Abstract
Spondylometaphyseal dysplasia (SMD) is characterized by developmental changes in long bones and vertebrae. It has large phenotypic diversity and multiple genetic causes, including a recent link to novel variants in the extracellular matrix (ECM) protein fibronectin (FN), a regulator of ECM assembly and key link between the ECM and proper cell function. We identified a patient with a unique SMD, similar to SMD with corner fractures. The patient has been followed over 19 years and presents with short stature, genu varum, kyphoscoliosis, and pectus carinatum. Radiography shows metaphyseal changes that resolved over time, vertebral changes, and capitular avascular necrosis. Whole exome sequencing identified a novel heterozygous FN1 variant (p.Cys97Trp). Using mass spectroscopy, mutant FN was detected in plasma and in culture medium of primary dermal fibroblasts isolated from the patient, but mutant protein was much less abundant than wild type FN. Immunofluorescence and immunoblotting analyses show that mutant fibroblasts assemble significantly lower amounts of FN matrix than wild type cells, and mutant FN was preferentially retained within the endoplasmic reticulum. This work highlights the importance of FN in skeletal development, and its potential role in the pathogenesis of a subtype of SMD.
Keywords: Extracellular matrix, fibronectins, osteochondrodysplasia, spondylometaphyseal dysplasia, chondrogenesis
Graphical Abstract

INTRODUCTION
The development of endochondral bone to form the vertebrate skeleton involves cell rearrangements and differentiation, remodeling of the extracellular matrix (ECM), localized growth factor signaling, and changes in gene expression.1,2 Tightly controlled changes in expression and assembly of the ECM components fibronectin (FN), collagens, and proteoglycans occur throughout chondrogenesis, in the growth plate, and at ossification sites.3,4 Defects in bone development cause skeletal dysplasias, which have been categorized into 42 groups based on phenotypic presentation and underlying pathophysiology.5 Increasingly, genomic and exomic sequencing data have expanded our understanding of the types of genes and mutations that can cause skeletal dysplasias.5 One type of dysplasia, the spondylometaphyseal dysplasia (SMD) group, is characterized radiographically as affecting the structure of the spine and metaphyses of long bones.6 There are multiple types that differ in prevalence, etiology, and associated defects, such as corner fractures (MIM 184255) or cone-rod dystrophy (MIM 608940).5 Mutations in genes encoding the ECM proteins collagen types II and X cause SMD,7–9 but the genetic defects for many SMDs are unknown.
FN has recently been genetically linked to an SMD subtype, SMD with corner fractures (MIM 184255).10 FN is a major component of most ECMs, forming a critical link between cells and structural components of the ECM.11,12 Fibronectin is secreted as a dimer and is assembled into polymeric fibrils by cell surface integrin receptors. Assembly depends on FN-FN interactions via the N-terminal assembly domain containing the major FN binding site.11 The resulting FN fibrillar network surrounds, connects to, and influences cells. Importantly, this FN network is essential for the incorporation of collagens, growth factors, and other ECM components into the matrix.13–16 The historical lack of FN mutations that lead to disease is thought to be a manifestation of FN’s essential role in early development;17 homozygous null mutations in the FN gene are lethal.17,18 Therefore, the discovery of novel FN mutations linked to disease presents the opportunity to study the mechanisms through which a FN1 point mutation affects protein, cell, and developmental functions. Here, we present the case of a patient with SMD and identify a novel point mutation in the FN1 gene. We show that primary cells from the affected patient secrete both wild type and mutant FNs but assemble a reduced FN matrix.
MATERIALS AND METHODS
The local institutional review board approved this study (Hadassah-Hebrew University Hospital) in accordance with the World Medical Association Declaration of Helsinki. Informed consent was obtained from the parents of the affected patient and later the patient himself.
Whole Exome Sequencing
For the affected patient and his parents, we performed whole exome sequencing using standard procedures. In brief, following DNA extraction from whole blood, sequencing was implemented on exon targets isolated by capture with the Agilent SureSelect Human All Exon V4 (50 Mb) Kit (Agilent Technologies). DNA was sheared, ligated to adaptors, and purified for PCR amplification and products were captured according to the manufacturer’s instructions. Sequences (100-bp paired-end) were generated on a HiSeq2500 (Illumina, San Diego, California, USA). Read alignment and variant calling were performed with DNAnexus (Palo Alto, California, USA) using default parameters with the human genome assembly hg19 (GRCh37) as reference. Probability calculations performed by 10 in silico prediction programs suggest that the identified FN1 mutation could have deleterious effects on protein function (Supplementary Figure S2A).
