Abstract
To mitigate the deleterious effects of abiotic stress, the use of plant growth-promoting bacteria along with diazotrophic bacteria has been increasing. The objectives of this study were to investigate the key enzymes related to nitrogen and carbon metabolism in the biological nitrogen fixation process and to elucidate the activities of these enzymes by the synergistic interaction between Bradyrhizobium and plant growth-promoting bacteria in the absence and presence of salt stress. Cowpea plants were cultivated under axenic conditions, inoculated with Bradyrhizobium and co-inoculated with Bradyrhizobium sp. and Actinomadura sp., Bradyrhizobium sp. and Bacillus sp., Bradyrhizobium sp. and Paenibacillus graminis, and Bradyrhizobium sp. and Streptomycessp.; the plants were also maintained in the absence (control) and presence of salt stress (50 mmolL−1 NaCl). Salinity reduced the amino acids, free ammonia, ureides, proteins and total nitrogen content in nodules and increased the levels of sucrose and soluble sugars. The co-inoculations responded differently to the activity of glutamine synthetase enzymes under salt stress, as well as glutamate synthase, glutamate dehydrogenase aminating, and acid invertase in the control and salt stress. Considering the development conditions of this experiment, co-inoculation with Bradyrhizobium sp. and Bacillus sp. in cowpea provided better symbiotic performance, mitigating the deleterious effects of salt stress.
Keywords: BNF, Plant growth-promoting bacteria, Glutamine synthetase, Acid invertase, Salinity
Introduction
Biological nitrogen fixation (BNF) involves a complex interaction with a host plant and represents an environmentally clean nitrogen pathway in agricultural systems.1 In this way, the inoculation of legumes with rhizobia favors the increase in and the availability of nitrogen in commercial legume production.2 This process can be affected by physical, chemical and biological factors, and it is more frequent in legumes in symbiosis with fixing bacteria.3 However, it is necessary to observe the combination and compatibility of the bacterial strains involved.4 Co-inoculation involving strains of Bradyrhizobium sp. with plant growth-promoting bacteria (PGPB) can have positive effects by increasing nutrient mobilization, mainly that of N and C.5
PGPB can improve the growth and development of plant species via several mechanisms, such as nitrogen fixation, phosphate solubilization, phytohormone synthesis and increased iron uptake by the production of siderophores.6 Moreover, PGPB can also benefit plants by exhibiting protective action against pathogens, diseases and environmental stresses.7 In addition, PGPB can colonize the roots of plant species and create a favorable microenvironment for plant development. Thus, the inoculation of agricultural species with PGPB represents a very promising technique.
When subjected to abiotic stress such as drought and salt stress, plants develop mechanisms of tolerance or adaptation such as osmotic adjustment; these mechanisms allow plants to maintain their development even under stress conditions.8 Recent advances in molecular studies have yielded insights into the signaling networks of plant-microbe interactions that contribute to salt tolerance. The beneficial effects of plant growth-promoting rhizobacteria involve boosting key physiological processes including water and nutrient uptake, photosynthesis, and source-sink relationships that promote growth and development.9
Osmotic adjustment with organic or inorganic solutes carried out by plant species subjected to salinity allows those plants to maintain a continuous soil water uptake.10 In addition, plants can use other mechanisms when subjected to salinity stress, such as the control of the absorption of Na+ by roots or the accumulation and selective compartmentalization of excess ions in the vacuole.11
Several studies have shown that co-inoculation of legume species with rhizobia and PGPB has a beneficial effect on the growth and development of plants.12 In this context, we hypothesized that co-inoculation with Bradyrhizobium sp. and PGPB optimizes the development and the BNF of cowpea by mitigating the deleterious effects of salt stress. Metabolites and key enzymes of carbon and nitrogen metabolism were evaluated in cowpea inoculated with Bradyrhizobium sp. and co-inoculated with Bradyrhizobium sp. and PGPB in the presence and absence of salt stress.
Materials and methods
Experimental design and statistical analysis
The experimental design consisted of a randomized block in a 5 × 2 + 1 factorial scheme. There were five bacterial combinations (one inoculation with Bradyrhizobium sp. and four co-inoculations with Bradyrhizobium sp. and PGPB), two salinity levels (0 and 50 mmol L−1 NaCl), and an absolute control (uninoculated plants without nitrogen and without NaCl). Four blocks and two replicates per block were used. The data were subjected to analysis of variance (ANOVA) with a significance level of 5% by the F test, and the means were compared by the Tukey test at 5% probability. A contrast analysis was performed to evaluate the effects of inoculation treatments versus the absolute control in the presence and absence of salinity. All statistical analyses were performed using the SASM-Agri 8.1 program.13
Production of microorganisms and the preparation of inoculants
The rhizobia and PGPB strains used in the experiment (Table 1) were multiplied under controlled conditions for the production of inoculants. The following media were used to purify and multiply microorganisms: yeast mannitol agar (YMA) and yeast mannitol (YM)14 at pH 6.5 for Bradyrhizobium sp., dextrose yeast glucose sucrose (DYGS)15 at pH 6.0 for Bacillus sp., trypticase soy agar (TSA) and tryptic soy broth (TSB) at pH 7.3 for Paenibacillus graminis, and arginine yeast and agar (AYA)16 at pH 6.4 for Actinomadura sp. and Streptomyces sp. The rhizobia inoculants were incubated on a rotary shaker (200 rpm) at 28 °C for 96 h, and the PGPB inoculants were maintained on a rotary shaker (200 rpm) at 30 °C for 48–96 h depending on the bacterial strain. These bacterial strains showed promising results in previous studies, and their behavior was tested in cowpea in the presence and absence of salinity stress.12
Table 1.
