Abstract
The leguminous inoculation with nodule-inducing bacteria that perform biological nitrogen fixation is a good example of an “eco-friendly agricultural practice”. Bradyrhizobium strains BR 3267 and BR 3262 are recommended for cowpea (Vigna unguiculata) inoculation in Brazil and showed remarkable responses; nevertheless neither strain was characterized at species level, which is our goal in the present work using a polyphasic approach. The strains presented the typical phenotype of Bradyrhizobium with a slow growth and a white colony on yeast extract-mannitol medium. Strain BR 3267 was more versatile in its use of carbon sources compared to BR 3262. The fatty acid composition of BR 3267 was similar to the type strain of Bradyrhizobium yuanmingense; while BR 3262 was similar to Bradyrhizobium elkanii and Bradyrhizobium pachyrhizi. Phylogenetic analyses based on 16S rRNA and three housekeeping genes placed both strains within the genus Bradyrhizobium: strain BR 3267 was closest to B. yuanmingense and BR 3262 to B. pachyrhizi. Genome average nucleotide identity and DNA–DNA reassociation confirmed the genomic identification of B. yuanmingense BR 3267 and B. pachyrhizi BR 3262. The nodC and nifH gene analyses showed that strains BR 3267 and BR 3262 hold divergent symbiotic genes. In summary, the results indicate that cowpea can establish effective symbiosis with divergent bradyrhizobia isolated from Brazilian soils.
Keywords: Vigna unguiculata, Rhizobia, MLSA, DDH, ANI
Introduction
Bacteria collectively known as rhizobia form an important part of the soil microbiota and perform biological nitrogen fixation (BNF) through nitrogenase activity when in symbiosis with leguminous plants. This ecological phenomenon has great biotechnological impact on biomass and grain production. For example, in the soybean crop production in Brazil it is estimated to save over US$ 10 billion annually by the use of Bradyrhizobium inoculation instead of chemical fertilization.1
The Brazilian Ministry of Agriculture has a list of rhizobial strains recommended for more than 50 leguminous grain-producing crops, forage and green manure.2 In recent years efforts have been made to better characterize these strains recommended for inoculation in Brazil and at least five new species have been described.3, 4, 5, 6
Brazil is the world's third leading cowpea producer, with an estimated production of 500,000 tons per year.7 The crop yield varies from 400 to 2000 kg ha−1, depending on the system and the region of cultivation.7 This yield has been rising in recent years with improvements in cultivation management, such as the inoculation of seeds with nitrogen-fixing bacteria, a practice applied to approximately 100,000 hectares.
The strains BR 3267 and BR 3262 are considered as “elite” for inoculation of cowpea plants in Brazil2 and several studies under controlled and field conditions have shown that both strains make significant contributions to crop yields, including more than 50% N accumulation via BNF.8 BR 3267 strain was isolated from the semiarid northeastern region of the country, using cowpea as trap plants, while BR 3262 strain was isolated from an Atlantic Forest area in southeastern Brazil using the same strategy.1, 9 Although these strains were isolated more than a decade ago, they were only partially characterized through assessment of the growth rate, colony morphology and 16S rRNA phylogeny,11 but not classified at the species level.
Zilli and colleagues11 have characterized the 16S rRNA genes of the BR 3267 and BR 3262 strains and concluded that both are members of the genus Bradyrhizobium. This genus was created in the early 1980s to accommodate root nodule-inducing bacteria with slow growth on media containing mannitol and yeast extract.12 Since the turn of the century, two major subgroup divisions (I and II) have been recognized within this genus based on DNA–DNA hybridization.12, 13, 14 The Bradyrhizobium japonicum and Bradyrhizobium elkanii were the first species assigned to subgroup I and II, respectively.15, 16 According to Zilli and colleagues,10 BR 3267 clustered within the subgroup B. japonicum and BR 3262 within the subgroup B. elkanii. However, in the light of current knowledge, those results can be considered inconclusive because new Bradyrhizobium species was recently described. Thus, further investigation employing the latest molecular techniques is needed for the correct positioning of these strains.
In the past five years, the use of housekeeping genes as powerful phylogenetic markers for bacteria has led to the description of over 15 new species within the genus Bradyrhizobium17 and enabled the separation of genetically close strains into different species. Examples are the definition of the species Bradyrhizobium diazoefficiens based on B. japonicum and Bradyrhizobium pachyrhizi from B. elkanii.3, 18 Furthermore, new methods for genome-to genome comparison, like Average Nucleotide Identity (ANI) and Genome Blast Distance Phylogeny (GBDP) have been introduced and are contributing to improve bacterial taxonomy.19, 20
Therefore, the goal of our study was to characterize at a finer the taxonomic level both BR 3267 and BR 3262 strains using a polyphasic approach.
Materials and methods
Strains used
The cowpea strains BR 3267 and BR 3262, and the type strain B. elkanii USDA 76T were obtained from the Johanna Döbereiner Biological Resource Center (CRB-JD, Embrapa Agrobiologia, Seropédica-Rio de Janeiro, Brazil). The type strains B. pachyrhizi PAC 48T (=LMG 24246T) and Bradyrhizobium yuanmingense CCBAU 10071T (=LMG 21827) were obtained from the LMG Culture Collection (Belgium). The strains were grown on yeast extract-mannitol agar medium (YMA) and were incubated at 28 °C21 for seven days until they reached sufficient colony growth levels to observe morphological features and purity.
