Abstract
Gnathostome jaws derive from the first pharyngeal arch (PA1), a complex structure constituted by Neural Crest Cells (NCCs), mesodermal, ectodermal and endodermal cells. Here, to determine the regionalized morphogenetic impact of Dlx5/6 expression, we specifically target their inactivation or overexpression to NCCs. NCC-specific Dlx5/6 inactivation (NCC∆Dlx5/6) generates severely hypomorphic lower jaws that present typical maxillary traits. Therefore, differently from Dlx5/6 null-embryos, the upper and the lower jaws of NCC∆Dlx5/6 mice present a different size. Reciprocally, forced Dlx5 expression in maxillary NCCs provokes the appearance of distinct mandibular characters in the upper jaw. We conclude that: (1) Dlx5/6 activation in NCCs invariably determines lower jaw identity; (2) the morphogenetic processes that generate functional matching jaws depend on the harmonization of Dlx5/6 expression in NCCs and in distinct ectodermal territories. The co-evolution of synergistic opposing jaws requires the coordination of distinct regulatory pathways involving the same transcription factors in distant embryonic territories.
Introduction
The gnathostome skull is characterized by the presence of articulated, muscularized, asymmetric jaws, capable to support predatory behaviours and feeding functions such as mastication and swallowing1,2. During development, upper and lower jaws derive from the maxillary and mandibular processes of the first pharyngeal arch (PA1). Hox-free Neural Crest Cells (‘NCCs’) emigrating from the mesencephalic and anterior rhombencephalic neural folds3–13 colonize the PA1 and give rise to most facial cartilages, bones and tendons. Before migration, NCCs lack the topographic information needed to unfold jaw morphogenetic processes14. In the course of migration and after reaching their final destination in the craniofacial buds, NCCs contact mesodermal, ectodermal and endodermal cells. Morphogenetic cues exchanged between these different cell populations result in the formation of functionally and morphologically different upper and lower jaws capable to operate in synergy to assure feeding15–17.
Surgical deletion and grafting of different regions of the embryonic anterior foregut endoderm and ectoderm have demonstrated that these epithelia convey to NCCs the topographic cues needed for PAs patterning and facial morphogenesis12,14,18. The molecular nature of these instructive signals is not completely elucidated, but experimental evidence suggests the involvement of bone morphogenetic proteins (BMPs), FGFs, endothelin-1 (Edn1), Notch/Delta and Sonic hedgehog19.
In the mouse, loss of Edn1 and/or endothelin receptor type-A (Ednra)20,21 result in the transformation of the lower jaw into a structure presenting major morphological hallmarks of an upper jaw such as absence of MC, zygomatic arch-like structures and vibrissae. These observations indicate that Edn1 signalling plays a major role in the specification of lower jaw identity22,23. It must be noted, however, that, after Edn1-pathway inactivation, upper jaw-like structures are much smaller than those found in the normal upper jaw20,23. A further demonstration of the importance of Edn1-signalling for the establishment of jaw patterning comes from the constitutive activation of Ednra in upper jaw NCCs where its ligand, Edn1, is not normally present22. We reported that ectopic Ednra activation induces the transformation of maxillary into mandibular structures with duplicated MCs and dermatocranial jaws constituted by four, opposing dentary-like bones. In the same study we obtained a similar transformation forcing the expression of Hand2, a downstream target of the Edn1 pathway, in the Ednra-positive domain24. It appears that Edn1 drives lower jaw development through a calmodulin-CamKII-histone deacetylase cascade that de-represses Mef2c, which then transactivates Dlx5/6 expression25,26.
Dlx homeobox genes are vertebrate homologues of Drosophila distal-less; these genes play a fundamental role in specifying dorsoventral PA1 patterning1,27. The six gnathostome Dlx genes are organized in bigenic tandems and are all expressed in NCCs of craniofacial primordia in spatially and temporally restricted patterns. Dlx1 and Dlx2 are present in both maxillary and mandibular NCCs while Dlx5 and Dlx6 are expressed only in mandibular NCCs. The targeted simultaneous inactivation of Dlx5 and Dlx628,29 in all cells of the developing embryo, including NCCs and epithelial cells, results in the coordinated transformation of upper and lower jaws generating a symmetric mouth in which each individual jaw presents a morphology similar to the others transposed along sagittal and horizontal planes.
The transformations in jaw identity induced by the inactivation of Edn1 signalling are accompanied by the down regulation of Dlx5 and Dlx6 in NCCs20,22,23,30; however, a territory of Edn-1 independent Dlx5/6 expression is consistently maintained in the proximal PA122,31. Edn1 is expressed in the epithelium and mesodermal core of the mandibular part of PA1, whereas Ednra is almost exclusively expressed by NCCs suggesting that altered Edn1 signalling affects Dlx5 and Dlx6 expression only in NCCs and not in other cell types. Given the difference between the lower jaw morphology obtained in Edn1/Ednra mutants and Dlx5/6 mutants, these findings suggest that the Edn1-dependent activation of Dlx5/6 in NCCs is necessary to specify mandibular identity, but is insufficient to generate a normal lower jaw morphology17,20,22,23,30. Even if Dlx5 and Dlx6 are not expressed by maxillary NCCs, after constitutive inactivation of these genes, maxillary components are also affected28,29. To explain this observation it has been hypothesized that two Dlx5/6-dependent signalling centres are present at the extremities of both the mandibular and maxillary processes; the so-called “Hinge and Caps” model of jaw organization2,32,33. This model predicts the presence of two opposing morphogen gradients, one emanating from the region of the upper/lower jaw articulation (hinge) and one from the distal extremities of PA1 (caps). By lineage analysis, we have shown that the maxillary arch epithelium harbours a cellular contingent derived from frontonasal Dlx5-expressing progenitors suggesting that transient Dlx5/6 expression could program these epithelial cells to provide the cues needed for maxillary arch morphogenesis34.
Here, we address the morphogenetic role of Dlx5/6 expression in NCCs. To this end we first invalidate Dlx5 and Dlx6 specifically in NCCs and then we force ectopic expression of Dlx5 in maxillary NCCs that do not normally express this gene. Our findings confirm that Dlx5/6 expression in NCCs is necessary to specify mandibular identity and has an organizer role for craniofacial myogenesis. However, NCC-limited Dlx5/6 expression is insufficient to determine normal jaw morphogenesis; coordination of Dlx5/6 expression in NCCs and in other cellular components is essential for the development and evolution of a functional mouth.