Bradyrhizobium sp. and plant growth promoting bacteria used in the experiment.
| Microorganisms | Access code | Origin |
|---|---|---|
| Bradyrhizobium sp. | UFLA 03-84 | Pasture soil (Jí-Paraná, Rondônia-Brazil) |
| Actinomadura sp. | 183-EL | Caatinga rhizosphere (Pernambuco-Brazil) |
| Paenibacillus graminis | MC 04.21 | Corn rhizosphere (Zea mays) in Cerrado soil-Brazil |
| Bacillus sp. | IPACC11 | Stalks sugarcane (Saccharum officinarum) of forest zone of Pernambuco-Brazil |
| Streptomyces sp. | 212 | Rhizosphere of Arugula (Eruca sativa) |
Plant culture and treatments
The experiment was conducted under axenic conditions in a greenhouse (air temperature of 31 °C, air humidity of 60% and a day length of 9 h). The seeds of cowpea cultivar ‘IPA 206’ were disinfested17 and sown in Leonard pots containing washed (pH 6.5) and autoclaved (1 h; 120 °C; 101 kPa) sand. At the time of sowing, the seeds were inoculated using 1.0 mL of the bacterial suspension (108 CFU mL−1) with Bradyrhizobium sp. or co-inoculated with 1.0 mL of the bacterial suspension containing Bradyrhizobium sp. and 1.0 mL of the bacterial suspension containing a strain of PGPB (107 CFU mL−1). Co-inoculations were formulated in accordance with the information in Table 1.
Throughout the experiment, the plants were irrigated by capillarity with an N-free nutrient solution,18 as modified by Silveira et al.19 Thinning was carried out at four days after germination (DAG), and two plants per pot (experimental unit) were maintained. The plants were subjected to salt stress at 15 DAG, in which 50 mmol L−1 sodium chloride (NaCl) was added to the nutrient solution. The nutrient solutions (pH of 6.5) were placed in Leonard jars and changed weekly. At the time, the substrate was washed with distilled water, and the pH and electrical conductivity (EC) of the drainage were measured to match the pH and EC of the vessel; the EC values were 0.99 mS cm−1 and 5.6 mS cm−1 for the control and stress treatments, respectively.
At 37 DAG, the roots containing nodules were immersed in liquid nitrogen and then kept at -80 °C. For transport, the nodule-containing roots frozen in liquid N2 were stored in thermal containers containing dry ice. The frozen nodules were lyophilized, macerated and stored in a desiccator until their biochemical determination.
Biochemical determination
Nitrogen metabolism analyses included the following: total nitrogen,20 free ammonia,21 N-α-amino-soluble acids (free amino acids),22 leghemoglobin,23 ureides,24 proline,25 soluble proteins,26 glutamine synthetase,27 glutamate synthase,28 and glutamate dehydrogenase aminating.29 Carbon metabolism analyses included sucrose,30 total soluble sugars,31 acid invertase and neutral invertase activity.32
Results
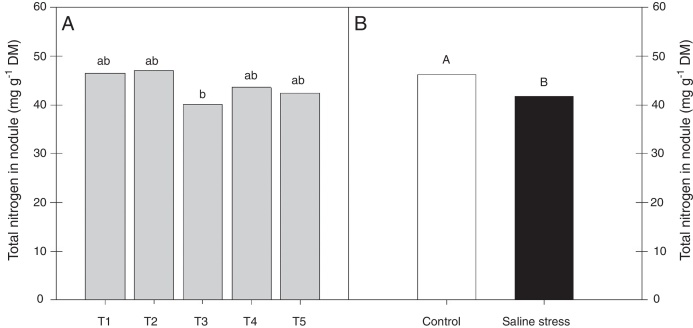
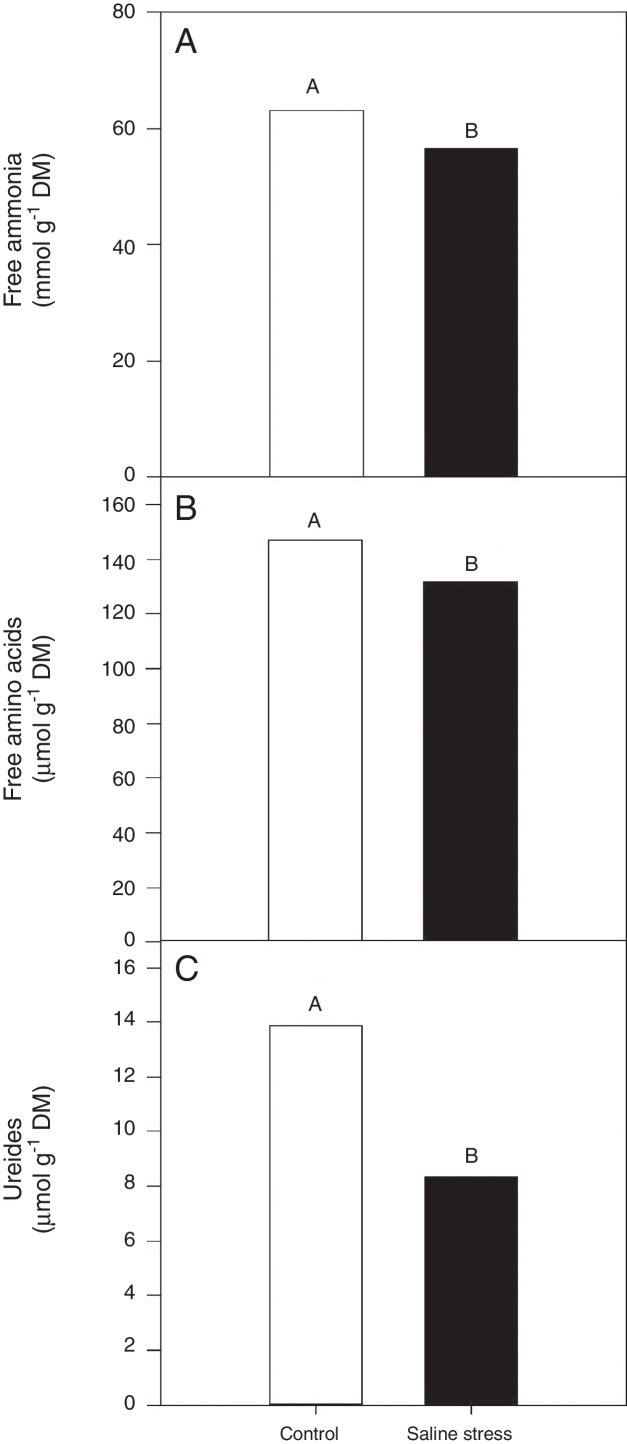
Only the cowpea plants co-inoculated with Bradyrhizobium sp. and P. graminis (T3) showed low nitrogen content in the nodules, regardless of whether they were grown in the presence or absence of salt stress (Fig. 1A). When comparing cultivation conditions, the plants cultivated in the absence of salt stress showed higher nitrogen content in their nodules, regardless of inoculation. The total nitrogen in the nodules of the cowpea plants was affected by the presence of salt stress, as the plants grown under this condition showed a 9.7% decrease in this variable (Fig. 1B). The ammonia and free amino acids content in the nodules decreased by 10.4% when the plants were cultivated under salt stress in relation to the plants grown under control conditions, regardless of the bacterial combination used in the inoculation (Fig. 2A and B, respectively). The ureides content was 39.8% lower in the plants that were cultivated under salt stress than in the plants grown under control conditions (Fig. 2C).