Phenotypic and physiologic characterizations
Inoculum preparation for both BR 3267 and BR 3262 strains was carried out using YMA medium at 28 °C. Carbon source utilization was assessed with Biolog GN2 microplates (Biolog Inc., Hayward, CA) following the manufacturer's instructions, except that cell concentration was adjusted to 5 on the McFarland scale. Plates were incubated in the dark at 28 °C for ten days. The biochemical features were assessed using API 20 NE strips (bioMérieux, Marcy-L’Etoile, France) following a standard protocol that uses a saline solution (0.85% NaCl) for bacterial suspension. Additionally, the tolerance to abiotic stress, such as temperature, pH and salinity (NaCl), was determined by examining the growth in YMA medium. The temperature tolerance was evaluated at 15, 20, 25, 28, 30, 32 and 37 °C, and the pH tolerance was tested in a range from 4 to 10. The salinity tolerance was examined at 28 °C in YMA medium supplemented with 0.1, 0.3, 0.5, 1.0, 1.5, 2.0 or 2.5% (w/v) of NaCl. The resistance to antibiotics was determined using YMA medium and the disk diffusion method for ampicillin (25 μg), chloramphenicol (50 μg), erythromycin (30 μg), gentamicin (10 μg), kanamycin (30 μg), neomycin (10 μg), penicillin (10 μg), streptomycin (10 μg) and tetracycline (30 μg). All tests were run in triplicate.
Fatty acid composition
The BR 3267 and BR 3262 and type strains B. elkanii USDA 76T, B. pachyrhizi PAC 48T and B. yuanmingense CCBAU 10071T type strains were characterized based on whole-cell fatty acids, derived using the methyl esters’ method (FAME) and analyzed with a Hewlett Packard gas chromatograph fitted with a fused silica capillary column (25 m × 0.2 mm internal diameter). The type strains B. elkanii USDA 76T, B. pachyrhizi PAC 48T and B. yuanmingense CCBAU 10071T were chosen because they were the closest related strains to BR 3262 and BR 3267 respectively, based on EzTaxon identification search (http://www.ezbiocloud.net/eztaxon/identify). ChemStation A.09.01 software [1206] and a MIDI Microbial Identification System 4.0 (Sherlock TSBA Library, MIDI ID, Inc., Newark, DE, USA) were used for the fatty acid identification.
DNA extraction, PCR amplification and sequencing of PCR fragments
Pure cultures of Bradyrhizobium strains were grown on YM medium for 4 days at 28 °C under stirring of 120 rpm. Then, 2 mL of cell suspension were centrifuged (12,000 × g), and the total genomic DNA was extracted using a Wizard® Genomic DNA Purification Kit (Promega, USA) following the manufacturer's instructions. The recA gene was amplified by PCR using the primers TSrecAf and TSrecAr, and the glnII gene was amplified with the primers TSglnIIf and TSglnIIr.22 The primers NodCfor540 and NodCrev1160 were used to amplify the symbiotic nodC gene, as described by Sarita et al.23 For the nifH, the primers NifHF/NifHI were used following the methods described by Laguerre et al.24 DNA sequences of the PCR products were obtained for both strands with the same primers used for gene amplifications. The 16S rRNA sequences of the BR 3267 (Gene Bank A.no. AY649439) and BR 3262 (A.no. AY649430) strains obtained in a previous study10 were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). For the gyrB gene, the sequences were retrieved from draft genome sequences of the BR 3267 and BR 3262 strains (Ac. no. KT005410 and KT005409, respectively).
Phylogenetic analysis
Forward and reverse readings of the 16S rRNA, recA, glnII, nodC and nifH gene fragments were edited and assembled using DNABASER (http://www.dnabaser.com). Multiple sequence alignments were performed using ClustalW through MEGA 6.0.25 Maximum likelihood (ML) phylogenetic reconstructions were completed using MEGA 6 for single gene sequences (16S rRNA, recA, glnII, gyrB, nodC and nifH), and a multilocus sequence analysis (MLSA) was performed using the concatenated sequences of the housekeeping genes recA, glnII and gyrB. The distance matrices were calculated using the Kimura two-parameter substitution model,26 and the robustness of the tree nodes was evaluated with a bootstrap analysis27 using 500 pseudoreplicates. The software default parameters were considered in all of the analyses. Sequences of rhizobial type strains used for alignment and phylogenetic analysis were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/), and a Microvirga vignae BR 3299 strain28 was used as the outgroup in the ML analysis of the 16S rRNA gene.
DNA–DNA relatedness and average nucleotide identity (ANI)
The DNA–DNA hybridization (DDH) among bacterial strains was determined based on the thermal denaturation temperatures of hybrid and homologous genomic DNA, as described by Gonzalez and Saiz-Jimenez,29, 30 except that the DNA concentration was adjusted to 2 μg per reaction. The experiments were performed in 96-well optical plates with appropriate optical adhesives in three replicates and including wells without DNA as negative control. For DNA hybridization, a Bio-Rad MyCicler thermocycler was used, and for fluorimetric measurements during the denaturation stage an Applied Biosystems 7500 Real-Time system was used. Thermal conditions for hybridization consisted on a denaturation step of 99 °C for 10 min, followed by an annealing period of 8 h at 79° C (optimum temperature for renaturation – TOR).29, 31 It was followed by progressive 10-min steps, each at 1.8 °C below the previous one, until 25 °C when it was hold for 30 min before refrigeration to 4 °C. The fluorescence was measured using a denaturation ramp settled at the step and hold mode. Heating rate was 0.2 °C s−1 with fluorescence decreasing measurement at each 0.2 °C step, during a 12 s hold, and between 25 and 99.9 °C.29 The DDH experiments were performed between strains BR 3267 and BR 3262 and their phylogenetically related type strains based on the 16S rRNA gene similarities. BR3267 was compared against B. yuanmingense CCBAU 10071T, and BR 3262 was compared against B. pachyrhizi PAC 48T and B. elkanii USDA 76T.
The ANI estimation was performed with a JSpecies platform version 1.2.1,32 and MUMmer was used for genome alignment.33 All system requirements (BLAST and MUMmer) were downloaded and installed locally in a system Linux. Bacterial genome were retrieved from NCBI database as fasta file and used to ANI calculation, considering JSpecies default parameters. The sequenced genome of strain BR 3262 (Ac.no. LJYE00000000.1)34 was compared to genomes of the strains USDA 76T (B. elkanii – GenBank A.no. ARAG00000000.1) and PAC48T (B. pachyrhizi – GenBank A.no. LFIQ00000000.1). The genome of the strain BR 3267 (A.no. LJYF00000000.1)35 was compared to that of the strain CCBAU 35157 (B. yuanmingense – A.no. AJQL00000000.1).