Mass Spectrometry Analysis of FN
Blood was drawn by standard methods, and anti-coagulated with EDTA. Plasma was separated by centrifugation and used to isolate plasma FN using gelatin-Sepharose affinity chromatography.19 FN was purified by a similar procedure from culture media conditioned by cells for 4 days. FN samples were separated by SDS-PAGE under reducing conditions, bands were visualized with Coomassie Brilliant Blue dye, and excised gel bands were submitted for mass spectrometry (MS). Protein in gel bands was digested using 1.5 μg trypsin (Promega).20 Samples were dried completely in a speedvac and resuspended with 15 μl of 0.1% formic acid pH 3. 5 μl was injected per run using an Easy-nLC 1200 UPLC system. Samples were loaded directly onto a 40 cm long 75 um inner diameter nano capillary column packed with 1.9 μm C18-AQ (Dr. Maisch, Germany) mated to metal emitter (Thermo Scientific, USA) in-line with Thermo Fusion Lumos. The mass spectrometer was operated in data dependent mode with the 120,000 resolution MS1 scan (400–1800 m/z) in the Orbitrap followed by up to 20 MS/MS scans in the ion trap. Dynamic exclusion list was invoked to exclude previously sequenced peptides for 120 s if sequenced within the last 30 s.
Raw files were searched using MSAmanada,21 Sequest HT,22 and Byonic23 algorithms and validated using the Percolator algorithm24 within the Proteome Discoverer 2.1 suite (Thermo Scientific, USA). 10 ppm MS1 and 0.6 Da MS2 mass tolerances were specified. Caramidomethylation and propionamidation of cysteine and oxidation of methionine were specified as dynamic modifications. The resulting msf file was used to construct a spectral library (percolator peptide q-value > 0.95) and extract MS1 ion chromatographs in Skyline for analysis.25,26 Peptide abundance was calculated as the sum of areas-under-the-curve of MS1 chromatographs (intensity of peptide signal versus retention time) for peptides containing either the wild type C amino acid and the modifications described, or variant W amino acid and modifications. The abundance of C97W peptides was adjusted for the relative ionization efficiency of C97W compared to WT peptides, as determined by relative abundance of common peptides from recombinant proteins. The percent of mutant FN was calculated as the adjusted abundance of C97W peptides divided by the sum of adjusted C97W and WT peptide abundances in that sample (Supplementary Figure S2D).
Isolation and Culture of Dermal Fibroblasts
A skin biopsy was obtained from the affected patient and FNC97W/+ dermal fibroblasts were isolated from the biopsy by standard procedures. FNC97W/+ cells were maintained in MEM with 20% fetal bovine serum (FBS, Hyclone). GM03349 dermal fibroblasts were obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research. These cells were isolated from an apparently healthy ten-year-old male and were maintained in MEM with 15% FBS and non-essential amino acids. For experiments, cells were seeded in media prepared with serum depleted of FN by gelatin-Sepharose chromatography.19
Immunofluorescence Staining and Imaging
Cells grown on coverslips were fixed in 3.7% formaldehyde in PBS and, as noted for certain experiments, cells were permeabilized by treatment with 0.5% TritonX-100. FN was visualized using R184 rabbit anti-FN antiserum (raised against III1–6 domains of FN) and Alexa Fluor 488- or Alexa Fluor 568-conjugated goat anti-rabbit IgG in 2% ovalbumin in PBS. Co-staining with BiP was done sequentially, and visualized with an anti-BiP antibody27 and Alexa Fluor 488-goat anti-mouse IgG. Matrix assembly was measured as the total mean fluorescence value for a field of cells. For each condition, 10 images were taken across 2 biological replicates, minimum value was subtracted out as background signal, and Student’s t-test was applied for statistical significance. Representative images were adjusted equivalently using FIJI.
Immunoblotting
Cells were lysed in SDS buffer (4% SDS, 20mM Tris-HCl pH 8.8, 2 mM EDTA, and protease inhibitor cocktail (Roche)). For cell lysates, total protein concentration was measured by BCA assay (Pierce Chemical Co.), and an equal amount of total protein was loaded. Equal protein loading was confirmed by probing blots with an anti-GAPDH antibody (Cell Signaling Technology). For culture media samples, cells were tryspinized and counted, and loading volume was adjusted for cell number. Normalized samples were reduced and separated by SDS-PAGE, transferred to nitrocellulose, then detected by immunoblotting using mouse anti-human FN hybridoma supernatant (HFN7.1, Developmental Studies Hybridoma Bank) and HRP-conjugated goat anti-mouse IgG (Pierce Chemical Co.). To biotinylate ECM proteins, cells were incubated in freshly prepared 1 mg / mL sulfo-NHS-biotin (Pierce Chemical Co.) in PBS for 30 minutes, followed by quenching with five washes in 25 mM Tris-buffered saline, pH 7.45. Cells were lysed in SDS buffer, reduced, separated, and transferred as described, and biotinylated proteins were detected using HRP-conjugated Streptavidin. FN is the most abundant protein in fibroblast ECM in culture,28 such that it is the most prominent band detected by this analysis. Blots were developed using ECL (Thermo Scientific), films were scanned, and bands within the linear range were quantified using FIJI.29 Samples from at least 3 biological replicates were analyzed, and Student’s t-test was applied for statistical significance.
RESULTS
Case Report
The affected male is the 5th of 7 children born to healthy, unrelated parents, with no family history of skeletal dysplasia. Mild intrauterine growth restriction was noted during pregnancy, which was otherwise normal. He was born by spontaneous vaginal delivery after 38 weeks of gestation, birth weight 2300 g, below 3rd percentile. There were no developmental problems in early infancy. Around the age of 1 year, short stature was noted. He started independent walking at the age of 1 year and 3 months. Subsequently, there was progressive asymmetric bowing of the legs and he developed a waddling gait. When he first presented to us at age 4, his height was 87.5 cm, below 3rd percentile.