Fig. 1.
Nitrogen total nodules (*CV = 10.42%) of cowpea plants cv. “IPA 206” inoculated with Bradyrhizobium sp. (T1) or co-inoculated Bradyrhizobium sp. and Actinomadura sp. (T2), Bradyrhizobium sp. and Paenibacillus graminis (T3), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5) (A), without and with salt stress induction (0 and 50 mmol L−1 NaCl) (B). Means followed the same letter lower (bacterial combinations) and capital (cultivation conditions) not differ statistically (p < 0.05) according to Tukey's test. *Variation's coefficient.
Fig. 2.
Free ammonia (A) (*CV = 14.57%), free amino acids (B) (CV = 14.57%), ureides (C) (CV = 33.47%) and shoot dry matter (D) (CV = 18.17%) in cowpea plants nodules grown without and with saline stress (0 and 50 mmol L−1 NaCl), inoculated with Bradyrhizobium sp. (T1) or co-inoculated Bradyrhizobium sp. and Actinomadura sp. (T2), Bradyrhizobium sp. and Paenibacillus graminis (T3), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5). Means followed the same letter not differ statistically (p < 0.05) according to Tukey's test. *Variation's coefficient.
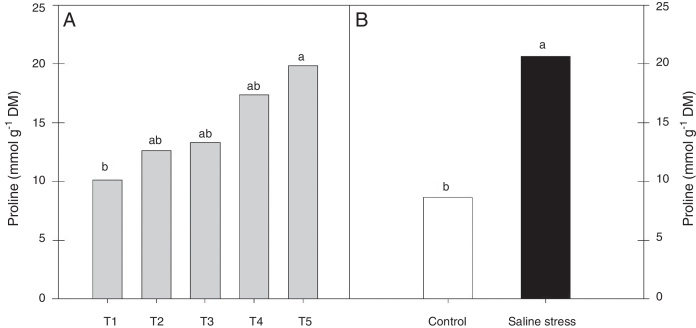
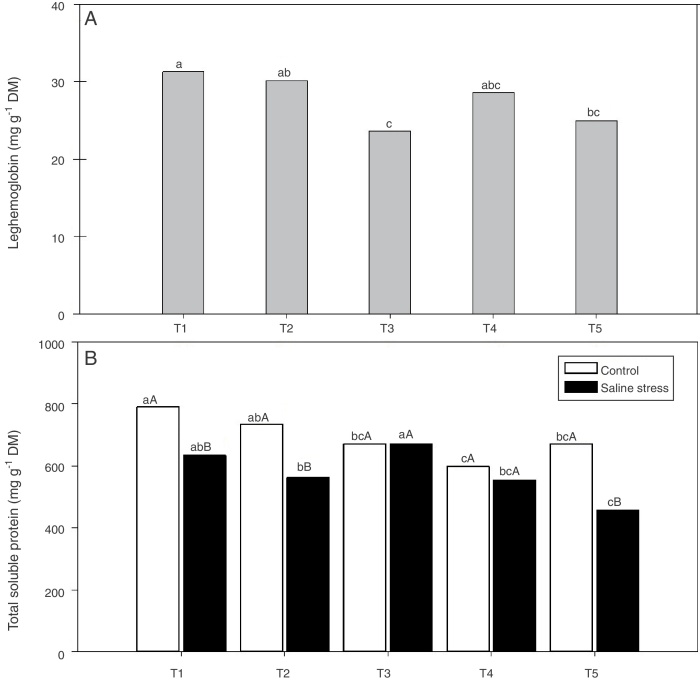
Regarding the response of the proline content in the nodules of plants, co-inoculation with Bradyrhizobium sp. and Streptomyces sp. (T5) differed from the inoculation with Bradyrhizobium sp. (T1) (Fig. 3A); the proline content in the nodules of cowpea increased when the plants were subjected to salt stress (Fig. 3B). The plants that were inoculated with Bradyrhizobium sp. and co-inoculated with Bradyrhizobium sp. and Actinomadura sp. (T2) or Bradyrhizobium sp. and Bacillus sp. (T4) showed increased leghemoglobin contents, regardless of whether the plants were grown in the presence or absence of salt stress. On the other hand, compared with inoculation with Bradyrhizobium sp. (T1), co-inoculation with Bradyrhizobium sp. and P. graminis (T3) or Bradyrhizobium sp. and Streptomyces sp. (T5) reduced the leghemoglobin content (Fig. 4A).
Fig. 3.
Proline (*CV = 39.18%) in cowpea nodules inoculated with Bradyrhizobium sp. (T1) or co-inoculated Bradyrhizobium sp. and Actinomadura sp. (T2), Bradyrhizobium sp. and Paenibacillus graminis (T3), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5) (A), without and with salt stress induction (0 and 50 mmol L−1 NaCl) (B). Means followed the same letter lower (bacterial combinations) and capital (cultivation conditions) not differ statistically (p < 0.05) according to Tukey's test. *Variation's coefficient.
