroblast growth factor 2 is required for epithelial recovery, but not for pulmonary fibrosis, in response to bleomycin. American journal of respiratory cell and molecular biology. 2015;52:116–128. doi: 10.1165/rcmb.2014-0184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi ES, Williams ST, Lee H, et al. Keratinocyte growth factor ameliorates radiation- and bleomycin-induced lung injury and mortality. The American journal of pathology. 1996;149:1963–1970. [PMC free article] [PubMed] [Google Scholar]
- 24.Gupte VV, Ramasamy SK, Reddy R, et al. Overexpression of fibroblast growth factor-10 during both inflammatory and fibrotic phases attenuates bleomycin-induced pulmonary fibrosis in mice. American journal of respiratory and critical care medicine. 2009;180:424–436. doi: 10.1164/rccm.200811-1794OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimbori C, Bellaye PS, Xia J, et al. Fibroblast growth factor-1 attenuates TGF-beta1-induced lung fibrosis. The Journal of pathology. 2016;240:197–210. doi: 10.1002/path.4768. [DOI] [PubMed] [Google Scholar]
- 26.Ramos C, Montano M, Becerril C, et al. Acidic fibroblast growth factor decreases alpha-smooth muscle actin expression and induces apoptosis in human normal lung fibroblasts. American journal of physiology Lung cellular and molecular physiology. 2006;291:L871–879. doi: 10.1152/ajplung.00019.2006. [DOI] [PubMed] [Google Scholar]
- 27.Joannes A, Brayer S, Besnard V, et al. FGF9 and FGF18 in idiopathic pulmonary fibrosis promote survival and migration and inhibit myofibroblast differentiation of human lung fibroblasts in vitro. American journal of physiology Lung cellular and molecular physiology. 2016 doi: 10.1152/ajplung.00185.2015. ajplung 00185 02015. [DOI] [PubMed] [Google Scholar]
- 28.Justet A, Joannes A, Besnard V, et al. FGF9 prevents pleural fibrosis induced by intra-pleural adenovirus injection in mice. American journal of physiology Lung cellular and molecular physiology. 2017 doi: 10.1152/ajplung.00508.2016. ajplung 00508 02016. [DOI] [PubMed] [Google Scholar]
- 29.Sato-Matsubara M, Matsubara T, Daikoku A, et al. Fibroblast growth factor 2 (FGF2) regulates cytoglobin expression and activation of human hepatic stellate cells via JNK signaling. The Journal of biological chemistry. 2017;292:18961–18972. doi: 10.1074/jbc.M117.793794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee HT, Lee JG, Na M, et al. FGF-2 induced by interleukin-1 beta through the action of phosphatidylinositol 3-kinase mediates endothelial mesenchymal transformation in corneal endothelial cells. The Journal of biological chemistry. 2004;279:32325–32332. doi: 10.1074/jbc.M405208200. [DOI] [PubMed] [Google Scholar]
- 31.Lee HT, Kay EP. FGF-2 induced reorganization and disruption of actin cytoskeleton through PI 3-kinase, Rho, and Cdc42 in corneal endothelial cells. Mol Vis. 2003;9:624–634. [PubMed] [Google Scholar]
- 32.Cushing MC, Mariner PD, Liao JT, et al. Fibroblast growth factor represses Smad-mediated myofibroblast activation in aortic valvular interstitial cells. FASEB J. 2008;22:1769–1777. doi: 10.1096/fj.07-087627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen PY, Qin L, Tellides G, et al. Fibroblast growth factor receptor 1 is a key inhibitor of TGFbeta signaling in the endothelium. Sci Signal. 2014;7:ra90. doi: 10.1126/scisignal.2005504. [DOI] [PubMed] [Google Scholar]
- 34.Chen PY, Qin L, Barnes C, et al. FGF regulates TGF-beta signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep. 2012;2:1684–1696. doi: 10.1016/j.celrep.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Correia AC, Moonen JR, Brinker MG, et al. FGF2 inhibits endothelial-mesenchymal transition through microRNA-20a-mediated repression of canonical TGF-beta signaling. J Cell Sci. 2016;129:569–579. doi: 10.1242/jcs.176248. [DOI] [PubMed] [Google Scholar]