Results
Generation of mouse models with deregulation of Dlx5/6 expression in NCCs
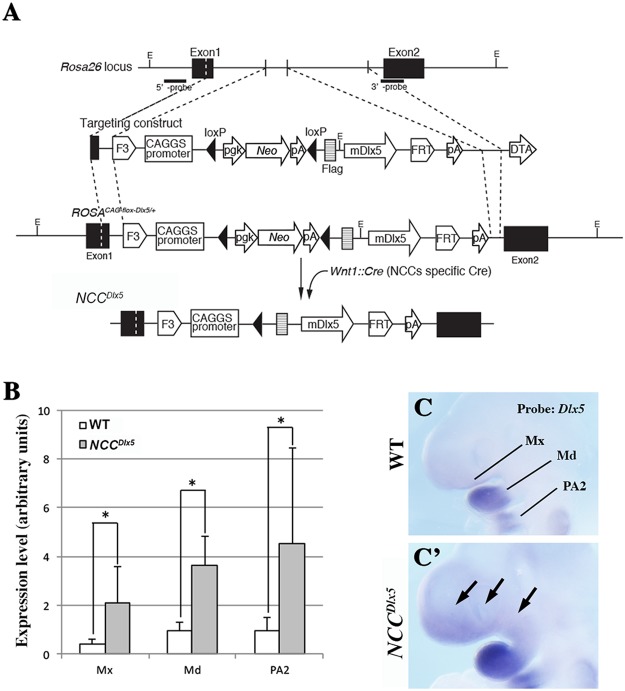
To induce ectopic expression of Dlx5 in the NCC lineage, we crossed ROSACAG-flox-Dlx5/+ mice, which express Dlx5 in a Cre-dependent manner, with Wnt1Cre mice35 to obtain the NCCDlx5 mouse line in which Dlx5 should be expressed in premigratory neural crest cells (Fig. 1A). The expression of Dlx5 in the pharyngeal arches was analyzed by quantitative RT-PCR (Fig. 1B) and whole-mount in situ hybridization (Fig. 1C,C’). RT-PCR was performed on dissected maxillary, mandibular and second pharyngeal (PA2) arches from E10.5 control and NCCDlx5 embryos. In all NCCDlx5 samples, Dlx5 expression was higher than that found in the corresponding controls (Fig. 1B). By in situ hybridization, ectopic Dlx5 expression was detected in the presumptive maxillary and nasal regions, well beyond the normal territory of expression. Dlx5 expression was also strongly increased in NCCs colonizing the mandibular process and PA2 while no significant difference could be observed in ectodermal PA1 expression at this stage. (Fig. 1C,C’). This pattern is consistent with the distribution of lacZ expression in Wnt-1::Cre/R26R mice36 suggesting that indeed Dlx5 expression has been activated and maintained ectopically in NCCs.
Figure 1.
Generation of NCCDlx5 mice. (A) Strategy for conditional expression of Dlx5 from the Rosa26 locus and generation of NCCDlx5 mice. Probes for genotyping are indicated as 5′- and 3′- probes. E, EcoRI. (B) Comparison of Dlx5 mRNA levels in the maxillary arch (Mx), mandibular arch (Md) and PA2 of control and NCCDlx5 E10.5 embryo. Messenger RNA levels were estimated by qRT-PCR. The values showed on the graph were mean ± SD of 5 duplicated samples. Statistics: Mann-Whitney U test using R-software (version 3.1.3). *p < 0.01. (C,C’) Whole mount in situ hybridization for Dlx5 on control and NCCDlx5 E9.5 mice. Black arrows: areas of ectopic Dlx5 expression.
To inactivate Dlx5/6 in NCCs, Dlx5/6flox/flox mice, in which the homeodomain-encoding regions of both Dlx5 and Dlx6 are flanked by lox sequences37, were crossed with Wnt1Cre-ERT2/+ mice in which tamoxifen exposure induces Cre-recombinase activity in cells of the developing neural tube, in migrating NCCs and in dopaminergic neurons, but not in other cell types. To cover most of the period of neural crest delamination and migration38 Dlx5/6flox/flox::Wnt1Cre-ERT2/+ timed pregnant dams received two intraperitoneal injections of tamoxifen at embryonic developmental days E6 and E7, generating NCC∆Dlx5/6 embryos. The need to inactivate both Dlx5 and Dlx6 in NCCs derives from the fact that these two closely related genes are redundant in defining lower jaw identity: both need to be inactivated to transform the identity of the lower jaw into an upper jaw28,29.
Craniofacial defects observed after deregulation of Dlx5/6 expression in NCCs
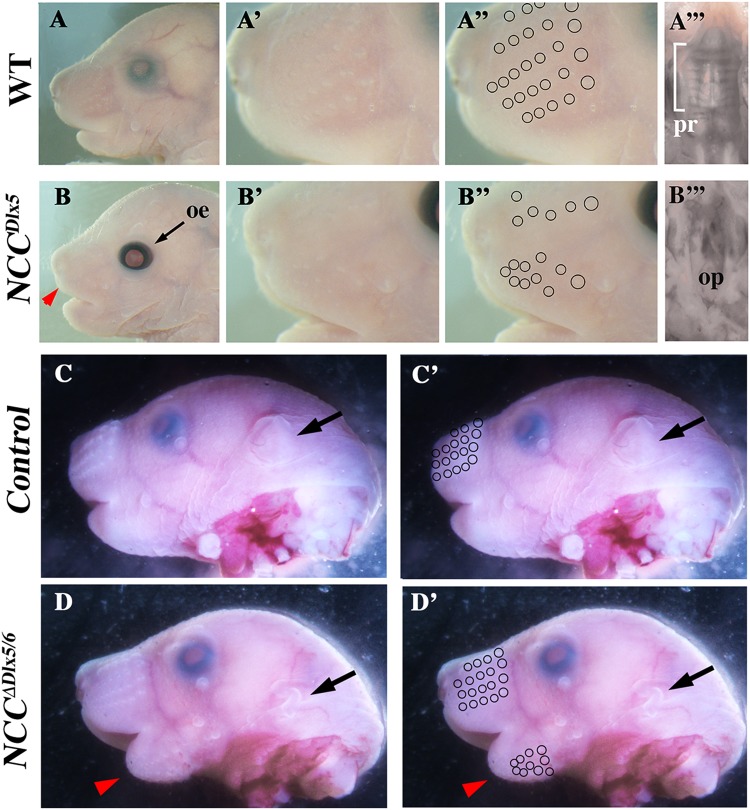
At E18.5, NCCDlx5 foetuses presented a fully penetrant phenotype characterized by a short snout, open eyelids, misaligned vibrissae (Fig. 2A-B”) correctly located in the distal territory of the snout, and a cleft palate with no obvious signs of palatine rugae (Fig. 2A”’,B”’ and Fig. 3D”). On the contrary, E18.5 NCC∆Dlx5/6 foetuses (Fig. 2C-D’) presented a marked mandibular retrognathism, with vibrissae present on both upper and lower jaws. In the upper jaw of NCC∆Dlx5/6 foetuses, vibrissae were arranged in 5 rows as in control littermates, while ectopic vibrissae in the lower jaw were very close to each other and did not present a recognizable pattern; in these animals some vibrissae developed close to the midline. The external ear of E18.5 NCC∆Dlx5/6 foetuses was hypomorphic (black arrow in Fig. 2C-D’) while the palate and the eyelids did not present any obvious malformation.