Fig. 4.
Leghemoglobin (*CV = 15.51%) (A) and total soluble proteins (*CV = 39.18%) (B) in cowpea nodules inoculated Bradyrhizobium sp. (T1) Bradyrhizobium sp. and Actinomadura sp. (T2), Bradyrhizobium sp. and Paenibacillus graminis (T3), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5), independent of the cultivation condition and without and with salt stress induction (0 and 50 mmol L−1 NaCl), respectively. Means followed the same letter lower not differ statistically (p < 0.05) according to Tukey's test. *Variation's coefficient.
When the cowpea plants were grown under the control conditions, the nodules of plants inoculated with Bradyrhizobium sp. (T1) and co-inoculated with Bradyrhizobium sp. and Actinomadura sp. (T2) showed higher soluble protein content; in the presence of 50 mmol L−1 NaCl, the soluble protein content increased in the nodules of plants inoculated with Bradyrhizobium sp. (T1) and co-inoculated with Bradyrhizobium sp. and Bacillus sp. (T4) (Fig. 4B). The Bradyrhizobium sp. and P. graminis (T3) and the Bradyrhizobium sp. and Bacillus sp. (T4) co-inoculated plants as well as the control plants did not exhibit reduced soluble protein contents in the nodules when cultivated under salt stress (Fig. 4B). When comparing the cultivation conditions, the nodules of plants co-inoculated with Bradyrhizobium sp. and P. graminis (T3) and Bradyrhizobium sp. and Bacillus sp. (T4) presented higher soluble protein contents when the plants were grown under salt stress compared to those grown under control conditions. These bacterial combinations allowed the production of soluble proteins in the nodules in an equivalent manner even when subjected to salt stress. The plants that were co-inoculated with the other treatments (T2 and T5) showed decreased soluble protein contents when grown in the presence of NaCl (Fig. 4B).
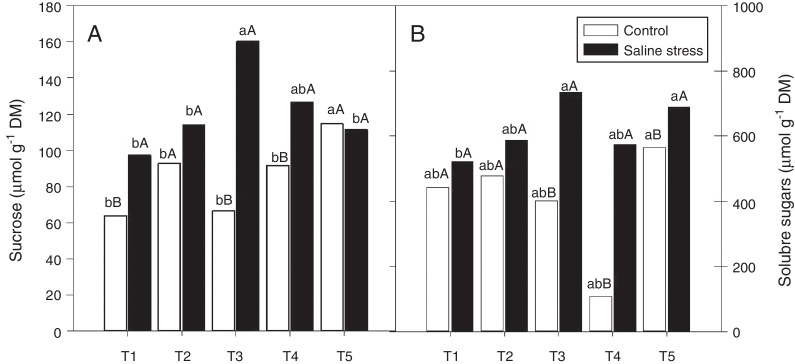
The nodules of plants co-inoculated with Bradyrhizobium sp. and Streptomyces sp. (T5) presented increased sucrose content when the plants were cultivated under control conditions; however, when 50 mmol L−1 NaCl was added to the nutrient solution, the co-inoculations that provided increased sucrose content in nodules were Bradyrhizobium sp. and P. graminis (T3) and Bradyrhizobium sp. and Bacillus sp. (T4) (Fig. 5A). When comparing the effect of salinity on the co-inoculations, it was observed that the plants inoculated with Bradyrhizobium sp. (T1) and co-inoculated with Bradyrhizobium sp. and P. graminis (T3) and Bradyrhizobium sp. and Bacillus sp. (T4) showed increases in sucrose content of 151.7%, 240.2% and 138.2%, respectively, when the plants were subjected to salt stress compared to those grown under control conditions (Fig. 5A).
Fig. 5.
Sucrose soluble (*CV = 18.52%) (A) and sugars soluble (CV = 14.62%) (B) in cowpea plants inoculated with Bradyrhizobium sp. (T1) or co-inoculated Bradyrhizobium sp and Actinomadura sp. (T2), Bradyrhizobium sp. and Paenibacillus graminis (T3), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5), without and with salt stress induction (0 and 50 mmol L−1 NaCl) (B). Means followed the same letter lower (bacterial combinations) and capital (cultivation conditions) not differ statistically (p < 0.05) according to Tukey's test. *Variation's coefficient.
There was no difference between the inoculated bacterial combinations regarding the soluble sugar content in the plants grown under control conditions. When the plants were cultivated in the presence of NaCl, the soluble sugar content increased in the plants co-inoculated with Bradyrhizobium sp. and Actinomadura sp. (T2), Bradyrhizobium sp. and P. graminis (T3), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5) (Fig. 5B). When comparing saline and control conditions, the soluble sugar content increased by 182.9%, 528.7% and 121.9% in the plants co-inoculated with Bradyrhizobium sp. and P. graminis (T3), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5), respectively, when those plants were grown under salt stress (Fig. 5B).