- 36.Kawai-Kowase K, Sato H, Oyama Y, et al. Basic fibroblast growth factor antagonizes transforming growth factor-beta1-induced smooth muscle gene expression through extracellular signal-regulated kinase 1/2 signaling pathway activation. Arterioscler Thromb Vasc Biol. 2004;24:1384–1390. doi: 10.1161/01.ATV.0000136548.17816.07. [DOI] [PubMed] [Google Scholar]
- 37.Chen PY, Qin L, Li G, et al. Fibroblast growth factor (FGF) signaling regulates transforming growth factor beta (TGFbeta)-dependent smooth muscle cell phenotype modulation. Sci Rep. 2016;6:33407. doi: 10.1038/srep33407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papetti M, Shujath J, Riley KN, et al. FGF-2 antagonizes the TGF-beta1-mediated induction of pericyte alpha-smooth muscle actin expression: a role for myf-5 and Smad-mediated signaling pathways. Invest Ophthalmol Vis Sci. 2003;44:4994–5005. doi: 10.1167/iovs.03-0291. [DOI] [PubMed] [Google Scholar]
- 39.Valencik ML, McDonald JA. Codon optimization markedly improves doxycycline regulated gene expression in the mouse heart. Transgenic research. 2001;10:269–275. doi: 10.1023/a:1016601928465. [DOI] [PubMed] [Google Scholar]
- 40.Perl AK, Tichelaar JW, Whitsett JA. Conditional gene expression in the respiratory epithelium of the mouse. Transgenic research. 2002;11:21–29. doi: 10.1023/a:1013986627504. [DOI] [PubMed] [Google Scholar]
- 41.Sandbo N, Kregel S, Taurin S, et al. Critical role of serum response factor in pulmonary myofibroblast differentiation induced by TGF-beta. American journal of respiratory cell and molecular biology. 2009;41:332–338. doi: 10.1165/rcmb.2008-0288OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandbo N, Lau A, Kach J, et al. Delayed stress fiber formation mediates pulmonary myofibroblast differentiation in response to TGF-beta. American journal of physiology Lung cellular and molecular physiology. 2011;301:L656–666. doi: 10.1152/ajplung.00166.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coffin JD, Florkiewicz RZ, Neumann J, et al. Abnormal bone growth and selective translational regulation in basic fibroblast growth factor (FGF-2) transgenic mice. Mol Biol Cell. 1995;6:1861–1873. doi: 10.1091/mbc.6.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattey DL, Dawes PT, Nixon NB, et al. Transforming growth factor beta 1 and interleukin 4 induced alpha smooth muscle actin expression and myofibroblast-like differentiation in human synovial fibroblasts in vitro: modulation by basic fibroblast growth factor. Ann Rheum Dis. 1997;56:426–431. doi: 10.1136/ard.56.7.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ronnov-Jessen L, Petersen OW. Induction of alpha-smooth muscle actin by transforming growth factor-beta 1 in quiescent human breast gland fibroblasts. Implications for myofibroblast generation in breast neoplasia. Lab Invest. 1993;68:696–707. [PubMed] [Google Scholar]
- 46.Coffey E, Newman DR, Sannes PL. Expression of fibroblast growth factor 9 in normal human lung and idiopathic pulmonary fibrosis. J Histochem Cytochem. 2013;61:671–679. doi: 10.1369/0022155413497366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugahara K, Iyama K, Kuroda MJ, et al. Double intratracheal instillation of keratinocyte growth factor prevents bleomycin-induced lung fibrosis in rats. The Journal of pathology. 1998;186:90–98. doi: 10.1002/(SICI)1096-9896(199809)186:1<90::AID-PATH137>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 48.Deterding RR, Havill AM, Yano T, et al. Prevention of bleomycin-induced lung injury in rats by keratinocyte growth factor. Proc Assoc Am Physicians. 1997;109:254–268. [PubMed] [Google Scholar]
- 49.Guo J, Yi ES, Havill AM, et al. Intravenous keratinocyte growth factor protects against experimental pulmonary injury. The American journal of physiology. 1998;275:L800–805. doi: 10.1152/ajplung.1998.275.4.L800. [DOI] [PubMed] [Google Scholar]