Figure 2.
External appearances of NCCDlx5 and NCC∆Dlx5/6 mouse heads at E18.5. Craniofacial appearance of control (A-A”’), NCCDlx5 (B-B”’) and NCC∆Dlx5/6 (C-D’) mice. NCCDlx5 mice have open eyelids (oe), a shortened snout (red arrowhead in B), misaligned vibrissae (open circles in A”, B”) and an open palate (op in B”’) with no sign of palatine rugae (pr in A”’). NCC∆Dlx5/6 mouse heads present a severe mandibular retrognathism (red arrows in D, D’) with vibrissae well-aligned in the upper jaw, but also appearing ectopically in the lower jaw (open circles in C’, D’). The external ear (black arrow in C-D’) was severely hypomorphic in NCC∆Dlx5/6 mice.
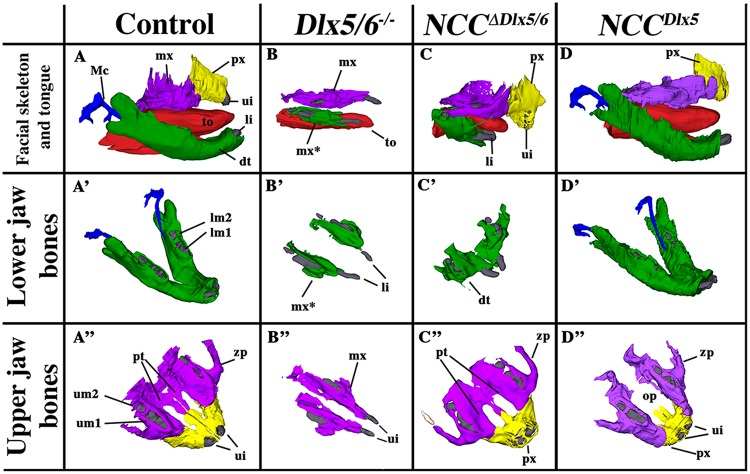
Figure 3.
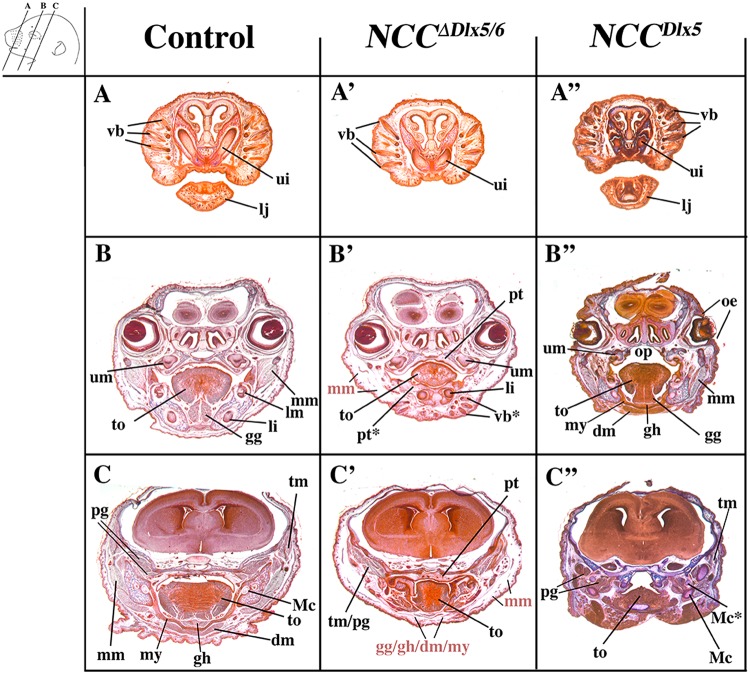
Selected views of bone and cartilagineous elements from 3D reconstructions of control, Dlx5/6−/−, NCC∆Dlx5/6 and NCCDlx5 E18.5 mouse foetuses. (A-A”) Skeletal elements from normal mouse embryonic heads. (B-B”) After constitutive Dlx5/6 inactivation, both the upper and the lower jaws are transformed generating similar bony elements which give rise to a highly symmetric mouth in which each jaw (upper/lower, right/left) presents a morphology similar to the others transposed along sagittal or dorso/ventral reflection planes. Incisors (li, ui) are short and not in direct contact with the transformed dentary (mx*) or maxillary (mx) bones. The premaxillary bone is absent. (C-C”) NCC∆Dlx5/6 heads present a severely shortened dentary bone (green in C,C’) which fuses in the midline and displays well developed lateral processes reminiscent of maxillary bones. Maxillary and premaxillary (px) bones are relatively well-developed with a closed palate (pt) and normal zygomatic processes (zp). (D-D”) NCCDlx5 mice present an almost normal dentary bone with a reduced coronoid process (see Fig. 6) and well developed Meckelian cartilage (D’). In NCCDlx5 mice maxillary bones are abnormal, they present an open palate (op), and protrude anteriorly, the zygomatic process of the maxilla (zp) is malformed, the premaxillary bone (px) is reduced. Abbreviations: dt dentary bone; li, lower incisor; lm1, lm2, lower molars 1 and 2; Mc, Mekelian cartilage; mx, maxillary bones; mx*, transformed dentary bone; op, open palate; pt, palate; px, premaxillary bone; ui, upper incisor; to, tongue; zp, zygomatic process.