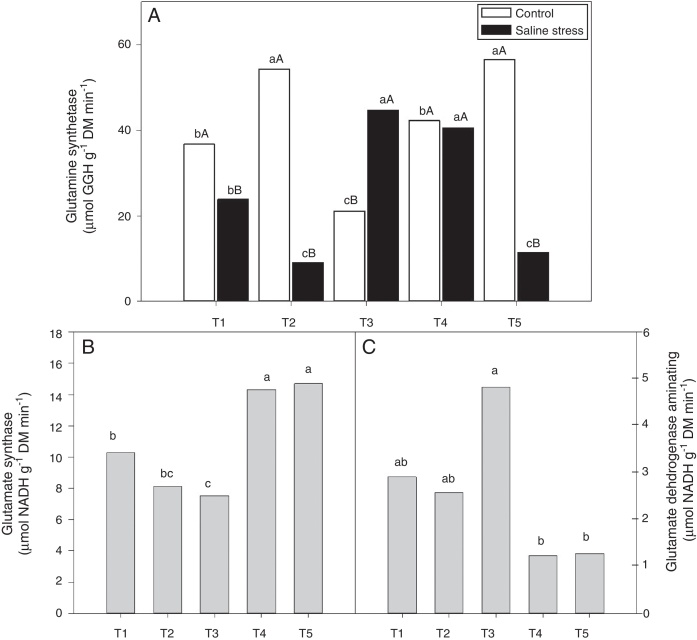
The activity of glutamine synthetase (GS) increased in plants co-inoculated with Bradyrhizobium sp. and Actinomadura sp. (T2) and Bradyrhizobium sp. and Streptomyces sp. (T5), under control conditions. Under salt stress conditions, the highest GS activity in the plant nodules was exhibited by the plants co-inoculated with Bradyrhizobium sp. and P. graminis (T3) and Bradyrhizobium sp. and Bacillus sp. (T4) (Fig. 6A). When evaluating the effect of stress for each inoculation, only the plants co-inoculated with Bradyrhizobium sp. and Bacillus sp. (T4) showed no reduction in GS activity when those plants were cultivated under salt stress compared to those grown under control conditions. On the other hand, the plants co-inoculated with Bradyrhizobium sp. and P. graminis (T3) showed higher GS activity when cultivated under salt stress than when cultivated under control conditions (Fig. 6A).
Fig. 6.
Activity of glutamine synthetase (*CV = 14.48%) (A), activity of glutamate synthase (B) (*CV = 13.59%) and glutamate dehydrogenase aminating (C) (*CV = 19.59%) on nodules of cowpea plants inoculated Bradyrhizobium sp. (T1) or co-inoculated Bradyrhizobium sp. and Actinomadura sp. (T2), Bradyrhizobium sp. and Paenibacillus graminis (T3), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5), without and with salt stress induction (0 and 50 mmol L−1 NaCl), and independent of the cultivation condition, respectively. Means followed the same letter lower (bacterial combinations) and capital (cultivation conditions) not differ statistically (p < 0.05) according to Tukey's test. *Variation's coefficient.
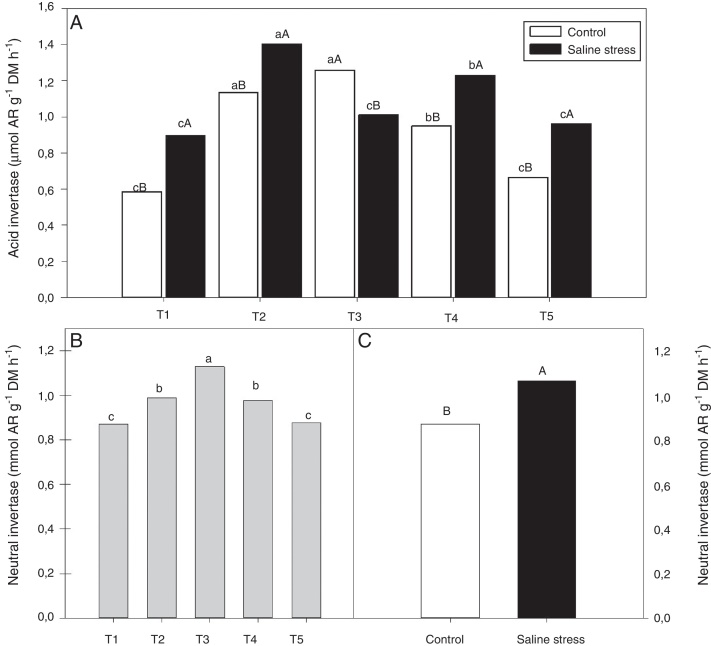
The nodules of plants co-inoculated with Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5) showed increased activity of the enzyme glutamate synthase (GOGAT), regardless of cultivation conditions (Fig. 6B). Similarly, glutamate dehydrogenase (GDH) activity increased in the nodules of plants of cowpea inoculated with Bradyrhizobium sp. (T1) and co-inoculated with Bradyrhizobium sp. and Actinomadura sp. (T2) and Bradyrhizobium sp. and P. graminis (T3), regardless of cultivation conditions (Fig. 6C). The acid invertase activity increased in the nodules of plants co-inoculated with Bradyrhizobium sp. and Actinomadura sp. (T2) and Bradyrhizobium sp. and P. graminis (T3) when those plants were grown in the absence of salt stress. On the other hand, when the plants were grown under salt stress conditions, the highest activity of acid invertase occurred in response to co-inoculation with Bradyrhizobium sp. and Actinomadura sp. (T2) (Fig. 7A).
Fig. 7.
Activity acid invertase (*CV = 7.87%) (A) and activity neutral invertase (*CV = 4.52%) (B and C) in nodules in cowpea inoculated Bradyrhizobium sp. (T1) or co-inoculated Bradyrhizobium sp. and Actinomadura sp. (T2), Bradyrhizobium sp. and Paenibacillus graminis (T3), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5), without and with salt stress induction (0 and 50 mmol L−1 NaCl). Means followed the same letter lower (bacterial combinations) and capital (cultivation conditions) not differ statistically (p < 0.05) according to Tukey's test. *Variation's coefficient.
When evaluating the different cultivation conditions, it was observed that the plants inoculated with Bradyrhizobium sp. (T1) and co-inoculated with Bradyrhizobium sp. and Actinomadura sp. (T2), Bradyrhizobium sp. and Bacillus sp. (T4) and Bradyrhizobium sp. and Streptomyces sp. (T5) exhibited increased acid invertase activity in their nodules when cultivated using nutrient solution supplemented with 50 mmol L−1 NaCl (Fig. 7A). The neutral invertase activity increased in the nodules of cowpea plants co-inoculated with Bradyrhizobium sp. and P. graminis (T3), regardless of cultivation conditions; in addition, this co-inoculation exhibited the highest sucrose content in the presence of salt stress. When comparing cultivation conditions, the plants subjected to salt stress showed higher neutral invertase activity in their nodules than did the plants grown under control conditions (Fig. 7B).