Results
Phenotypic and physiologic features
Both BR 3267 and BR 3262 strains can grow in a pH range of 4–10, tolerate up to 0.5% NaCl in the medium and present normal growth in a temperature range from 15 to 32 °C, and BR 3267 could also grow up to 37 °C (Table 1). Likewise, both strains showed positive reactions to enzyme urease and nitrate reductase. Their susceptibilities to different antibiotics were variable (Table 1). BR 3267 was sensitive to streptomycin and tetracycline, while BR 3262 was tolerant. With respect to the carbon sources evaluated with the Biolog kit, BR 3262 strain was able to use 33 of the 95 sources tested, while BR 3267 strain was able to use more than 50.
Table 1.
Phenotypic features of the cowpea Bradyrhizobium strains BR 3267 and BR 3262 as characterized by API 20NE and Biolog GN2 microplates. Data obtained from 5 days duplicate read mean values (+ = positive; − = negative, w = weak).
| Phenotypic feature | BR 3267 | BR 3262 | Phenotypic feature | BR 3267 | BR 3262 | Phenotypic feature | BR 3267 | BR 3262 |
|---|---|---|---|---|---|---|---|---|
| Enzymatic reaction | pH 4 | + | + | Ester | ||||
| Catalase | + | − | pH 10 | + | + | Methyl pyruvate | + | − |
| Hydrolysis of gelatine | + | − | 1% NaCl | − | − | Mono-methyl-succinnate | + | − |
| Hydrolysis of esculin | + | + | Carbohydrates | Amino acids | ||||
| β-galactosidase | w | − | d-Arabitol | + | − | d-Alanine | − | + |
| Resistance to (μg) | α-d-Glucose | + | − | l-Alanine | − | + | ||
| Ampicillin (25) | − | + | d-Mannose | + | − | Glycyl-l-aspartic acid | + | − |
| Chloramphenicol (50) | − | + | d-Psicose | + | − | l-Leucine | + | − |
| Kanamycin (30) | + | − | d-Sorbitol | + | − | l-Phenylalanine | + | − |
| Neomycin (10) | + | − | Carboxylic acids | l-Proline | + | − | ||
| Penicillin (10) | − | + | d-Galacturonic acid | + | − | d-Serine | + | − |
| Streptomycin (10) | − | + | d-Glucosaminic acid | + | − | l-Threonine | + | − |
| Tetracycline (30) | − | + | α-Ketovaleric acid | + | − | γ-Aminobutyric acid | + | − |
| Erythromycin (30) | + | + | d,l-Lactic acid | + | − | Polymer | ||
| Gentamicin (10) | + | + | Malonic acid | + | − | Dextrin | − | + |
| Growth at | Quinic acid | + | − | Amide | ||||
| 15 °C | + | + | d-Saccharic acid | + | − | Glucuronamide | + | − |
| 32 °C | + | + | Sebacic acid | + | − | Amine | ||
| 37 °C | + | − | Succinic acid | + | − | Phenylethylamine | + | − |
Both strains BR 3267 and BR 3262 were positive to: Oxidase, Urease, Nitrate reduction, Hydrolysis of esculin, l-Arabinose, l-Fucose, d-Galactose, d-Mannitol, l-Rhamnose, Tween 40, Tween 80, Acetic acid, Citric acid, Formic acid, d-Galactonic acid lactone, d-Gluconic acid, α-Hydroxybutyric acid, β-Hydroxybutyric acid, γ-Hydroxybutyric acid, p-Hydroxyphenylacetic acid, α-Ketobutyric acid, α-Ketoglutaric acid, Propionic acid, Glycerol, l-Alanyl-glycine, l-Asparagine, l-Aspartic acid, l-Glutamic acid, Glycyl-l-glutamic acid, l-Pyroglutamic acid, l-Serine, Succinamic acid, l-Alaninamide and Bromo succinic acid; and negative to: Tryptophan deaminase, Glucose fermentation, Arginine dihydrolase, N-Acetyl-d-galactosamine, N-Acetyl-d-glucosamine, Adonitol, d-Cellobiose, i-Erythritol, d-Fructose, Gentiobiose, m-Inositol, α-d-Lactose, Lactulose, Maltose, d-Melibiose, β-Methyl-d-Glucoside, d-Raffinose, Sucrose, d-Trehalose, Turanose, Xylitol, α-Cyclodextrin, Glycogen, cis-Aconitic acid, d-Glucuronic acid, Itaconic acid, 2,3-Butanediol, d,l-α-Glycerol phosphate, Glucose-1-phosphate, Glucose-6-phosphate, l-Histidine, Hydroxy-l-proline, l-Ornithine, Urocanic acid, Inosine, Uridine, Thymidine, Putrescine and 2-Aminoethanol.
Fatty acid composition
The fatty acid composition was analyzed for the BR 3262 and BR 3267 strains and for B. elkanii USDA 76T, B. pachyrhizi PAC 48T and B. yuanmingense CCBAU 10071T, the closest type strains based on EzTaxon Identify search of the 16S rRNA gene (http://www.ezbiocloud.net/eztaxon/identify). The fatty acid compositions of BR 3267 and BR 3262 were similar to those of other strains within Bradyhizobium genus, especially because of the presence of 16:00 in approximately 15% and a summed feature 8 (18:1 w7c) between approximately 70 and 80%.40 BR 3262 has a composition close to those of B. elkanii USDA 76T and B. pachyrhizi PAC 48T, with the presence of 19:0 cyclo w8c of 15.98% and a summed feature 8 (18:1 w7c) of 69.32. Instead, BR 3267 and B. yuanmingense CCBAU 10071T did not present 19:0 cyclo w8c and had a summed feature 8 of 86.42 and 84.63%, respectively.