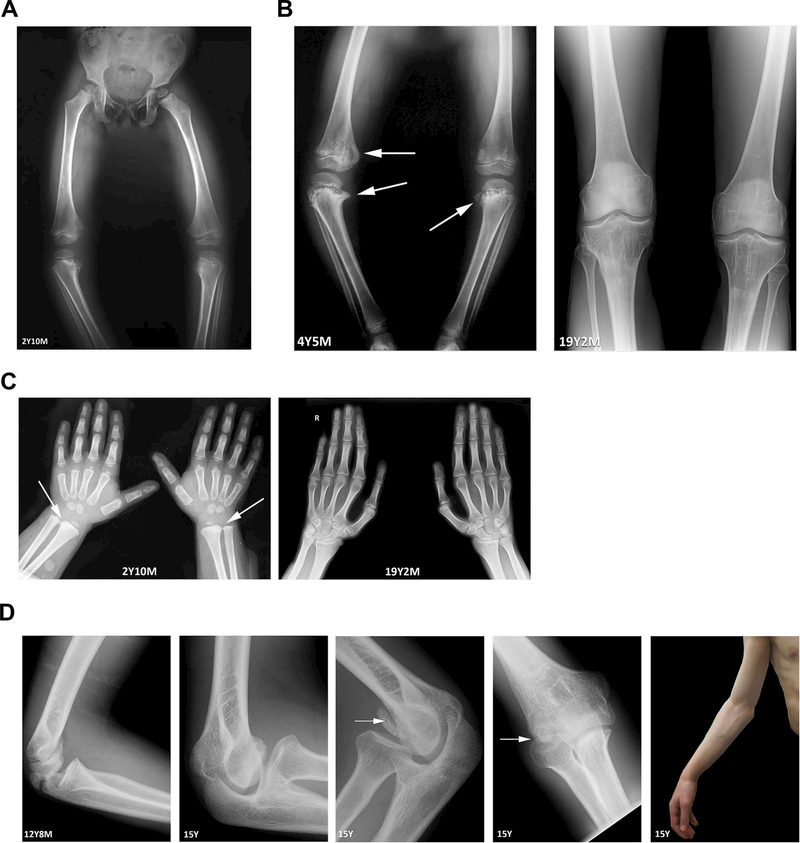
Radiographic studies began at age 2 years 10 months and demonstrated severe progressive kyphoscoliosis (Figure 1A) and bilateral genu varum (Figure 2A). Major vertebral changes were later noted, including multiple ovoid vertebral bodies (Figure 1B), anterior vertebral wedging, and platyspondyly affecting the cervical and thoracolumbar spine. Pectus carinatum was noted, as a part of the thoracic spine deformity (Figure 1C). Peripheral metaphyseal bone fragments, or corner fractures, were noted on the radii at age 2 years 10 months (Figure 2C, left), and in the knee (Figure 2A,B, left). Corner fractures resolved spontaneously by skeletal maturity (Figure 2B,C right). Avascular necrosis of the capitulum was noted incidentally, starting with capitulum surface irregularities and ultimately resulting in total bilateral capitulum necrosis and intra articular loose body formation (Figure 2D). The patient exhibits painless though limited elbow extension (Figure 2D) in the presence of a full range of flexion and prosupination.
Figure 1: Radiographs of spine and chest.
(A) Progressive kyphoscoliosis is shown in images at age 2 years 10 months (left) and at skeletal maturity at age 19 years 2 months (right). (B) Vertebral changes, namely, ovoid bodies and anterior wedging, are visible at 3 years 11 months and 5 years 6 months. (C) Pectus carinatum at skeletal maturity is most likely a result of spinal and vertebral changes.
Figure 2: Radiographs of long bones and joints.
(A) Bilateral genu varum, first noted at approximately 1 year of age, was first imaged at age 2 years 10 months. (B) Bilateral medial corner fractures and beaking in the distal femur and proximal tibiae (left, arrows) were first noted at age 4 years 5 months. These changes are reversed after surgical intervention and upon skeletal maturity (right). (C) Bilateral radial corner fractures in the wrist, first imaged at age 2 years 10 months (left, arrows) are no longer present upon skeletal maturity at age 19 years 2 months (right). (D) Boney changes (left) and limited extension (far right) of the elbow are suggestive of avascular necrosis.
The patient underwent multiple surgeries to address long bone dysplasia, and in all cases the post-operative course, bone formation following osteotomy, and healing processes were uneventful. This observation suggests that subperiosteal and endosteal bone formation remains unaffected. At age 6 years, he underwent bilateral corrective osteotomy and lengthening of both his tibiae, resulting in significant correction to the bow-leggedness on the right and complete correction on the left. At age 11 years 10 months, a tension band plate was inserted in the proximal right tibia and removed 18 months later to resolve residual tibial varus. At age 16 years 5 months, his height was below 1st percentile for age. Both tibiae were lengthened again, and he gained 7 cm in height, though his final height is 143 cm at skeletal maturity, below 1st percentile. No pathologic fractures were reported, suggesting that the bone is healthy once formed, and that remodeling and the endosteum are unaffected.