The craniofacial phenotypes resulting from either up- or down-regulation of Dlx5/6 in NCCs were analysed by 3D reconstruction after serial sectioning and imaging of the heads at E18.5 and compared to normal littermates or to heads of foetuses of the same age in which Dlx5 and Dlx6 had been constitutively inactivated (Dlx5/6−/− foetuses)28,34 (Figs 3 and 4 and Supplementary Figs 1–4 for 3D pdf files). As already described28,29,34, after constitutive Dlx5/6 inactivation, both lower and upper jaws are transformed and become similar to each other resulting in a mirror symmetry of the mouth respect to both the sagittal and the dorso/ventral planes. In Dlx5/6−/− mice, the lower jaw acquires morphological and molecular hallmarks of an upper jaw28,29,39 while the premaxillary bone is not recognizable (Fig. 3B-B”). Inactivation of Dlx5/6 only in NCCs (NCC∆Dlx5/6 embryos) resulted in severely reduced and virtually unrecognizable dentary bone, while maxillary bones, palate and premaxillary bone presented a relatively normal morphology (Fig. 3C’,C”). At variance with what observed in control and Dlx5/6−/− embryos, the transformed dentary bones of NCC∆Dlx5/6 mice were fused along the midline and presented medio-lateral processes with a general structure reminiscent of maxillary bones and palate. The NCC∆Dlx5/6 transformed dentary supported two lower incisors which, in Dlx5/6−/− embryos, are not enclosed in the transformed bone. Although morphologically distinct form the upper jaws, the lower jaws of NCC∆Dlx5/6 embryos presented further hallmarks of maxillary identity with well developed vibrissae (Figs 2C,C’ and 5B,B’) and absence of a recognizable Meckelian cartilage (Fig. 4C,C’ and 5C,C’). Remarkably the infraorbital foramen, a major trait of the mammalian maxilla was present in both the upper and lower jaws of Dlx5/6−/− and NCC∆Dlx5/6 embryos (Supplementary Figs 1–3) reinforcing the notion of a switch in identity.
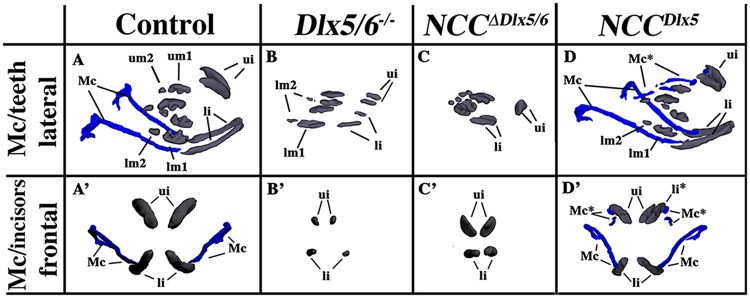
Figure 4.
Teeth complement and Meckelian cartilage from 3D reconstructions of control, Dlx5/6−/−, NCC∆Dlx5/6 and NCCDlx5 E18.5 mouse foetuses. Lateral (A–D) and frontal (A’–D’) views of teeth (grey) and Mekelian cartilage (blue) from control (A,A’), Dlx5/6−/− (B,B’), NCC∆Dlx5/6 (C,C’) and NCCDlx5 E18.5 mouse foetuses. The Meckelian cartilage (Mc) is absent in Dlx5/6−/− and NCC∆Dlx5/6 foetuses and is well formed in NCCDlx5 foetuses where supernumerary Meckel-like cartilage bars are present in the upper jaw (Mc*). In Dlx5−/− foetuses the incisors (ui, li) are short and straight and are not supported by bony elements. In NCC∆Dlx5/6 foetuses the incisors are also short, but converge towards the midline while in NCCDlx5 foetuses lower incisor are apparently normal while upper incisors are longer than normal and often duplicated with the second incisor (li*) supported by the transformed maxillary bone suggesting, therefore, that it might represent a transformed lower incisor. Abbreviations: li, lower incisor; lm1, lm2, lower molars 1 and 2; li*, transformed upper incisor; Mc, Meckelian cartilage; Mc*, duplicated Meckelian cartilage; ui, upper incisor.
Figure 5.
Representative cranial frontal sections of E18.5 control, NCC∆Dlx5/6 and NCCDlx5 mouse foetuses. The planes of section are indicated in the upper left insert. Compared to control foetuses (A–C), NCC∆Dlx5/6 foetuses (A’–B’) present ectopic vibrissae (vb*) in the lower jaw (B’), reduced and disorganized tongue musculature (to), absence of masseter, digastric, mylohyoid, geniohyoid and genioglossus muscles while the temporal and pterygoid muscles (tm, pg) form but show defective attachments on the transformed lower jaws. The midline fusion of the transformed dentary bone of NCC∆Dlx5/6 foetuses gives rise to a palate-like structure in the lower jaw (pt*) with folds reminiscent of palatine rugae. NCCDlx5 foetuses (A”–C”) present open eyelids (oe), an open palate (op), ectopic Meckelian-like cartilages, relatively normal tongue and associated musculature and mispatterned masticatory muscles adapted to the transformed skeletal elements. Abbreviations: dm, digastric muscle; gg, genioglossus muscle; gh, geniohyoid muscle; li, lower incisor; lm, lower molar; lj, lower jaw; Mc, Mekelian cartilage; Mc*, duplicated Mekelian cartilage; mm, masseter muscle; mx maxillary bones; mx*, transformed dentary bone; my, mylohyoid muscle; oe, open eyelids; op, open palate; pg, lateral and medial pterygoid muscles; pt palate; pt*, ectopic palate-like structure resulting from midline fusion of lower jaws; px, premaxillary bone; ui, upper incisor; um, upper molar; tm, temporal muscle; to, tongue; vb, vibrissae; vb*, ectopic vibrissae. Abbreviations in light red represent muscles that did not differentiate and are replaced by loose mesenchymal tissue.