Phylogenetic analyses
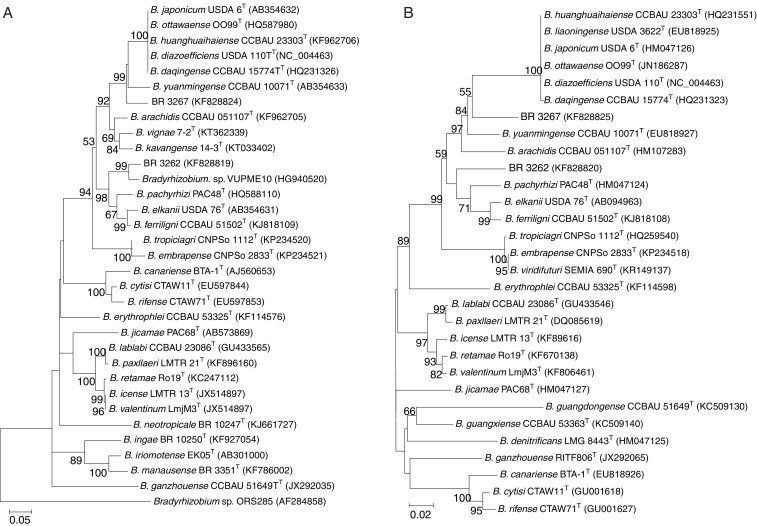
The Maximum Likelihood (ML) phylogenetic analysis performed on the 16S rRNA (1227 bp) gene placed the cowpea strains BR 3267 and BR 3262 into subgroups I and II, respectively, within the genus Bradyrhizobium (Fig. 1). The closest strain to BR 3267 was B. yuanmingense CCBAU 10071T (99.5% 16S rRNA similarity), with 67% ML bootstrap support, followed by Bradyrhizobium subterraneum 58 2-1T (99.6% 16S rRNA similarity) in a separated branch with 62% bootstrap. In contrast, BR 3262 was placed within subgroup II of Bradyrhizobium in a phylogenetic linage together with B. pachyrhizi PAC48T (100% 16S rRNA similarity), B. elkanii USDA 76T (99.9%) and Bradyrhizobium tropiciagri CNPSo 1112T (99.2%), but with nodes lower than <60% of bootstrap. The Bradyrhizobium ferriligni CCBAU 51502T was the closest taxon to the clade of B. tropiciagri–B. elkanii–B. pachyrhizi – BR 3262, as shown by the high ML bootstrap support (91%, Fig. 1); it shares 97.8% of 16S rRNA similarity with BR 3262.
Fig. 1.
Maximum Likelihood phylogenetic tree based on 16S rRNA gene sequences showing relationships between the strains BR 3262 and BR 3267 and the type species (T) of the genus Bradyrhizobium. Bootstrap values were inferred from 500 replicates and are indicated at the tree nodes when ≥50%. GenBank accession numbers are provided in parenthesis. Microvirga vignae BR3299T was included as the outgroup. The bar represents two estimated substitutions per 100 nucleotide positions.
We used aligned sequences from the recA (359 bp), glnII (505 bp) and gyrB (552 bp) housekeeping genes, which resulted in a concatenated sequence of 1416 bp, to better assess the phylogenetic affiliation of the BR 3267 and BR 3262 strains. ML phylogenetic trees of single and concatenated genes (Fig. 2) were generated and showed high congruence; therefore, we presented only the concatenated one. The concatenated phylogenetic analysis revealed a clear affiliation between BR 3267 and B. yuanmingense CCBAU 10071T, with high confidence bootstrap values of the 100%, in a lineage that was distinct from the closest soybean isolates Bradyrhizobium daqingense CCBAU 15774T48 and Bradyrhizobium huanghuaihaiense CCBAU 23303T.49 On the other hand, BR 3262 was phylogenetically close to B. pachyrhizi PAC48T, which formed a lineage with high confidence values (bootstrap ML of 98%) and separated from the closed taxa B. ferriligni CCBAU 51502T and B. elkanii USDA76T. Additionally, pairwise comparisons of the concatenated sequences revealed that BR 3267 shared 99% sequence identity with B. yuanmingense CCBAU 10071T, followed by Bradyrhizobium liaoningense USDA 3622T (95%) and B. daqingense CCBAU 15774T (94.5%). Likewise, BR 3262 was similar to B. pachyrhizi PAC48T (98.6%), followed by B. elkanii USDA 76T (96.9%) and B. ferriligni CCBAU 51502T (95.5%). In an independent analysis of the recA, glnII and a concatenated recA-glnII sequences (data not shown), BR 3267 and BR 3262 were far from the newest proposal species Bradyrhizobium kavangense 14-3T,50 Bradyrhizobium vignae 7-2T51 and B. subterraneum 58-2-1T52 isolated from African soils.
Fig. 2.
Unrooted maximum likelihood phylogenetic tree based on three concatenated sequences (recA, glnII, gyrB) showing relationships between the strains BR 3262 and BR 3267 and type species (T) of the genus Bradyrhizobium. The phylogenetic tree was built using the Kimura two-parameter method. Bootstrap values were inferred from 500 replicates and are indicated at the tree nodes when ≥50%. GenBank accession numbers are provided in parenthesis. The bar represents two estimated substitutions per 100 nucleotide positions.