Joints of the hips, ankles, shoulders, feet, and hands were not affected and continued to develop with a normal radiological appearance, as did the skull, small tubular bones (phalanges) of the fingers and toes, and pelvis (Supplementary Figure S1). Intramembranous bone formation appears to be relatively unaffected, as demonstrated in the relatively normal head circumference (25th percentile in circumference at age 13 years). Shoulders, wrists and hands have a full range of motion.
Identification and characterization of the FN1 mutation
Whole exome sequencing of affected patient RNA yielded 50.5 million mapped reads with 85-fold mean coverage. Following reads alignment, variant calling, and filtration, 29 remaining variants were heterozygous, and all but one were inherited from one of the parents. The remaining variant was Hg19.Chr2:216298171 G>C, NM_212482.2. c.291C>G p.Cys97Trp in FN1 (MIM 135600). We verified the finding by Sanger sequencing in the affected patient, and confirmed its absence from the samples obtained from his parents. The variant was neither present in any of the 138,600 individuals whose exome analyses were deposited at gnomAD nor within an in-house database comprising more than 2000 exomes. This variant resides in exon 3 of 46, and replaces a highly conserved cysteine at position 97 of 2478 with a tryptophan (FNC97W). In wild type FN, C97 forms a disulfide bond with C125.30
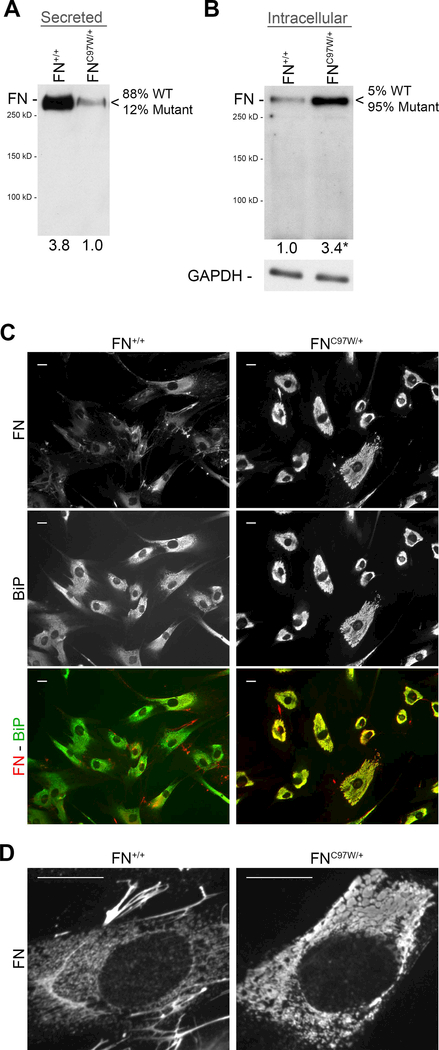
To determine the effects of FNC97W on cell functions, primary dermal fibroblasts were derived from the affected patient (FNC97W/+ cells). Genomic DNA and total RNA were extracted from these cells and used as templates for PCR or RT-PCR amplification, respectively. Sequencing of PCR products confirmed that FNC97W/+ cells are heterozygous for the mutation at the genomic and mRNA levels (Supplementary Figure S2B,C). To determine the proportions of secreted wild type and mutant FN’s, FN isolated from FNC97W/+ cell culture medium was analyzed by mass spectrometry. Peptides containing the single mutant amino acid change were detectable, but were significantly less abundant than wild type peptides (~88% WT, 12% mutant; Supplementary Figure S2D).
Mass spectrometry was also used to determine the abundance of mutant FN relative to wild type in vivo. Plasma FN was isolated from the blood of the patient. Plasma FN is produced and secreted by hepatocytes and thus is a source that is distinct from dermal cells. Peptides containing the single mutant amino acid change were detectable, but were significantly less abundant than wild type peptides (~77% WT, 23% mutant; Supplementary Figure S2D).