- 50.Jeon SG, Lee CG, Oh MH, et al. Recombinant basic fibroblast growth factor inhibits the airway hyperresponsiveness, mucus production, and lung inflammation induced by an allergen challenge. J Allergy Clin Immunol. 2007;119:831–837. doi: 10.1016/j.jaci.2006.12.653. [DOI] [PubMed] [Google Scholar]
- 51.Ohura T, Nakajo T, Moriguchi T, et al. Clinical efficacy of basic fibroblast growth factor on pressure ulcers: case-control pairing study using a new evaluation method. Wound Repair Regen. 2011;19:542–551. doi: 10.1111/j.1524-475X.2011.00726.x. [DOI] [PubMed] [Google Scholar]
- 52.Yao C, Yao P, Wu H, et al. Acceleration of wound healing in traumatic ulcers by absorbable collagen sponge containing recombinant basic fibroblast growth factor. Biomedical materials. 2006;1:33–37. doi: 10.1088/1748-6041/1/1/005. [DOI] [PubMed] [Google Scholar]
- 53.Fu X, Shen Z, Chen Y, et al. Recombinant bovine basic fibroblast growth factor accelerates wound healing in patients with burns, donor sites and chronic dermal ulcers. Chinese medical journal. 2000;113:367–371. [PubMed] [Google Scholar]
- 54.Pan RL, Xiang LX, Wang P, et al. Low-molecular-weight fibroblast growth factor 2 attenuates hepatic fibrosis by epigenetic down-regulation of Delta-like1. Hepatology. 2015;61:1708–1720. doi: 10.1002/hep.27649. [DOI] [PubMed] [Google Scholar]
- 55.Wang K, Lai C, Li T, et al. Basic fibroblast growth factor protects against influenza A virus-induced acute lung injury by recruiting neutrophils. J Mol Cell Biol. 2017 doi: 10.1093/jmcb/mjx047. [DOI] [PubMed] [Google Scholar]
- 56.Lee BJ, Moon HG, Shin TS, et al. Protective effects of basic fibroblast growth factor in the development of emphysema induced by interferon-gamma. Exp Mol Med. 2011;43:169–178. doi: 10.3858/emm.2011.43.4.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawago M, Yoshimasu T, Tabata Y, et al. Intrapleural administration of gelatin-embedded, sustained-release basic fibroblast growth factor for the regeneration of emphysematous lungs in rats. J Thorac Cardiovasc Surg. 2014;147:1644–1649. doi: 10.1016/j.jtcvs.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 58.Morino S, Toba T, Tao H, et al. Fibroblast growth factor-2 promotes recovery of pulmonary function in a canine models of elastase-induced emphysema. Exp Lung Res. 2007;33:15–26. doi: 10.1080/01902140601113070. [DOI] [PubMed] [Google Scholar]
- 59.Morino S, Nakamura T, Toba T, et al. Fibroblast growth factor-2 induces recovery of pulmonary blood flow in canine emphysema models. Chest. 2005;128:920–926. doi: 10.1378/chest.128.2.920. [DOI] [PubMed] [Google Scholar]
- 60.Chang SS, Yokomise H, Matsuura N, et al. Novel therapeutic approach for pulmonary emphysema using gelatin microspheres releasing basic fibroblast growth factor in a canine model. Surg Today. 2014;44:1536–1541. doi: 10.1007/s00595-014-0864-x. [DOI] [PubMed] [Google Scholar]
- 61.Phan SH. Fibroblast phenotypes in pulmonary fibrosis. American journal of respiratory cell and molecular biology. 2003;29:S87–92. [PubMed] [Google Scholar]
- 62.Sandbo N, Ngam C, Torr E, et al. Control of myofibroblast differentiation by microtubule dynamics through a regulated localization of mDia2. The Journal of biological chemistry. 2013;288:15466–15473. doi: 10.1074/jbc.M113.464461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandbo N, Dulin N. Actin cytoskeleton in myofibroblast differentiation: ultrastructure defining form and driving function. Transl Res. 2011;158:181–196. doi: 10.1016/j.trsl.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kubo E, Shibata S, Shibata T, et al. FGF2 antagonizes aberrant TGFbeta regulation of tropomyosin: role for posterior capsule opacity. Journal of cellular and molecular medicine. 2017;21:916–928. doi: 10.1111/jcmm.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greenberg RS, Bernstein AM, Benezra M, et al. FAK-dependent regulation of myofibroblast differentiation. FASEB J. 2006;20:1006–1008. doi: 10.1096/fj.05-4838fje. [DOI] [PubMed] [Google Scholar]