DNA–DNA relatedness and average nucleotide identity
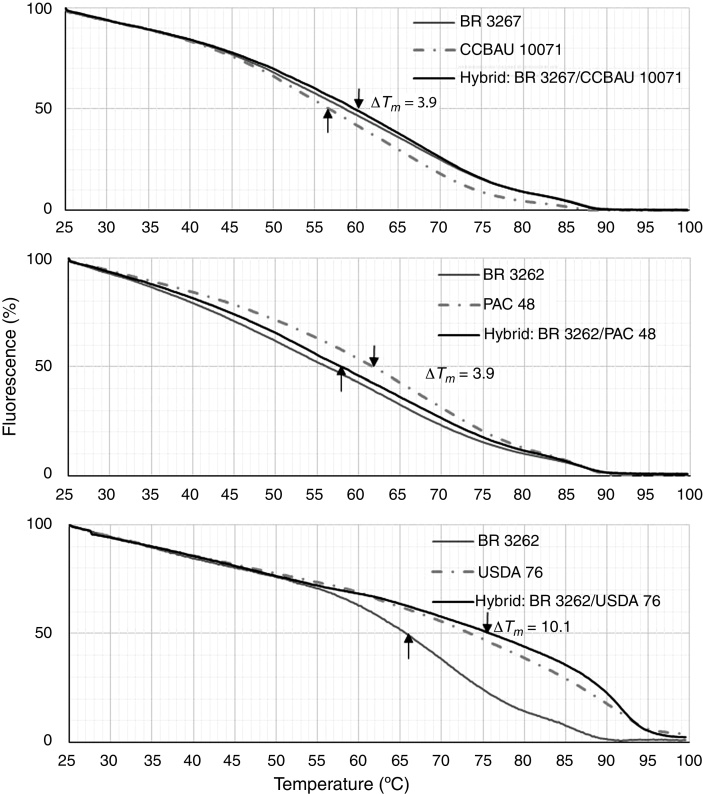
After obtaining the phylogenetic results, we established the contrasts for the analysis of the DNA–DNA homology following the protocol described by Gonzales and Saiz-Jimenez.29 BR 3262 was compared with B. elkanii USDA 76T and B. pachyrhizi PAC 48T (the closest strains according to MLSA), and BR 3267 was compared with B. yuanmingense CCBAU 10071T (also the closest strain in the MLSA). The hybridization of BR 3262 × B. elkanii USDA 76T and B. elkanii USDA 76T × B. pachyrhizi PAC 48T presented ΔTm values equal to 10.1 °C and 3.9 °C, respectively, and the hybridization of BR 3267 × B. yuanmingense CCBAU 10071T presented a value of 3.9 °C (Fig. 3).
Fig. 3.
Melting curves generated with Applied Biosystems 7500 Real-Time System for ΔTm determination between homologous and hybrid DNA. (a) Comparison between strain BR 3267 and B. yuanmingense CCBAU 1007T; (b) between BR 3262 and B. pachyrhizi PAC 48T; (C) between BR 3262 and B. elkanii USDA 76T.
For the ANI calculation, we used the available genomes of B. elkanii USDA 76T and B. pachyrhizi PAC 48T for comparison with BR 3262 and, B. yuanmingense CCBAU 3515754 for comparison with BR 3267 because no genome was available for the type strain of B. yuanmingense CCBAU 10071T. The ANI between strain BR 3262 and B. elkanii USDA 76T and between strain BR 3262 and B. pachyrhizi PAC 48T was 94.6 and 95.3, respectively. On another hand the ANI in the comparison of BR 3267 and B. yuanmingense CCBAU 35157 was 96.5.
Phylogeny based on the nodC and nifH symbiotic genes
Maximum Likelihood phylogenetic analyses of the symbiotic genes nodC and nifH separated the BR 3267 and BR 3262 strains into two divergent phylogenetic lineages (Fig. 4A for nodC, Fig. 4B for nifH). For both nodC and nifH, BR 3267 formed an independent branch that clustered together with B. yuanmingense CCBAU 10071T and the soybean type strains that represent the symbiovar glycinearum. In contrast, BR 3262 was grouped in an early branch linked to the related strains B. pachyrhizi PAC48T, B. elkanii USDA 76T and B. ferriligni CCBAU51502T for both symbiotic genes (Fig. 4A and B).
Fig. 4.
Unrooted maximum likelihood phylogenetic tree based on nodC (A) and nifH (B) genes showing relationships between the strains BR 3262 and BR 3267 and type species (T) and reference strains of the genus Bradyrhizobium. The phylogenetic tree was built using the Kimura two-parameter method. Bootstrap values were inferred from 500 replicates and are indicated at the tree nodes when ≥50%. GenBank accession numbers are provided in the parenthesis. The bar represents five or two estimated substitutions per 100 nucleotide positions.
Discussion
The analyses in the culture medium showed that both strains, BR 3267 and BR 3262, have similar behaviors in comparison to other members of the Bradyrhizobium genus regarding tolerance to temperature, pH and NaCl concentration in the culture medium. These traits indicate they are tolerant to different edapho-climatic factors and make them well adapted to the conditions of the tropical soils from which they were isolated. This adaptability is corroborated by their good performance in cowpea inoculations.8, 36, 37 The BR 3267 and BR 3262 strains were positive to urease and nitrate reductase enzymes reactions, which are apparently common characteristics among strains of the genus Bradyrhizobium.38, 39 It seems to be a common characteristic among Bradyrhizobium species of subgroup I (close to B. japonicum) to be sensitive to streptomycin and tetracycline, as showed by BR 3267. In contrast, BR 3262 was tolerant to these antibiotics, a common characteristic of Bradyrhizobium species belonging to subgroup II (close to B. elkanii).40 The 50 carbon sources used by BR 3267 points to greater access to different soil carbon compounds. The results of the fatty acid composition analysis corroborate previous findings that indicate that the main differences between subgroups I and II of Bradyrhizobium are the 19:0 cyclo w8c and the summed feature 8 (18:1 w7c), which indicate that BR 3267 belongs to the subgroup of B. japonicum and BR 3262 to the subgroup of B. elkanii.40, 41
When the 16S rRNA genes of strains BR 3267 and BR 3262 were first studied,11 it was observed that although BR 3267 could be grouped with B. japonicum, there were differences with respect to the B. japonicum type strain pattern. At the time, however, the species B. yuanmingense, which also belongs to subgroup I of Bradyrhizobium, had not yet been described; thus, there was no species indication. It was observed on the other hand that BR 3262 had a strong resemblance to B. elkanii by the 16S rRNA gene, which supported that the strain should be assigned to that species. At that time, the species B. pachyrhizi, which in terms of base composition of the 16S rRNA gene differs from strains B. elkanii USDA 76T only on the order of 0.1%, had not yet been described. 16S rRNA sequence analysis is not always fine enough for species assignment42, 43; in the genus Bradyrhizobium some species share almost identical 16S rRNA gene sequences. Examples are the type strains of the species B. japonicum and B. liaoningense,3 along with B. elkanii and B. pachyrhizi.18 Due to the high conservation of the 16S rRNA gene among members of the Bradyrhizobium genus,14, 15 several authors have recommended an MLSA of protein-encoding (i.e., housekeeping) genes.6, 22, 44, 45, 46, 47 In our case, the MLSA results were congruent to the 16S rRNA analysis and clearly indicate that BR 3267 and BR 3262 as members of the species B. yuanmingense and B. pachyrhizi, respectively (Fig. 2). The DNA–DNA homology results confirm the results obtained in the analysis of housekeeping genes, i.e., BR 3262 belongs to the B. pachyrhizi species, and BR 3267 is a member of the B. yuanmingense species. As previously shown, a ΔTm value equal to 5 °C can be used as the threshold to delineate a species, where lower values indicate equal species and higher values indicate different species.29 A ΔTm value of 5 °C corresponds to a homology value of approximately 70% obtained by traditional DNA–DNA hybridization techniques.29, 30 Moreover, we calculated the ANI, a robust method that replaces the traditional DNA–DNA hybridization techniques.19 This method involves comparing the sequences of two bacterial genomes, and a value of 95% is considered as the threshold.19, 53 In this case, when the homology is greater than 95%, the two strains belong to the same species; if the homology is lower, they belong to different species. Thus, the results of the comparisons again confirmed that BR 3262 belongs to B. pachyrhizi (ANI greater than 95%) and the same BR 3267 to B. yuanmingense (ANI greater than 95%).