Discussion
In this study, co-inoculation of cowpea with Bradyrhizobium sp. and PGPB provided a significant increase in nitrogen content, leghemoglobin content and proline content in the nodules, regardless of cultivation conditions. Leghemoglobin is a hemoprotein and is present in the nodules of legumes; its function is to transport oxygen to maintain the aerobic metabolism of bacteroids without inhibiting nitrogenase activity.33 Co-inoculation between Rhizobium and PGPB enabled an increase in shoot nitrogen content and maintained the soluble protein content even in plants under salt stress.
Under greenhouse conditions, evaluating nitrogen and carbon compounds as well as the activities of some enzymes of carbon and nitrogen metabolism, Rodrigues et al.,12 reported that cultivated cowpea plants co-inoculated with Bradyrhizobium sp. and P. graminis presented increased nitrogen contents in their nodules and that the nitrogen content did not present a significant difference between the studied treatments. The same results did not occur in this work, as co-inoculation with Bradyrhizobium sp. and P. graminis presented the lowest nitrogen content in the nodules, reinforcing the need for studies on co-inoculation and synergism among the bacteria involved.
The plants subjected to salt stress survived the 5.6 mS cm−1 electrical conductivity of the nutrient solution; indeed, this species is moderately tolerant to salinity and is able to tolerate saltwater irrigation with maximum electrical conductivity of 3.3 dS m−1.34 These survival results may also indicate the in vivo tolerance of the Bradyrhizobium sp. (UFLA 03-84) strain to salinity, since this strain was used to maintain control plants (without inoculation) under salt stress; however, these bacteria did not survive during the experiment (data not shown). These data corroborate the result of Nóbrega et al.,35 who revealed the in vitro tolerance of Bradyrhizobium sp. (UFLA 03-84) to be 30 g L−1 NaCl, which is considered highly tolerant to salinity, and indicate that this bacterial strain is well suited for inoculation tests that aim to increase the yield of cowpea under salinity conditions.
Given that bacteroids convert atmospheric nitrogen (N2) to ammonia (NH3), which is released and incorporated into amines or ureides and then exported to the host plant,36 it can be suggested that a reduction in total nitrogen, amino acids, ammonia and ureides contents in the nodules of cowpea plants subjected to salt stress indicates a reduction in nitrogenous compounds due to the sensitivity of biological nitrogen fixation to salinity. The symbiosis between Bradyrhizobium sp. and cowpea is sensitive to salt stress.37
Excess salts in the soil can reduce the availability of water to plants, causing ionic and osmotic stress, decreasing the water potential of the substrate, and reducing water availability.38 These phenomena cause the plant to reduce its water potential to a lower level than the soil is at in order to absorb water; this reduction occurs via the increase in the content of organic solutes with low molecular weight, commonly referred to as compatible solutes, which include amino acids, proline, betaines, carbohydrates, sucrose and sugars.39 An increase in compatible solutes is used as a strategy for osmotic adjustment and the prevention of both water loss and subsequent cell dehydration.40
Using the osmotic adjustment mechanism, when subjected to salt stress, cowpea plants increased the content of proline, sucrose and soluble sugars in their nodules. An increase in proline content is expected when plant osmoregulation occurs under stress. Thus, proline accumulates in several plants subjected to abiotic stresses, such as salt stress.39, 40 Nevertheless, some authors question the effectiveness of proline as an agent that can confer tolerance to salt stress.41 Many studies have shown that proline accumulation is a characteristic of environmental stress tolerance.42
There are two possible metabolic pathways for the incorporation of ammonium ions to form amino acids: the first involves the enzymes glutamate dehydrogenase (GDH) and glutamine synthetase (GS), and the second involves glutamine synthetase (GS) and glutamate synthase (GOGAT).43 The present study demonstrated that some co-inoculations are more efficient at maintaining GS activity under salt stress, indicating that those plants have a greater capacity to incorporate ammonium under saline conditions corresponding to an EC of 5.6 mS cm−1. The results show that co-inoculations elicit different metabolic pathways for the formation of amino acids; some plants prioritize the GS/GOGAT metabolic pathway for the incorporation of ammonium, while other co-inoculations elicit the alternative pathway GDH/GS of nitrogen assimilation. As observed, the GOGAT activity in the nodules was higher than the GDH activity, indicating that the GS/GOGAT metabolic pathway is the main pathway for the assimilation of ammonia. Plants use the GS and GOGAT enzymes to incorporate nitrogen fixation products in the form of amino acids (glutamine and glutamate); it has been suggested that genotypes efficient at nitrogen synthesis incorporate ammonium into amino acids via the GS and GOGAT enzymes.1
Regarding the activity of invertases, some co-inoculations responded differently under salt stress conditions; those that were able to maintain invertase activity at a high level could also maintain levels of carbon skeletons without major changes in the cowpea nodules, thus maintaining the BNF performed by bacteroids.44 There are two isoforms of the acid invertase enzyme: one is involved in sucrose synthesis away from sink tissues, is in the apoplast and is bound to the cell wall, establishing a sucrose concentration gradient from source to sink tissues; the other isoform is related to sugar storage, osmotic regulation and responses to abiotic stresses. Neutral or alkaline invertase is considered a maintenance enzyme; it is located in the cytoplasm and is involved in the degradation of sucrose when the activities of cell wall acid invertase and sucrose synthase are low, suggesting neutral invertase involvement in the degradation and accumulation of sucrose in the vacuole.45
Co-inoculations with Bradyrhizobium sp. and Actinomadura sp. and Bradyrhizobium sp. and P. graminis were more efficient at not altering the levels of carbon skeletons under control conditions. However, when the plants were subjected to salt stress, co-inoculation with Bradyrhizobium sp. and Actinomadura sp. maintained the invertase activity at a high level, maintaining the levels of carbon skeletons without major changes in cowpea nodules, thus maintaining the BNF performed by bacteroids.44 In this study, it can be verified that the synergistic interaction between diazotrophic bacteria and PGPB can provide increased nodulation, resulting in positive effects by increasing BNF via nitrogen–carbon metabolism in cowpea plants under salt stress conditions.