Effects of the FNC97W mutation on FN matrix assembly
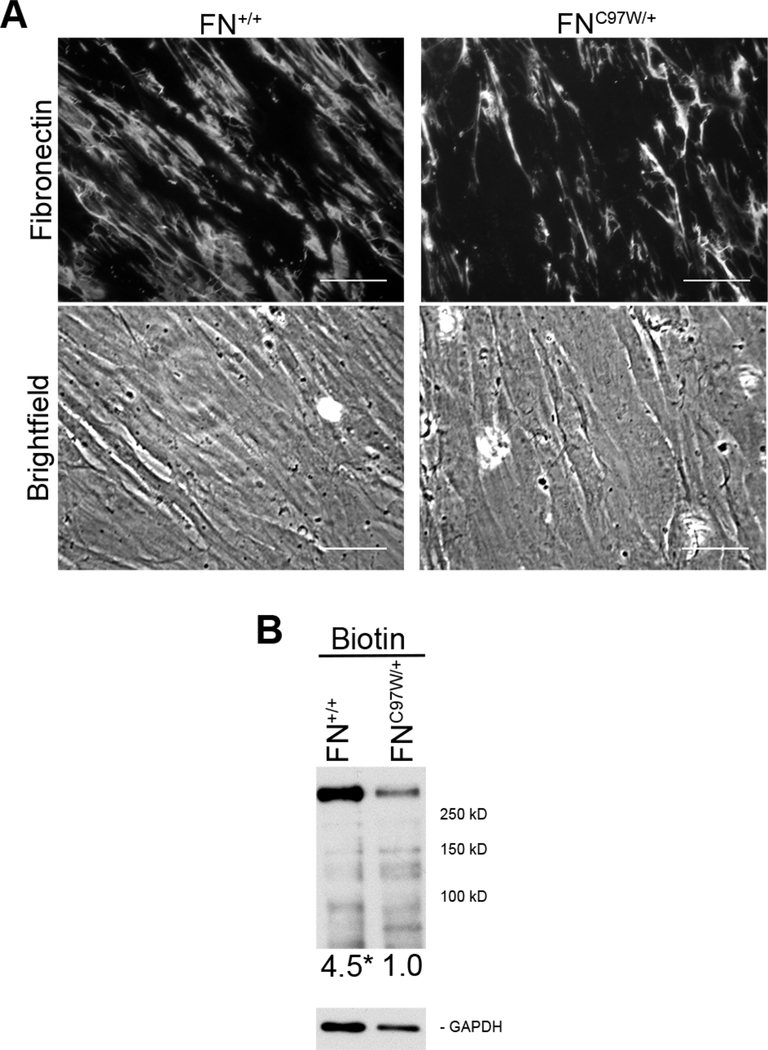
The FNC97W mutation resides in the N-terminal assembly domain that mediates FN-FN interactions essential for FN fibril formation.11 To compare the impact of this mutation on the phenotype of FNC97W/+ cells, we used age- and gender-matched FN+/+ dermal fibroblasts (GM03349, Coriell Repository). FN+/+ and FNC97W/+ cells demonstrate similar morphologies (Supplementary Figure S3A) and grow at similar rates (Supplementary Figure S3B). We compared these cells for the ability to assemble a FN matrix using indirect immunofluorescence staining. FNC97W/+ cell matrix had visibly fewer fibrils than the matrix assembled by wild type cells (Figure 3A). Quantitative analysis of the mean fluorescence intensity indicates that the FNC97W/+ cell matrix is only 70% of the FN matrix of the wild type control. To measure total FN protein in the matrix, ECM proteins were labeled in situ with a membrane-impermeable biotin cross-linking agent and FN levels were then quantified in total cell lysates (Figure 3B). FN at ~250 kDa is the predominant ECM protein produced by cultured fibroblasts28, and this band exactly co-migrates with FN by immunoblot of identical samples run side-by-side on the same blot (data not shown). Quantification of FN levels showed that FN in FN+/+ cell ECM is increased at least 4.5-fold compared to FN in the FNC97W/+ ECM. These ECM analyses show that mutant cells have a defect in FN matrix assembly. This assembly defect is not due to deficient cell assembly mechanisms since FNC97W/+ cells assemble normal levels of FN matrix when provided with an exogenous supply of wild type FN (data not shown).
Figure 3: Primary FNC97W/+ cells assemble reduced FN matrix compared to primary FN+/+ cells.
(A) The FN matrix of wild type (FN+/+) and mutant (FNC97W/+) cells was visualized by immunofluorescence microscopy after staining with R184 anti-FN polyclonal antiserum (top). Equivalent cell densities are visible by phase microscopy (bottom). Total fluorescence averaged over 10 images was 191+34 for FNC97W/+ cells and 267+54 for FN+/+ cells (SD, p < 0.01). Scale bar = 40 μm. (B) Extracellular proteins of confluent cells were biotinylated using a cell-impermeable crosslinker, 125 ng of total protein was separated by SDS-PAGE, and blots were probed with HRP-coupled streptavidin (top). Relative levels of biotinylated FN are, on average, at least 4.5 for FNC97W/+ versus 1.0 for FN+/+ (p<0.01), after normalization to GAPDH as a loading control (bottom).
To investigate the mechanism that limits matrix assembly, we analyzed the distribution of mutant versus wild type FN subunits by FNC97W/+ cells. FN+/+ cells secreted at least 3.8-fold higher levels of FN into the culture medium (Figure 4A). Combined with the mass spectrometry analyses of secreted FN, these results demonstrate that the mutant protein is secreted but at significantly lower levels than the wild type FN, leading to an overall lower level of total FN in the cell culture medium.
Figure 4: Altered subcellular distribution of FN in primary FNC97W/+ cells.