Symbiotic genes related to nodulation (nod) and nitrogen fixation (nif) processes hold no taxonomic information for species definition.17 However, in our study, they showed good phylogenetic correspondence to the core genome ones. Interestingly, nodC and nifH gene phylogenetic analyses have been used to predict the host range nodulation capacity and for the delineation of symbiovars, i.e., groups of rhizobial strains with similar symbiotic behaviors concerning nodulation and nitrogen fixation capacity with legumes.55 For the nodC gene, for example, a BLAST search in the NCBI database (http://blast.ncbi.nlm.nih.gov/) showed the Arachis hypogae strains CCBAU 53380 (Ac. no. KC509232) and CCBAU 51663 (Ac. no. KC509217), isolated in China, as the Bradyrhizobium strains that had the closest similarity (98%) with BR 3267, indicating that peanut might be a possible host for this latter strain. Instead, in the nodC gene, BR 3262 had 95% similarity with the cowpea strain Bradyrhizobium sp. VUMPE10 (Fig. 4A), isolated from an European soil and proposed as a new symbiovar (sv. vignae) within the Bradyrhizobium genus.56 These data indicate that cowpea nodulates with very effective Bradyrhizobium strains that are holding divergent symbiotic genes which might represent distinct symbiovars.
Cowpea can nodulate with a wide range of rhizobia genotypes, Bradyrhizobium being the major symbiont group. Thies and colleagues57 proposed that cowpea's promiscuity for nodulation with the bradyrhizobial population may have a limit. However, a global Bradyrhizobium collection for cowpea nodulation has not yet been well analyzed to identify true cowpea symbionts in light of the symbiovar concept. It is not well understood whether any Bradyrhizobium group is specialized to produce nodules and to fix nitrogen specifically in cowpea.
Conclusion
The results of the identifications of B. yuanmingense BR 3267 and B. pachyrhizi BR 3262 can be considered reliable. Furthermore, analysis of the nodC and nifH genes indicates that these two strains are similar to strains isolated on other continents. Strain BR 3262 possibly belongs to the vignae symbiovar, which was recently assigned for strains isolated from cowpea in Europe. Both strains hold similar symbiotic genomes with Bradyrhizobium strains outside of South America.
Apparently, cowpea is able to establish effective symbiosis with divergent bradyrhizobia strains isolated from Brazilian soils, confirming its promiscuous capacity for nodulation.
Conflicts of interest
The authors declare they have no conflicts of interest.
Acknowledgements
This study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). We would like to thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes) for the scholarships, likewise to CNPq for the fellowship of research productivity (PQ).
Associate Editor: Fernando Andreote
References
- 1.Hungria M., Mendes I.C. Nitrogen fixation with soybean: the perfect symbiosis? In: de Bruijn F.J., editor. Biological Nitrogen Fixation. John Wiley & Sons, Inc.; Hoboken, NJ: 2015. pp. 1009–1024. [Google Scholar]
- 2.Brasil Ministério da Agricultura Pecuária e Abastecimento. Instrução normativa n° 13 de 24 de março de 2011 . Diário Oficial da União da República Federativa do Brasil; 2011. Aprova as normas sobre especificações, garantias, registro, embalagem e rotulagem dos inoculantes destinados à agricultura, bem como as relações dos microorganismos autorizados e recomendados para produção de inoculantes no Brasil, na forma dos Anexos I, II, III, desta Instrução Normativa; pp. 3–7. Seção 1. [Google Scholar]
- 3.Delamuta J.R.M., Ribeiro R.A., Ormeño-Orrillo E., Melo I.S., Martínez-Romero E., Hungria M. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum Group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int J Syst Evol Microbiol. 2013;63:3342–3351. doi: 10.1099/ijs.0.049130-0. [DOI] [PubMed] [Google Scholar]
- 4.Delamuta J.R.M., Ribeiro R.A., Ormeño-Orrillo E. Bradyrhizobium tropiciagri sp. nov. and Bradyrhizobium embrapense sp. nov., nitrogen-fixing symbionts of tropical forage legumes. Int J Syst Evol Microbiol. 2015;65:4424–4433. doi: 10.1099/ijsem.0.000592. [DOI] [PubMed] [Google Scholar]
- 5.Helene L.C.F., Delamuta J.R.M., Ribeiro R.A. Bradyrhizobium viridifuturi sp. nov., encompassing nitrogen-fixing symbionts of legumes used for green manure and environmental services. Int J Syst Evol Microbiol. 2015;65:4441–4448. doi: 10.1099/ijsem.0.000591. [DOI] [PubMed] [Google Scholar]
- 6.Menna P., Barcellos F.G., Hungria M. Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on multilocus sequence analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD and dnaK genes. Int J Syst Evol Microbiol. 2009;59:2934–2950. doi: 10.1099/ijs.0.009779-0. [DOI] [PubMed] [Google Scholar]
- 7.Filho F.R.F. Embrapa Meio Norte; Teresina: 2011. Feijão-Caupi no Brasil: Produção, melhoramento genético, avanços e desafios. 81 pp. [Google Scholar]
- 8.Alcantara R.M.C.M., Xavier G.R., Rumjanek N.G., Rocha M.M., Carvalho J.S. Symbiotic efficiency in parents of Brazilian cultivars of the cowpea. Rev Ciênc Agron. 2014;45:1–9. [Google Scholar]