In Brazil, few studies exist on inoculation, specifically on co-inoculation between rhizobia and PGPB for leguminous species, as well as the beneficial or protective effects that these inoculations can promote for plants under stress. Considering that the great natural biodiversity of Brazilian biomes is little explored, the potential of bacterial diversity for inoculants has yet to be deeply studied.6
Conclusions
Inoculation with Bradyrhizobium sp. (UFLA 03-84) and co-inoculations with Bradyrhizobium sp. and Actinomadura sp. (UFLA 03-84 and 183-EL), Bradyrhizobium sp. and Bacillus sp. (UFLA 03-84 and IPACC11), and Bradyrhizobium sp. and Streptomyces sp. (UFLA 03-84 and 212) were more efficient at nitrogen fixation, even when the plants were grown under conditions of 50 mmol L−1 NaCl. The combinations of the microorganisms Bradyrhizobium sp. and Bacillus sp. (UFLA 03-84 and IPACC11) and Bradyrhizobium sp. and Streptomyces sp. (UFLA 03-84 and 212) elicited the GS/GOGAT pathway to incorporate ammonium, even when the plants were subjected to salinity stress. Overall, co-inoculations with Bradyrhizobium sp. and Actinomadura sp. (UFLA 03-84 and 183-EL) and Bradyrhizobium sp. and P. graminis (UFLA 03-84 and MC 04.21) benefited the carbon metabolism in cowpea plants, even when cultivated under salinity stress.
Conflicts of interest
The authors declare no conflicts of interest.
Associate Editor: Ieda Mendes
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bjm.2018.01.007.
Appendix B. Supplementary data
The following are the supplementary data to this article:
References
- 1.Udvardi M., Poole P.S. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol. 2013;64:781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 2.Clarke V.C., Loughlin P.C., Day D.A., Smit P.M.C. Transport processes of the legume symbiosome membrane. Front Plant Sci. 2014:5. [Google Scholar]
- 3.Rodrigues A.C., Antunes J.E.L., Costa A.F., Oliveira J.P., Figueiredo M.V.B. Interrelationship of Bradyrhizobium sp. and plant growth-promoting bacteria in cowpea: survival and symbiotic Performance. J Microbiol. 2013;51:49–55. doi: 10.1007/s12275-013-2335-2. [DOI] [PubMed] [Google Scholar]
- 4.Figueiredo M.V.B., Bonifacio A., Rodrigues A.C., Araujo F.F. 1st ed. Vol 1. Springer; Media Singapore: 2016. Plant growth-promoting rhizobacteria: key mechanisms of action; pp. 23–37. (Microbial-mediated Induced Systemic Resistance in Plants). [Google Scholar]
- 5.Arora N.K., Verma M., Mishra J. Rhizobial bioformulations: past, present and future. In: Mehnaz S., editor. Vol 2. 2017. pp. 69–99. (Rhizotrophs: Plant Growth Promotion to Bioremediation. Microorganisms for Sustainability). [Google Scholar]
- 6.Oliveira A.L.M., Costa K.R., Ferreira D.C. Aplicações da biodiversidade bacteriana do solo para a sustentabilidade da agricultura. Biochem Biotechnol Rep. 2014;3(1):56–77. [Google Scholar]
- 7.Souza R., Ambrosini A., Passaglia L.M.P. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet Molec Biol. 2015;38(4):401–419. doi: 10.1590/S1415-475738420150053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hossain M.A., Wani S.H., Bhattacharjee S., Burritt D.J., Tran L.S.P. Drought Stress Toler Plants. 2016:1. [Google Scholar]
- 9.Ilangumaran G., Smith D.L. Plant growth promoting rhizobacteria in amelioration of salinity stress: a systems biology perspective. Front Plant Sci. 2017;8:1768. doi: 10.3389/fpls.2017.01768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coelho J.B.M., Barros M.F.C., Bezerra Neto E., Souza E.R. Ponto de murcha permanente fisiológico e potencial osmótico de feijão caupi cultivado em solos salinizados. Revista Brasileira de Engenharia Agrícola e Ambiental. 2014;18(7):708–713. [Google Scholar]
- 11.Gupta B., Huang B. Mechanism of salinity tolerance in plants: physiological. biochemical, and molecular characterization. Int J Genomics. 2014:2014. doi: 10.1155/2014/701596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues A.C., Antunes J.E.L., Medeiros V.V., De França Barros B.G., Figueiredo M.D.V.B. Resposta da coinoculação de bactérias promotoras de crescimento em plantas e Bradyrhizobium sp. em caupi. Biosci J. 2012;28(1):196–202. [Google Scholar]
- 13.Canteri M.G., Althaus R.A., Virgens Filho J.S., Giglioti E.A., Godoy C.V. SASM – Agri: Sistema para análise e separação de médias em experimentos agrícolas pelos métodos Scott-Knott, Tukey e Duncan. Revista Brasileira de Agrocomputação. 2001;1(2):18–24. [Google Scholar]
- 14.Vincent J.M. International Biological Programme; London: 1970. A Manual for the Practical Study of Root Nodule Bacteria. 164 p. [IBP Handbook, 15] [Google Scholar]
- 15.Rodrigues Neto J., Malavolta Júnior V.A., Victor O. Meio simples para o isolamento e cultivo de Xanthomonas campestris pv. citri tipo B. Summa Phytopathol. 1986;12(1–2):16. [Google Scholar]
- 16.Nonomura H., Ohara Y. Distribution of soil actinomycetes. VI. A culture method effective for both preferential isolation and enumeration of Microbispora and Streptosporangium strains in soil. J Ferment Technol. 1969;47:463–469. [Google Scholar]
- 17.Hungria M., Araujo R.S. Embrapa-Serviço de Produção e Informação; Brasília: 1994. Manual de métodos empregados em estudos de microbiologia agrícola. [Google Scholar]