(A) Wild type (FN+/+) and mutant (FNC97W/+) cells were plated for 24 hours, then media were immunoblotted for human FN with HFN7.1 monoclonal antibody. Band quantification demonstrated at least 3.8-fold more FN secreted by FN+/+ cells compared to FNC97W/+ cells. The relative percentage of wild type (WT) and mutant peptides determined by mass spectrometry is shown (see Supplementary Figure S2D). (B) Subconfluent cells were lysed after 24 hours in culture to enrich for cellular proteins over ECM proteins. 1 μg of lysate was probed for human FN (top) or GAPDH as a loading control (bottom). Intracellular FN level was, on average, at least 3.4-fold higher in FNC97W/+ cells compared to FN+/+ cells (p<0.01). (C) Wild type FN+/+ (left) and mutant FNC97W/+ (right) cells were seeded at a low density to minimize matrix formation. Cells were fixed, permeabilized, and then stained with anti-FN antiserum (red) and anti-BiP monoclonal antibody to visualize the endoplasmic reticulum (green). Individual channels and merged signals are as indicated on the side. (D) High magnification imaging of cells from (C) stained for FN shows vesicles in FN+/+ cells (left) compared to large FN-positive inclusions in FNC97W/+ cells (right). Scale bar = 20 μm.
Mutations in ECM proteins can affect their ability to transit through the secretory pathway.9 Therefore, we compared the amounts of intracellular FN in wild type and mutant cells. Cell lysates were prepared with subconfluent cells that had not assembled a FN matrix, such that the majority of FN was intracellular, not assembled into an ECM. Immunoblots of these lysates with anti-FN antibodies show dramatically higher levels of FN in the mutant lysates than in wild type cells (Figure 4B). Mass spectrometric analysis of the intracellular FN fraction of FNC97W/+ cells showed that, in contrast to FN in cell culture medium and plasma, the vast majority of the intracellular FN contained the mutant peptide (~95% mutant, 5% wild type; Supplementary Figure S2D). To visualize differences in intracellular FN, subconfluent cells were permeabilized and immunostained. FNC97W/+ cells show a different pattern of intracellular FN from FN+/+ cells (Figure 4C). The intensity of intracellular FN staining is dramatically higher in mutant cells than in wild type, which show weaker, more diffuse intracellular staining. In addition, wild type cells have extracellular FN fibrils that are visible between adjacent cells, but fibrils were not detected in subconfluent FNC97W/+ cell cultures (Figure 4C, compare top left with top right). Co-staining with antibodies against BiP, an endoplasmic reticulum (ER) resident protein,27 shows colocalization of FN with BiP throughout FNC97W/+ cells (Figure 4C) indicating that all portions of the ER contain FNC97W/+ and suggesting a deficiency in FN secretion by mutant cells. In contrast, FN+/+ cells had less FN within the ER and regions that stained heavily for BiP but not FN (Figure 4C). When viewed at higher magnification, large intracellular inclusions containing FN are clearly visible in FNC97W/+ cells whereas FN-positive structures within FN+/+ cells were small and vesicular (Figure 4D). This phenotype is similar to the FN staining pattern observed when cells were treated with inhibitors of secretion.31 Together, these results show that mutant FN is not efficiently transported through the secretory pathway and that this preferential retention leads to an accumulation of mutant FN subunits in vesicular structures. Reduced FN secretion is likely to contribute to the observed reduction in FN matrix assembly by FNC97W/+ cells.
DISCUSSION
Proper assembly of the ECM is essential for normal tissue development. The FN matrix plays an essential role in development by serving as a foundational matrix to support the deposition of collagens and many other ECM proteins.13–16 Here, we characterize a novel FN1 mutation identified in an patient with a skeletal dysplasia that has features in common with several different types of SMD. Relative to age- and gender-matched controls, heterozygous primary dermal fibroblasts from this patient secrete and assemble reduced levels of FN, and preferentially retain mutant FN in the ER. FN levels in the blood are also reduced indicating a secretion defect in liver, the source of plasma FN. These results with patient-derived samples show alterations in FN production that may have systemic implications (from changes in plasma FN) as well as effects on the local environment of cells within tissues.
Mutations in type II collagen7 or FN10 have been identified in patients with SMD with corner fractures. Features in common with the patient described here include multiple vertebral changes, which contribute to abnormalities of the spine and chest, and metaphyseal changes that radiographically resemble corner fractures. As in other cases of SMD with corner fractures10,32, metaphyseal changes disappeared upon reaching skeletal maturity, reinforcing that these changes are most likely secondary ossification centers that are developmental in nature and not true fractures. Metaphyseal changes in the proximal humerus and developmental coxa vera are common in other cases of SMD linked to FN1 mutations,10 but were not observed in this patient. This patient also presents unique changes in the elbow, manifest as bilateral avascular necrosis, intra-articular loose body formation, and restricted range of motion. This is not a reported feature of other cases of SMD with corner fractures. It is unclear whether this joint defect results from pathologic changes in long bone metaphyses or from a different pathogenesis unique to this joint.