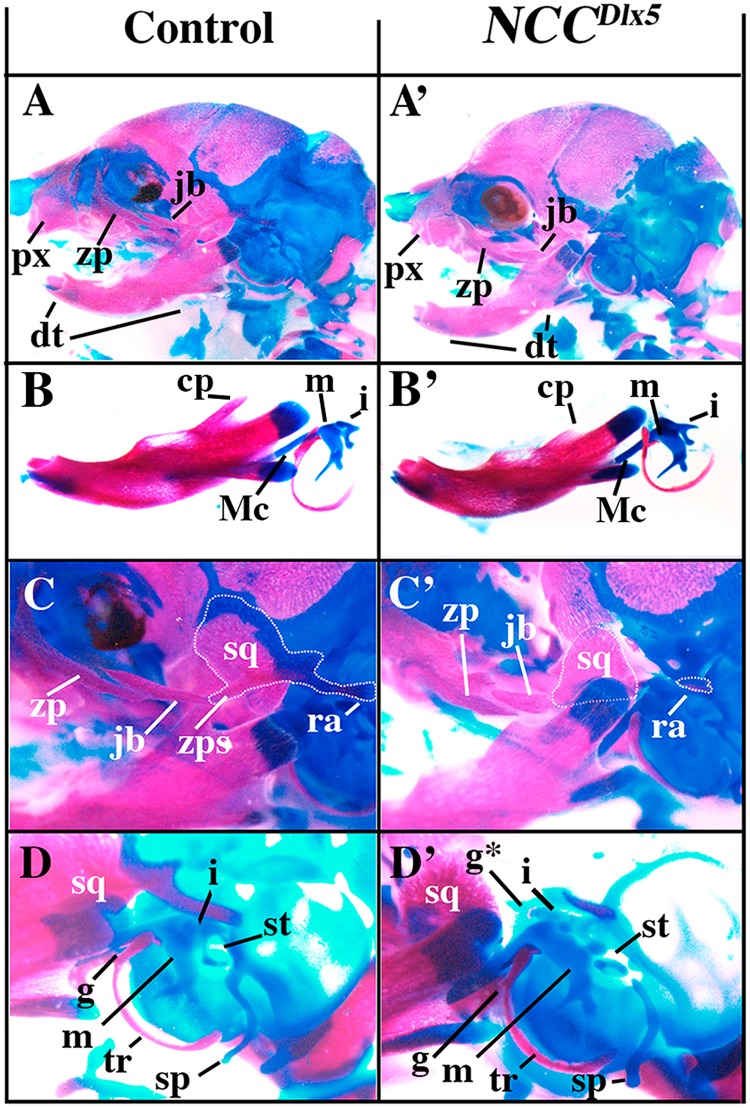
NCCDlx5 mice presented a relatively normal lower jaw, with a well-developed Meckelian cartilage and dentary bone (Figs 3D,D’, 4D,D’ and 6A-B’). Maxillary bones were hypomorphic with an open palate and smaller zygomatic and premaxillary bones (Figs 3D”, 5B”,C” and 6). We also observed the presence of fragmented cartilaginous rods converging towards the midline of the upper jaw (Figs 4D,D’ and 5C”) suggesting the presence of an ectopic Meckelian cartilage. Skeletal preparations of E18.5 NCCDlx5 foetuses (Figs 6 and 7) confirmed the malformation of upper jaw bones and cartilage components and the relatively normal appearance of lower jaw elements except for shortening of the coronoid process of the dentary bone (Fig. 6B,B’). The premaxillary bone was reduced (Figs 3A”,D” and 6A,A’), the zygomatic process of the maxilla was thicker and shorter (Fig. 6C,C’), the jugal bone was shorter (Fig. 6A,A’,C,C’) and the zygomatic and retroarticular processes of the squamosal were missing (Fig. 6C,C’). In NCCDlx5 embryos the incus seemed to have partially acquired a malleus-like identity as: (1) it short process was elongated, (2) a small ectopic bone, which could be interpreted as a duplicated gonial bone, appeared adjacent to the incus, and (3) the malleus-incus joint, which is normally of ball-and-socket type, was symmetric (Fig. 6D,D’; Supplementary. Fig. 5). The infraorbital foramen was not present in NCCDlx5 mice also suggesting an upper jaw transformation (Supplementary. Fig. 4).
Figure 6.
Craniofacial skeletal preparations from control and NCCDlx5 E18.5 foetuses. (A,A’) Lateral views of the head of control (A) and NCCDlx5 (A’) E18.5 foetuses. In NCCDlx5 mice the maxilla is deformed with thickening of the zygomatic process while the jugal and the premaxillary bones are shortened. (B,B’) Dentary bones of control (B) and NCCDlx5 (B’) E18.5 foetuses. The coronoid process (cp) is hypoplastic in NCCDlx5 mice while the Meckelian cartilage (Mc) and the malleus (m) are well-formed. (C,C’) Jaw joint region of control (C) and NCCDlx5 (C’) E18.5 mouse foetuses. The zygomatic and retroarticular processes (ra) of the squamosal (sq) are missing and the coronoid process of the dentary bone is hypoplastic in NCCDlx5 mice. (D,D’) Middle ear components of control (D) and NCCDlx5 (D’) E18.5 mouse foetuses. The incus (i) is deformed and dislocated from the stapes (st) fused to the styloid process (sp) in NCCDlx5 mice. Abbreviations: cp, coronoid process; dt, dentary bone; i, incus; jb, jugal bone; m, malleus; Mc, Meckel’s cartilage; pmx, premaxilla; ra, retroarticular process of squamosal; sp, styloid process; sq, squamosal; st, stapes; tr, ectotympanic ring; zp, zygomatic process of maxilla; zps, zygomatic process of squamosal.
Figure 7.
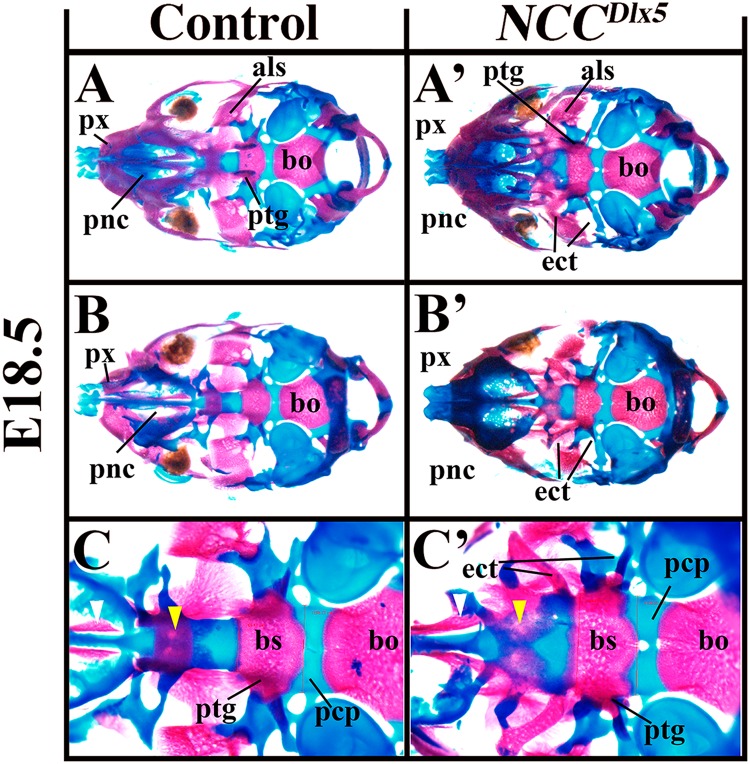
Cranial base deformity in NCCDlx5 mice. Ventral (A,A’,C,C’) and dorsal (B, B’) views of the cranial base of E18.5 control (A–C) and NCCDlx5 (A’,B’,C’) E18.5 mouse foetuses. In NCCDlx5 mice, palatal processes of the maxilla (white arrowhead in C,C’) and palatine (yellow arrowhead in C,C’) are defective. The paranasal cartilage (pnc), basisphenoid (bs) and the prechordal plate (pcp) are enlarged in width. Ectopic cartilaginous and osseous struts extended from the basisphenoid laterally and anteriorly, respectively (ect). Abbreviations: als, alisphenoidal bone; bo, basioccipital bone; bs, basisphenoidal bone; ect, ectopic structure; pcp, parachordal plate; px, premaxilla; pnc, paranasal cartilage; ptg, pterygoid.