- 18.Hoagland D.R., Arnon D.I. 1st ed. University of California; Berkeley: 1950. The Water Culture Method of Growing Plants Without Soil; p. 32. [Google Scholar]
- 19.Silveira J.A.G., Contado J., Rodrigues J., Oliveira J. Phosphoenolpyruvate carboxylase and glutamine synthetase activities in relation to nitrogen fixation in cowpea nodules. Revista Brasileira de Fisiologia Vegetal. 1998;10(1):19–23. [Google Scholar]
- 20.Baethgen W.E., Alley M.M. A manual colorimetric procedure for measuring ammonium nitrogen in soil and plant Kjeldahl digest. Commun Soil Sci Plant Anal. 1989;20(9–10):961–969. [Google Scholar]
- 21.Weatherburn M.W. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39(8):971–974. [Google Scholar]
- 22.Peoples M.B., Faizah A.W., Reakasem B., Herridge D.F. Australian Centre for International Agricultural Research; Canberra: 1989. Methods for Evaluating Nitrogen Fixation by Nodulated Legumes in the Field; p. 76. [Google Scholar]
- 23.Smagghe B.J., Hoy J.A., Percifield R. Review: correlations between oxygen affinity and sequence classifications of plant hemoglobin. Biopolymers. 2009;91(12):1083–1096. doi: 10.1002/bip.21256. [DOI] [PubMed] [Google Scholar]
- 24.Young E.W., Conway C.F. On the estimation of allantoin by the Rimini–Schryver reaction. J Biol Chem. 1942;142:839–853. [Google Scholar]
- 25.Bates L.S., Waldren R.P., Teare I.D. Rapid determination of free proline for water stress studies, Short Communication. Plant Soil. 1973;39:205–207. [Google Scholar]
- 26.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 27.Elliot W. Glutamine synthesis. Methods Enzymol. 1955;2:337–342. [Google Scholar]
- 28.Suzuki I., Cretin C., Omata T., Sugiyama T. Transcriptional and post-transcriptional regulation of nitrogen-responding expression of phosphoenolpyruvate carboxylase gene in maize. Plant Physiol. 1994;105:1223–1229. doi: 10.1104/pp.105.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coombs J., Hall D.O. Pergamon Press; New York: 1982. Techniques in Productivity and Photosynthesis; p. 171. [Google Scholar]
- 30.Van Handel E. Direct microdetermination of sucrose. Anal Biochem. 1968;22:280–283. doi: 10.1016/0003-2697(68)90317-5. [DOI] [PubMed] [Google Scholar]
- 31.Dubois M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 32.Zhu Y.J., Komor E., Moore P.H. Sucrose accumulation in the sugarcane stems regulated by the difference between the activities of soluble acid invertase and sucrose phosphate synthase. Plant Physiol. 1997;115:609–616. doi: 10.1104/pp.115.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navascués J., Pérez-Rontomé C., Gay M. Leghemoglobin green derivatives with nitrated hemes evidence production of highly reactive nitrogen species during aging of legume nodules. Proc Natl Acad Sci U S A. 2012;109(7):2660–2665. doi: 10.1073/pnas.1116559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silveira J.A.G., Cardoso B.B., Melo A.R.B., Viégas R.A. Salt-induced decrease in nitrate uptake and assimilation in cowpea plants. Revista Brasileira de Fisiologia Vegetal. 1999;11(2):77–82. [Google Scholar]
- 35.Nóbrega R.S.A., Motta J.S., Lacerda A.M., Moreira F.D.S. Tolerância de bactérias diazotróficas simbióticas à salinidade in vitro. Ciência e Agrotecnologia. 2004;28(4):899–905. [Google Scholar]
- 36.Figueiredo M.V.B., Martinez C.R., Burity H.A., Chanway C.P. Plant growth-promoting rhizobacteria for improving nodulation and nitrogen fixation in the common bean (Phaseolus vulgaris L.) World J Microbiol Biotechnol. 2008;24:1187–1193. [Google Scholar]
- 37.Medeiros R., Santos V., Araújo A., Oliveira Filho C. Estresse salino sobre a nodulação em feijão-caupi. Revista Caatinga. 2008;21(5):202–206. [Google Scholar]
- 38.Munns R., Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 39.Willadino L., Camara T.R. Tolerância das plantas à salinidade: aspectos fisiológicos e bioquímicos. Enciclopédia Biosfera. 2010;6(11):1–23. [Google Scholar]
- 40.Glick B.R. Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 2012:2012. doi: 10.6064/2012/963401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samavat S., Samavat S., Mafakheri S., Shakouri M.J. Promoting common bean growth and nitrogen fixation by the coinoculation of Rhizobium and Pseudomonas fluorescens isolates. Bulg J Agric Sci. 2012;18:387–395. [Google Scholar]
- 42.Rao A., Ahmad S.D., Sabir S.M. Potential biochemical indicators improve salt tolerance in fifteen cultivars of wheat (Triticuma estivum L.) from Pakistan. Int J Sci Eng Res. 2013;4:389–406. [Google Scholar]
- 43.Shan A.Y.K.V., Oliveira L.E.M., Bonome L.T.S., Mesquita A.C. Assimilação metabólica de nitrogênio em plântulas de seringueira cultivadas com nitrato ou amônio. Pesquisa Agropecuária Brasileira. 2012;47(6):754–762. [Google Scholar]
- 44.Ben Salah I., Slatni T., Gruber M. Relationship between symbiotic nitrogen fixation, sucrose synthesis and anti-oxidant activities in source leaves of two Medicago ciliaris lines cultivated under salt stress. Environ Exp Bot. 2011;70:166–173. [Google Scholar]
- 45.Yao S.G., Kodama R., Wang H., Ichii M., Taketa S., Yoshida H. Analysis of the rice SHORT-ROOT5 gene revealed functional diversification of plant neutral/alkaline invertase family. Plant Sci. 2009;176(5):627–634. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.