Our results suggest two potential mechanisms, not mutually exclusive, to explain the FN matrix deficiencies. The mutation identified here as well as other FN mutations related to SMD10 reside in the N-terminal assembly domain. FN interactions involving this domain are essential for fibril formation33 and may be altered in mutant FN. Deletion of any one of the five type I modules that make up this domain is sufficient to dramatically reduced FN binding and matrix incorporation.34 Such a deficiency in FN-FN binding would reduce the initiation of fibril formation as observed with the FNC97W/+ cells. The second mechanism derives from our evidence showing intracellular retention of mutant FN and a decrease in secreted FN by primary dermal fibroblasts. Secretion of mutant recombinant FN fragments by cells transduced with other FN1 point mutations was also decreased.10 Retention and secretion defects suggest that reduced matrix levels may be due to a reduced abundance of extracellular FN.
Although the decrease in secreted FN is significant, a reduction in FN alone or a haploinsufficiency is not sufficient to produce developmental defects. Heterozygous FN null mice have approximately half the level of plasma FN of wild type littermates, but the mice show no overt defects,17 indicating that half of normal FN production is sufficient to perform the essential FN matrix functions. However, the mutation may reduce FN levels by more than 50%. Indeed, FNC97W/+ in culture medium was much less than half that of the wild type control, suggesting the reduction in FN may be more significant than that observed in heterozygous knockout mice.
Mutations in FN have only recently been identified in patients with SMD, but previous studies suggest an important role for FN in skeletal development. FN expression increases significantly as mesenchymal cells aggregate in both limb and wing buds3. Cell aggregation is required to induce differentiation through up-regulation of Sox 9, the transcription factor that mediates chondrogenesis.35 Aggregation is dependent on FN matrix assembly and FN expression is sustained throughout chondrogenesis and in cartilaginous tissue.4 In human disease, chondrodysplasia and achondrogenesis can be caused by mutations in the diastrophic dysplasia sulfate transporter (DTDST), and our previous work has shown that DTDST-dependent sulfation of proteoglycans is required for FN matrix assembly36 and mesenchymal cell aggregation.37 The FN matrix is required for initiation of collagen fibrillogenesis16 and, since collagen matrices have key roles in skeletal development, insufficient FN matrix could perturb collagen assembly leading to a skeletal dysplasia.
The complex and incompletely understood relationship between FN, skeletal development, and its many key players suggests multiple models through which a reduced FN matrix might lead to skeletal dysplasia. Reduced FN matrix could lead to reduced collagen matrix, a known cause of skeletal dysplasia. Increased retention of mutant ECM protein has been suggested as a pathogenic mechanism in other skeletal dysplasias.9 The reduced production of FN could result in insufficient FN matrix assembly needed at the start of chondrogenesis for stem cell aggregation and differentiation. The fact that FN mutations linked to SMD are not embryonic lethal suggests that any affected processes are only delayed or otherwise altered, not entirely halted. Indeed, variations in presentation and specific defects may be due to differences in the amount of mutant FN secreted, whether plasma FN can compensate, or whether cell viability is compromised by mutations. The localization of long bone changes to the metaphyses, the location of proliferative chondrocytes, suggests this cell type as a particular target in the phenotype described here. However, exactly how a FN matrix affects these chondrocytes remains a question worthy of further study.
Supplementary Material
Acknowledgments:
The authors thank Dr. Tharan Srikumar, Manager of the Mass Spectrometry and Proteomics Core facility at Princeton University, for technical and scientific assistance. We also thank Mark Pinkerton, Michael Weaver, and Christina O’Halloran for technical assistance. This work was supported by NIH R01 CA160611 (to JES).
Footnotes
Conflict of Interest: Nothing to declare
REFERENCES
- 1.Tsang KY, Chan D, Cheah KSE. Fate of growth plate hypertrophic chondrocytes: death or lineage extension? Dev Growth Differ. 2015;57(2):179–192. doi: 10.1111/dgd.12203. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 3.Bobick BE, Chen FH, Le AM, Tuan RS. Regulation of the chondrogenic phenotype in culture. Birth Defects Res C Embryo Today. 2009;87(4):351–371. doi: 10.1002/bdrc.20167. [DOI] [PubMed] [Google Scholar]
- 4.Singh P, Schwarzbauer JE. Fibronectin and stem cell differentiation - lessons from chondrogenesis. J Cell Sci. 2012;125(16):3703–3712. doi: 10.1242/jcs.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonafe L, Cormier-Daire V, Hall C, et al. Nosology and classification of genetic skeletal disorders: 2015 revision. Am J Med Genet A. 2015;167A(12):2869–2892. doi: 10.1002/ajmg.a.37365. [DOI] [PubMed] [Google Scholar]
- 6.Krakow D, Rimoin DL. The skeletal dysplasias. Genet Med. 2010;12(6):327–341. doi: 10.1097/GIM.0b013e3181daae9b. [DOI] [PubMed] [Google Scholar]