- 9.Martins L.M.V., Xavier G.R., Rangel F.W. Contribution of biological nitrogen fixation to cowpea: a strategy for improving grain yield in the semi-arid region of Brazil. Biol Fertil Soils. 2003;38:333–339. [Google Scholar]
- 10.Zilli J.E., Ferreira E., Neves M., Rumjanek N. Efficiency of fast-growing rhizobia capable of nodulating cowpea. An Acad Bras Cienc. 1999;71:553–560. [Google Scholar]
- 11.Zilli J.E., Valicheski R.R., Rumjanek N.G., Simões-Araújo J.L., Freire Filho F.R., Neves M.C.P. Symbiotic efficiency of cowpea Bradyrhizobium strains in Cerrado soils. Pesqui Agropecu Bras. 2006;41:811–818. [Google Scholar]
- 12.Jordan D. NOTES: transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int J Syst Bacteriol. 1982;32:136–139. [Google Scholar]
- 13.Hollis A.B., Kloos W.E., Elkan G.H. DNA:DNA hybridization studies of Rhizobium japonicum and related Rhizobiaceae. J Gen Microbiol. 1981;123:215–222. [Google Scholar]
- 14.Willems A., Coopman R., Gillis M. Phylogenetic and DNA–DNA hybridization analyses of Bradyrhizobium species. Int J Syst Evol Microbiol. 2001;51:111–117. doi: 10.1099/00207713-51-1-111. [DOI] [PubMed] [Google Scholar]
- 15.Willems A., Doignon-Bourcier F., Goris J. DNA–DNA hybridization study of Bradyrhizobium strains. Int J Syst Evol Microbiol. 2001;51:1315–1322. doi: 10.1099/00207713-51-4-1315. [DOI] [PubMed] [Google Scholar]
- 16.Kuykendall L.D., Saxena B., Devine T.E., Udell S.E. Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can J Microbiol. 1992;38:501–515. [Google Scholar]
- 17.Peix A., Ramírez-Bahena M.H., Velázquez E., Bedmar E.J. Bacterial associations with legumes. Crit Rev Plant Sci. 2015;34:17–42. [Google Scholar]
- 18.Ramírez-Bahena M.H., Peix A., Rivas R. Bradyrhizobium pachyrhizi sp. nov. and Bradyrhizobium jicamae sp. nov., isolated from effective nodules of Pachyrhizus erosus. Int J Syst Evol Microbiol. 2009;59:1929–1934. doi: 10.1099/ijs.0.006320-0. [DOI] [PubMed] [Google Scholar]
- 19.Konstantinidis K.T., Tiedje J.M. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson C., Amaral G., Campeão M. Microbial taxonomy in the post-genomic era: rebuilding from scratch? Arch Microbiol. 2014;197:359–370. doi: 10.1007/s00203-014-1071-2. [DOI] [PubMed] [Google Scholar]
- 21.Vincent J.M. [Published for the] International Biological Programme [by] Blackwell Scientific; Oxford: 1970. A Manual for the Practical Study of Root-Nodule Bacteria. International Biological Programme. [Google Scholar]
- 22.Stepkowski T., Moulin L., Krzyzanska A., McInnes A., Law I.J., Howieson J. European origin of Bradyrhizobium populations infecting lupins and serradella in soils of Western Australia and South Africa. Appl Environ Microbiol. 2005;71:7041–7052. doi: 10.1128/AEM.71.11.7041-7052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarita S., Sharma P.K., Priefer U.B., Prell J. Direct amplification of rhizobial nodC sequences from soil total DNA and comparison to nodC diversity of root nodule isolates. FEMS Microbiol Ecol. 2005;54:1–11. doi: 10.1016/j.femsec.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 24.Laguerre G., Nour S.M., Macheret V., Sanjuan J., Drouin P., Amarger N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology. 2005;147:981–993. doi: 10.1099/00221287-147-4-981. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 28.Radl V., Simões-Araújo J.L., Leite J. Microvirga vignae sp. nov., a root nodule symbiotic bacterium isolated from cowpea grown in the semi-arid of Brazil. Int J Syst Evol Microbiol. 2014;64:725–730. doi: 10.1099/ijs.0.053082-0. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez J.M., Saiz-Jimenez C. A simple fluorimetric method for the estimation of DNA–DNA relatedness between closely related microorganisms by thermal denaturation temperatures. Extremophiles. 2005;9:75–79. doi: 10.1007/s00792-004-0417-0. [DOI] [PubMed] [Google Scholar]
- 30.Moreira A.P., Pereira N., Thompson F. Usefulness of a real-time PCR platform for G + C content and DNA–DNA hybridization estimations in vibrios. Int J Syst Evol Microbiol. 2001;61:2379–2383. doi: 10.1099/ijs.0.023606-0. [DOI] [PubMed] [Google Scholar]
- 31.De Ley J., De Smedt J. Improvements of the membrane filter method for DNA:rRNA hybridization. Antonie Van Leeuwenhoek. 1975;12:133–141. doi: 10.1007/BF02565064. [DOI] [PubMed] [Google Scholar]
- 32.Richter M., Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delcher A.L., Kasif S., Fleischmann R.D., Peterson J., White O., Salzberg S.L. Alignment of whole genomes. Nucl Acids Res. 1999;27:2369–2376. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simões-Araújo J.L., Leite J., Rouws L.M.F. Draft genome sequence of Bradyrhizobium sp. strain BR 3262, an effective microsymbiont recommended for cowpea inoculation in Brazil. Braz J Microbiol. 2016 doi: 10.1016/j.bjm.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simões-Araújo J.L., Leite J., Passos S.R., Xavier G.R., Rumjanek N.G., Zilli J.E. Draft genome sequence of Bradyrhizobium sp. strain BR 3267, an elite strain recommended for cowpea inoculation in Brazi. Braz J Microbiol. 2016 doi: 10.1016/j.bjm.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gualter R.M.R., Boddey R.M., Rumjanek N.G., Freitas A.C.R.d., Xavier G.R. Eficiência agronômica de estirpes de rizóbio em feijão-caupi cultivado na região da Pré-Amazônia Maranhense. Pesqui Agropecu Bras. 2011;46:303–308. [Google Scholar]