Incisor phenotypes
Lower incisor germs of normal E18.5 foetuses are elongated structures that develop within the dentary bone and converge medially towards the distal tip (Fig. 4A,A’ and Supplementary Fig. 1). Normal lower incisors form an occlusive pattern with upper incisor germs, which are supported by the premaxillary bone. In normal animals, upper incisors are much shorter and thicker than their lower jaw counterpart and also converge towards the midline (Fig. 4A,A’ and Supplementary Fig. 1). Remarkably, in E18.5 Dlx5/6−/− foetuses, the four incisor germs were not supported by any bone structure (dentary or premaxillary) and appeared as short rods of comparable length oriented along the proximo-distal axis not converging toward the midline (Figs 3B-B” and 4B,B’ and Supplementary Fig. 2). The four incisors germs of Dlx5/6−/− foetuses were, therefore, very similar to each other. The lack of supporting bones for Dlx5/6−/− incisors suggests that bones and incisors teeth can develop independently. In NCC∆Dlx5/6 E18.5 foetuses (Figs 3C’, 4C,C’ and Supplementary Fig. 3), lower incisors were constituted by short, straight rods oriented towards the midline and protruding for about two thirds of their length from the transformed dentary bone. Upper incisors germs of NCC∆Dlx5/6 mutants were short and pointed structures entirely supported by premaxillary bones; they were oriented ventrally along an oblique axis towards the midline.
Incisor morphology of NCCDlx5 E18.5 foetuses was particularly interesting (Fig. 4D,D’ and Supplementary Fig. 4). Lower incisors were apparently normal in shape and position; upper incisors were often duplicated: a relatively normal short upper incisor was supported by the premaxillary bone and was juxtaposed to a second, longer, upper incisor that was partially supported by the transformed maxillary bone (to visualize the site of insertion see Supplementary Fig. 4). When a single upper incisor was found, it was larger on the medio/lateral axis suggesting that it might represent the fusion of two parallel incisors. This finding could further support the view that over expression of Dlx5 in maxillary NCCs results in the transformation of maxillary into mandibular structures carrying an incisor.
Ectopic Dlx5 expression in NCCs affects the development of cranial base structures
NCCDlx5 mice presented changes in the cranial base, which is partially derived from NCCs40,41; on the contrary, NCC∆Dlx5/6 foetuses did not present obvious differences of these structures. In NCCDlx5 mice, midline structures of the cranial base including the paranasal cartilage, the presphenoid and the basisphenoid were larger than in control mice (Fig. 7A-C’). From the basisphenoid, ectopic cartilaginous and osseous struts extended laterally and anteriorly; the later ectopic process fused to the hypochiasmatic cartilage of mesodermal origin, disturbing the formation of the optic foramen (Fig. 7C,C’).
Dlx5/6 expression in neural crest cells is required for proper jaw muscularization
We have previously shown that Dlx5/6−/− mice display agenesis of masticatory muscles due to the lack of instructive cues from NCCs onto cephalic myogenic mesoderm precursors31. In E18.5 NCC∆Dlx5/6 foetuses, craniofacial muscularization was affected with a phenotype strongly resembling that observed in Dlx5/6−/− mice31. The masticatory masseter muscles failed to differentiate normally and were replaced by loose mesenchymal tissue (Fig. 5B’). The other masticatory muscles, temporal and pterygoids, were present dorsally but showed defective attachments on the transformed lower jaws (Fig. 5C’). The tongue and associated musculature were severely affected: the suprahyoid muscles (mylohyoid, digastric and geniohyoid) and the extrinsic genioglossus tongue muscles did not differentiate and were replaced by loose tissue containing few disorganized myofibers; the intrinsic tongue musculature was reduced, disorganized and remained as a vestigial medially-located structure disconnected from lower jaw bones (Fig. 5B’,C’).
In NCCDlx5 foetuses all craniofacial muscles could be identified, but their attachment points and their shape were changed to adapt to the transformed skeletal elements (Fig. 5B”,C”). These observations reinforce the notion that cephalic myogenic precursors require Dlx5/6-dependent morphogenetic instructions from NCCs for proper jaw muscle patterning and differentiation42.