- 7.Machol K, Jain M, Almannai M, et al. Corner fracture type spondylometaphyseal dysplasia: Overlap with type II collagenopathies. Am J Med Genet A. 2016;173(3):733–739. doi: 10.1002/ajmg.a.38059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikegawa S, Nishimura G, Nagai T, Hasegawa T, Ohashi H, Nakamura Y. Mutation of the type X collagen gene (COL10A1) causes spondylometaphyseal dysplasia. Am J Hum Genet. 1998;63(6):1659–1662. doi: 10.1086/302158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold WV, Fertala A. Skeletal diseases caused by mutations that affect collagen structure and function. Int J Biochem Cell Biol. 2013;45(8):1556–1567. doi: 10.1016/j.biocel.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 10.Lee CS, Fu H, Baratang N, et al. Mutations in Fibronectin Cause a Subtype of Spondylometaphyseal Dysplasia with “Corner Fractures”. Am J Hum Genet. 2017;101(5):815–823. doi: 10.1016/j.ajhg.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh P, Carraher C, Schwarzbauer JE. Assembly of Fibronectin Extracellular Matrix. Annu Rev Cell Dev Biol. 2010;26(1):397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzbauer JE, DeSimone DW. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb Perspect Biol. 2011;3(7):a005041–a005041. doi: 10.1101/cshperspect.a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velling T, Risteli J, Wennerberg K, Mosher DF, Johansson S. Polymerization of type I and III collagens is dependent on fibronectin and enhanced by integrins alpha 11beta 1 and alpha 2beta 1. J Biol Chem. 2002;277(40):37377–37381. doi: 10.1074/jbc.M206286200. [DOI] [PubMed] [Google Scholar]
- 14.Miller CG, Pozzi A, Zent R, Schwarzbauer JE. Effects of high glucose on integrin activity and fibronectin matrix assembly by mesangial cells. Mol Biol Cell. 2014;25(16):2342–2350. doi: 10.1091/mbc.E14-03-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dallas SL, Sivakumar P, Jones CJP, et al. Fibronectin regulates latent transforming growth factor-beta (TGF beta) by controlling matrix assembly of latent TGF beta-binding protein-1. J Biol Chem. 2005;280(19):18871–18880. doi: 10.1074/jbc.M410762200. [DOI] [PubMed] [Google Scholar]
- 16.Sottile J, Hocking DC. Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Mol Biol Cell. 2002;13(10):3546–3559. doi: 10.1091/mbc.E02-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119(4):1079–1091. [DOI] [PubMed] [Google Scholar]
- 18.Georges-Labouesse EN, George EL, Rayburn H, Hynes RO. Mesodermal development in mouse embryos mutant for fibronectin. Dev Dyn. 1996;207(2):145–156. doi:. [DOI] [PubMed] [Google Scholar]
- 19.Engvall E, Ruoslahti E. Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int J Cancer. 1977;20(1):1–5. [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 21.Dorfer V, Pichler P, Stranzl T, et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res. 2014;13(8):3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 23.Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics. 2012;Chapter 13:Unit13.20. doi: 10.1002/0471250953.bi1320s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spivak M, Weston J, Bottou L, Käll L, Noble WS. Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. J Proteome Res. 2009;8(7):3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schilling B, Rardin MJ, MacLean BX, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics. 2012;11(5):202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bole DG, Hendershot LM, Kearney JF. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol. 1986;102(5):1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao Y, Schwarzbauer JE. Stimulatory effects of a three-dimensional microenvironment on cell-mediated fibronectin fibrillogenesis. Journal of Cell Science. 2005;118(Pt 19):4427–4436. doi: 10.1242/jcs.02566. [DOI] [PubMed] [Google Scholar]
- 29.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potts JR, Bright JR, Bolton D, Pickford AR, Campbell ID. Solution structure of the N-terminal F1 module pair from human fibronectin. Biochemistry. 1999;38(26):8304–8312. doi: 10.1021/bi990202b. [DOI] [PubMed] [Google Scholar]
- 31.Ledger PW, Uchida N, Tanzer ML. Immunocytochemical localization of procollagen and fibronectin in human fibroblasts: effects of the monovalent ionophore, monensin. J Cell Biol. 1980;87(3 Pt 1):663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Currarino G, Birch JG, Herring JA. Developmental coxa vara associated with spondylometaphyseal dysplasia (DCV/SMD): “SMD-corner fracture type” (DCV/SMD-CF) demonstrated in most reported cases. Pediatr Radiol. 2000;30(1):14–24. doi: 10.1007/s002470050005. [DOI] [PubMed] [Google Scholar]
- 33.Schwarzbauer JE. Identification of the fibronectin sequences required for assembly of a fibrillar matrix. J Cell Biol. 1991;113(6):1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sottile J, Schwarzbauer J, Selegue J, Mosher DF. Five type I modules of fibronectin form a functional unit that binds to fibroblasts and Staphylococcus aureus. J Biol Chem. 1991;266(20):12840–12843. [PubMed] [Google Scholar]
- 35.Barna M, Niswander L. Visualization of cartilage formation: insight into cellular properties of skeletal progenitors and chondrodysplasia syndromes. Dev Cell. 2007;12(6):931–941. doi: 10.1016/j.devcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Galante LL, Schwarzbauer JE. Requirements for sulfate transport and the diastrophic dysplasia sulfate transporter in fibronectin matrix assembly. J Cell Biol. 2007;179(5):999–1009. doi: 10.1083/jcb.200707150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh P, Schwarzbauer JE. Fibronectin matrix assembly is essential for cell condensation during chondrogenesis. J Cell Sci. 2014;127(20):4420–4428. doi: 10.1242/jcs.150276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.