- 37.Silva Júnior E.B., Silva K., Oliveira S.S. Cowpea nodulation and production in response to inoculation with different rhizobia densities. Pesqui Agropecu Bras. 2014;49:804–812. [Google Scholar]
- 38.Mesa S., Alché J.D., Bedmar E., Delgado M.J. Expression of nir, nor and nos denitrification genes from Bradyrhizobium japonicum in soybean root nodules. Physiol Plant. 2004;120:205–211. doi: 10.1111/j.0031-9317.2004.0211.x. [DOI] [PubMed] [Google Scholar]
- 39.Mesa S., Göttfert M., Bedmar E.J. The nir, nor, and nos denitrification genes are dispersed over the Bradyrhizobium japonicum chromosome. Arch Microbiol. 2001;176:136–142. doi: 10.1007/s002030100305. [DOI] [PubMed] [Google Scholar]
- 40.Kuykendall L., Roy M., O’neill J., Devine T. Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol. 1988;38:358–361. [Google Scholar]
- 41.Tighe S.W., de Lajudie P., Dipietro K., Lindström K., Nick G., Jarvis B.D. Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium species using the Sherlock Microbial Identification System. Int J Syst Evol Microbiol. 2000;50:787–801. doi: 10.1099/00207713-50-2-787. [DOI] [PubMed] [Google Scholar]
- 42.Mignard S., Flandrois J. 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods. 2006;67:574–581. doi: 10.1016/j.mimet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasan R., Karaoz U., Volegova M. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLOS ONE. 2015;10:e0117617. doi: 10.1371/journal.pone.0117617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nzoué A., Miché L., Klonowska A., Laguerre G., de Lajudie P., Moulin L. Multilocus sequence analysis of bradyrhizobia isolated from Aeschynomene species in Senegal. Syst Appl Microbiol. 2009;32:400–412. doi: 10.1016/j.syapm.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Parker M. The spread of Bradyrhizobium lineages across host legume clades: from Abarema to Zygia. Microb Ecol. 2015;69:630–640. doi: 10.1007/s00248-014-0503-5. [DOI] [PubMed] [Google Scholar]
- 46.Rivas R., Martens M., de Lajudie P., Willems A. Multilocus sequence analysis of the genus Bradyrhizobium. Syst Appl Microbiol. 2009;32:101–110. doi: 10.1016/j.syapm.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Vinuesa P., Silva C., Werner D., Martínez-Romero E. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol Phylogenet Evol. 2005;34:29–54. doi: 10.1016/j.ympev.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 48.Wang J.Y., Wang R., Zhang Y.M. Bradyrhizobium daqingense sp. nov. isolated from nodules of soybean grown in Daqing City of China. Int J Syst Evol Microbiol. 2012;63:616–624. doi: 10.1099/ijs.0.034280-0. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y.M., Li Y., Chen W.F. Bradyrhizobium huanghuaihaiense sp. nov., an effective symbiotic bacterium isolated from soybean (Glycine max L.) nodules. Int J Syst Evol Microbiol. 2012;62:1951–1957. doi: 10.1099/ijs.0.034546-0. [DOI] [PubMed] [Google Scholar]
- 50.Grönemeyer J.L., Hurek T., Reinhold-Hurek B. Bradyrhizobium kavangense sp. nov., a symbiotic nitrogen-fixing bacterium from root nodules of traditional pulses in the Kavango region of Namibia. Int J Syst Evol Microbiol. 2015;65:4886–4894. doi: 10.1099/ijsem.0.000666. [DOI] [PubMed] [Google Scholar]
- 51.Grönemeyer J.L., Hurek T., Bünger W., Reinhold-Hurek B. Bradyrhizobium vignae sp. nov., a nitrogen-fixing symbiont isolated from effective nodules of Vigna and Arachis in Africa. Int J Syst Evol Microbiol. 2015;66:62–69. doi: 10.1099/ijsem.0.000674. [DOI] [PubMed] [Google Scholar]
- 52.Grönemeyer J.L., Chimwamurombe P., Reinhold-Hurek B. Bradyrhizobium subterraneum sp. nov., a symbiotic nitrogen-fixing bacterium from root nodules of groundnuts in Namibia. Int J Syst Evol Microbiol. 2015;65:324–327. doi: 10.1099/ijsem.0.000403. [DOI] [PubMed] [Google Scholar]
- 53.Goris J., Konstantinidis K., Klappenbach J., Coenye T., Vandamme P., Tiedje J. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 54.Tian C.F., Zhou Y.J., Zhang Y.M. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc Natl Acad Sci U S A. 2012;109:8629–8634. doi: 10.1073/pnas.1120436109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogel M.A., Ormeño-Orrillo E., Martinez Romero E. Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol. 2011;34:96–104. doi: 10.1016/j.syapm.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 56.Bejarano A., Ramírez-Bahena M.-H., Velázquez E., Peix A. Vigna unguiculata is nodulated in Spain by endosymbionts of Genisteae legumes and by a new symbiovar (vignae) of the genus Bradyrhizobium. Syst Appl Microbiol. 2014;37:533–5340. doi: 10.1016/j.syapm.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Thies J.E., Bohlool B.B., Singleton P.W. Subgroups of the cowpea miscellany: symbiotic specificity within bradyrhizobium spp. for Vigna unguiculata, phaseolus lunatus, Arachis hypogaea, and macroptilium atropurpureum. Appl Environ Microbiol. 1991;57:1540–1545. doi: 10.1128/aem.57.5.1540-1545.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]