Dlx5 overexpression in NCCs switches maxillary to mandibular transcriptomic signature
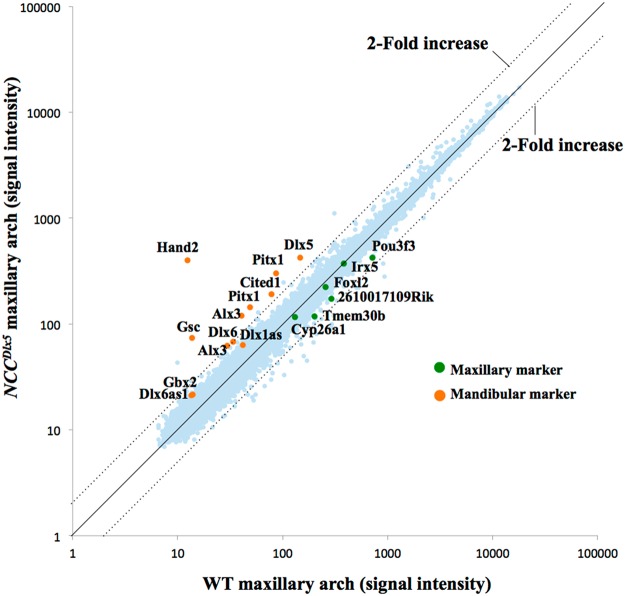
To identify regulatory pathways involved in determining jaw identity, we performed transcriptome analysis on PA tissues of E10.5 control and NCCDlx5 embryos. When the NCCDlx5 maxillary arch samples were compared to those from control maxillary arches, 12 and 21 genes were identified as increased or decreased by more than 2-fold, respectively (Supplementary Table 1). Scatter plot of the signal intensity showed that the upregulated genes corresponded to mandibular marker previously reported to be downstream of Dlx5/6 (Fig. 8)39. By contrast, maxillary marker genes reported to be upregulated in the transformed mandibular arch of Dlx5/6−/− embryos showed only small deviation from the diagonal (Fig. 8). To confirm this difference and further characterize the Dlx5-downstream genes, we then performed microarray analysis on PA tissues of E10.5 Dlx5/6−/− embryos and compared the results with genes deregulated in the NCCDlx5 maxillary arch. In the Dlx5/6−/− mandibular arch we identified 18 genes whose expression was increased and 45 genes whose expression was decreased by more than 2-folds (Supplementary Table 2). Remarkably, 10 of 12 genes up-regulated in the NCCDlx5 maxillary arch were also down-regulated in the Dlx5/6−/− mandibular arch suggesting a common set of downstream targets involved in lower jaw specification (Supplementary Table 2). By contrast, only 1 gene, Has2, a putative hyaluronan synthase, was shared between the up-regulated genes in Dlx5/6−/− mandibular arch and the down-regulated genes in NCCDlx5 maxillary arch (Supplementary Table 2).
Figure 8.
Scatter plot of signal intensities representing differential gene expression in the E10.5 control and NCCDlx5 maxillary arches. Points corresponding to known maxillary and mandibular arch markers are colored green and orange, respectively.
Discussion
In modern tetrapods, upper and lower jaws are, in general, distinct anatomical structures. Since many decades, however, it has been remarked that in basal reptiles and amphibians the upper and the lower jaws are essentially mirror images of each other43. The lower jaw of contemporary species is constituted by two mobile dentary bones that form around an early rod-like Meckelian cartilage and converge towards the centre where they fuse at the mandibular symphysis. The stationary upper jaw is mostly constituted by dermal bones that fuse along the midline and extend medio-laterally to form the palate and, together with premaxillary bones, the frontal part of the face.
During embryonic development both upper and lower jaws derive from the first pharyngeal arch (PA1) that is colonized by Hox-negative NCCs9,11,13 which give rise to most cartilages, bones and tendons of the jaws, while the associated musculature originates from the myogenic mesoderm. Endothelin-1 (Edn1) is a key signal at the origin of asymmetric jaw development. Binding of Edn1 to its receptor A (Ednra) in NCCs activates the expression of lower-jaw specific genes including Dlx5/6 and Hand217. Remarkably targeted inactivation of either Edn1, Ednra, Dlx5/6 or Hand2 result in the transformation of the lower jaw into new structures that present key morphological hallmarks of an upper jaw such as, for example, the presence of vibrissae and the absence of Meckelian cartilage20,23,44. It must be pointed out, however, that each of these mutations generates different lower jaw morphologies. Targeted constitutive inactivation of Dlx5/6 results in the simultaneous transformation of the upper and of the lower jaws giving rise to a symmetric mouth in which each of the four jaws (upper/lower, right/left) presents a morphology very similar to each of the others transposed along reflection planes28,29. We have shown that the Dlx5/6-dependent patterning of the upper jaw does not derive from their expression in mandibular NCCs but might depend on their transitory expression in an ectodermal, distal, signalling centre located in the lambdoid-junction area34.
Constitutive disruption of either (1) Edn1, (2) its receptor Ednra in NCCs or (3) the Edn1 downstream target Hand2 results in jaw phenotypes very different from those observed in Dlx5/6−/− embryos: the upper jaw does not present any obvious malformation while the lower jaw becomes small and hypomorphic and acquires, at the same time, upper-jaw characters such as the presence of vibrissae20,22,23,44,45. In Edn1, Ednra or Hand2 mutants the lower jaw is transformed, but the dorso-ventral symmetry of the mouth is not acquired. Conversely, activation of the Edn-signalling pathway in the PA1 maxillary NCCs contingent, where it is normally silent, results in the activation of Dlx5/6 and Hand2 leading to the transformation of upper jaws into dentary-like structures articulated with unaffected lower jaws presenting their own Meckelian cartilages17. Importantly, it has been shown that Dlx5/6-positive NCCs do not only contribute to the morphogenesis of bones and cartilages, but exert also an instructive role for the patterning and differentiation of the myogenic mesoderm; the constitutive inactivation of Dlx5/6 results in absence of masticatory muscles while in Ednra mutants, the Edn1-independent reactivation of Dlx5/6 in PA1 is sufficient to rescue muscular differentiation31. The sum of these results suggests that Dlx5/6 expression in different jaw precursors including, ad minima, NCCs and distal ectodermal cells, has a critical role in sculpturing vertebrate facial structures. Here, by genetically down- or up-modulating the expression of Dlx5/6 specifically in NCCs, we addressed the question of which morphological messages are conveyed by NCC-restricted Dlx5/6 expression and which other cues depend from their expression in other cell types.
We show that, if Dlx5/6 are expressed in either mandibular or maxillary NCCs the morphological and molecular signature of a lower jaw can be recognized while, in their absence, the jaws assume invariably a maxillary identity. In particular the main morphological elements which we have considered as typical traits of mandibular identity are: (1) the presence of a Meckelian cartilaginous rod, (2) the absence of midline fusion between the right and left jaws that are joined only at the distal symphysis, (3) the absence of an infraorbital foramen, (4) the presence of an elongated incisor, (5) the absence of a premaxillary bone, (6) the absence of vibrissae, (7) the presence of tongue and associated musculature and (8) the insertion of masticatory muscles. Maxillary identity on the contrary has been defined by the following characters: (1) the absence of a Meckelian cartilage, (2) the fusion of dermocranial bones along the midline to form a palate which might present palatine rugae, (3) the presence of an infraorbital foramen, (4) lateral extensions of the bones forming zygomatic-like structures, (5) the presence of a premaxillary bone, (6) the presence of vibrissae aligned in five regular lines, (7) short incisors supported by the premaxillary bone and not by other maxillary components, (8) the connection of the superficial masseter to the infraorbital foramen.