- 66.Hu B, Wu YM, Wu Z, et al. Nkx2. 5/Csx represses myofibroblast differentiation. American journal of respiratory cell and molecular biology. 2010;42:218–226. doi: 10.1165/rcmb.2008-0404OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramos C, Becerril C, Montano M, et al. FGF-1 reverts epithelial-mesenchymal transition induced by TGF-{beta}1 through MAPK/ERK kinase pathway. American journal of physiology Lung cellular and molecular physiology. 2010;299:L222–231. doi: 10.1152/ajplung.00070.2010. [DOI] [PubMed] [Google Scholar]
- 68.Koizumi K, Takano K, Kaneyasu A, et al. RhoD activated by fibroblast growth factor induces cytoneme-like cellular protrusions through mDia3C. Mol Biol Cell. 2012;23:4647–4661. doi: 10.1091/mbc.E12-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abe M, Sogabe Y, Syuto T, et al. Evidence that PI3K, Rac, Rho, and Rho kinase are involved in basic fibroblast growth factor-stimulated fibroblast-Collagen matrix contraction. Journal of cellular biochemistry. 2007;102:1290–1299. doi: 10.1002/jcb.21359. [DOI] [PubMed] [Google Scholar]
- 70.Dolivo DM, Larson SA, Dominko T. FGF2-mediated attenuation of myofibroblast activation is modulated by distinct MAPK signaling pathways in human dermal fibroblasts. J Dermatol Sci. 2017;88:339–348. doi: 10.1016/j.jdermsci.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ornitz DM, Xu J, Colvin JS, et al. Receptor specificity of the fibroblast growth factor family. The Journal of biological chemistry. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 72.MacKenzie B, Henneke I, Hezel S, et al. Attenuating endogenous Fgfr2b ligands during bleomycin-induced lung fibrosis does not compromise murine lung repair. American journal of physiology Lung cellular and molecular physiology. 2015;308:L1014–1024. doi: 10.1152/ajplung.00291.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *73.Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. Journal of clinical pathology. 1988;41:467–470. doi: 10.1136/jcp.41.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *74.Katoh K, Kano Y, Fujiwara K. Isolation and in vitro contraction of stress fibers. Methods Enzymol. 2000;325:369–380. doi: 10.1016/s0076-6879(00)25458-x. [DOI] [PubMed] [Google Scholar]
- *75.White SR, Fischer BM, Marroquin BA, et al. Interleukin-1beta mediates human airway epithelial cell migration via NF-kappaB. American journal of physiology Lung cellular and molecular physiology. 2008;295:L1018–1027. doi: 10.1152/ajplung.00065.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
siRNA sequences used for knockdown of FGFR1 and FGFR4
Figure S1. Human FGF2 overexpression in DTG mice in response to doxycycline
Figure S2. Continuous overexpression of FGF2 for 21 days following bleomycin reduces pulmonary fibrosis and collagen accumulation
Figure S3. FGF2 overexpression decreases collagen 3 and Pai-1 expression, but does not alter bleomycin-induced lung injury or epithelial gene expression
Figure S4. Doxycycline-induced overexpression of FGF2 is unaltered by bleomycin treatment
Figure S5. FGF2 overexpression beginning 7 days post-bleomycin decreases Pai-1 expression, but does not alter epithelial gene expression
Figure S6. The FGFR-specific tyrosine kinase inhibitor PD173074 blocks the effect of FGF2, but not TGFβ1 in primary human lung fibroblasts
Figure S7. Recombinant FGF2 stimulates fibroblast migration and proliferation, but does not alter effect of TGFβ1 in primary human lung fibroblasts
Figure S8. Efficient and selective knockdown of FGFR1 and FGFR4 using siRNA