Remarkably, all mandibular traits are invariably present when Dlx5/6 are expressed in NCCs either in the upper or in the lower jaw. Conversely, maxillary traits appear in the absence of Dlx5/6 expression in NCCs associated to a defective muscular system.
Our first conclusion is that NCC-limited expression of Dlx5/6 is necessary and sufficient to specify a mandibular identity and to maintain the myogenic program for the formation of muscularized jaws.
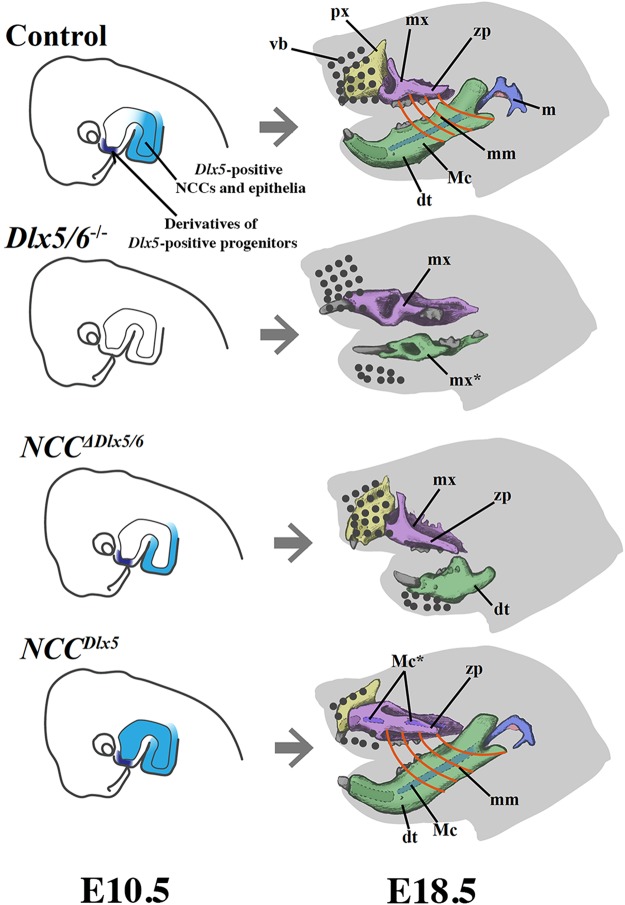
The second, striking, result of this study is that Dlx5/6 must be expressed in cellular contingents different from NCCs to permit the development of matching, functional jaws. Indeed, the inactivation of Dlx5/6 only in NCCs transforms the lower jaw into an hypomorphic upper jaw that is severely out of register from the existing maxilla. On the contrary, inactivation of Dlx5/6 in all cell types, including NCCs results in the transformation of both upper and lower jaws into similar structures that, if muscularized, could prefigure a functional mouth. It must be pointed out that transient expression of the transcription factors Dlx5/6 might be sufficient to change the developmental trajectory a cellular contingent and might confer the instructive capacity needed to modulate morphogenesis. In line with this concept, we have previously shown that the transient expression of Dlx5/6 in epithelial precursors deriving from the lambdoid junction (the intersection between presumptive maxillary and frontonasal bud-derived structures) might well be needed for correct morphogenesis of the upper jaw34. The main findings of this paper are summarized in Fig. 9.
Figure 9.
Summary diagram of the molecular (E10.5) and morphological (E18.5) alterations obtained after inactivation or induction of Dlx5/6 expression in NCCs. Abbreviations: dt, dentary bone; m, malleus; Mc, Meckel’s cartilage; mm, masseter muscle; mx, maxillary bones; px, premaxilla; vb, vibrissae; zp, zygomatic process. *Transformed ectopic elements.
It has been shown that FGF8 signals deriving from the mandibular PA1 epithelium are essential for jaw morphogenesis. Indeed, inactivation of mouse Fgf8 in PA1 epithelia results in the loss of most PA1 skeletal derivatives46. Interestingly, mandibular, but not maxillary ectomesenchyme is competent to respond to Fgf8 maintaining Dlx5 expression47,48. A possible interpretation of our data would be that Dlx5 expression in the lower jaw epithelium could be essential to maintain molecular signals such as Fgf8 and, in so doing, could permit correct lower jaw morphogenesis. Interestingly, at the end of gastrulation Dlx5 expression first appears in lateral bands of the embryonic/extraembryonic junction at the late-streak stage which precedes neural plate formation49. Later, at E7.25 expression of Dlx5 is detected in the rostral and in the posterior ectoderm. At even later stages, Dlx5 is found in NCC progenitors and in epithelial precursors of the ventral cephalic ectoderm (VCE). During migration, NCCs down-regulate Dlx5 expression and reactivate it only after arrival in the mandibular PA1 in response to an Edn1 signal. At E8.5 the NCCs-derived mesenchyme of the early PA1 is clearly Dlx5-positive50, however the epithelial component of PA1 does not express yet express Dlx549. The lower jaw epithelium seems to activate Dlx5 expression between E8.5, when the epithelium is still Dlx5-negative49, and E10.5, when the epithelial Dlx5 expression is comparable to that of the ectomesenchyme51, (see also Fig. 9). It seems plausible therefore that Dlx5 expression both in NCCs and in the PA1 mandibular epithelium is essential to permit correct lower jaw morphogenesis. VCE cells also down regulate Dlx5 expression, but their derivatives migrate then toward anterior parts of PA134 and are essential for correct maxillary development. It seems therefore that the integration of signals deriving from three Dlx5-positive cellular progenitors (NCCs, PA1 epithelial cells and VCE cells) is needed for the generation of functional, matching jaws. Dlx5 expression in these different territories might well depend on different regulatory mechanisms. An example of this differential regulation is the observation that the I56i enhancer is active in PA1 NCCs, but not in epithelial structures such as the VCE, the PA1 epithelium or the otic vesicle48.
Vital biological functions such as feeding or reproduction often depend on the interaction between complementary structures located either in the same individual (e.g. jaws) or in separate members of the same species (e.g. sexual organs). The coevolution of synergistic complementary structures in separate individuals or in different territories of the same organism has, therefore, been instrumental to ensure species survival. We show that, for the specific case of mouth development, the same cluster of genes is: (1) determining lower jaw identity and (2) tuning the morphogenetic process of both the upper and the lower jaw generating a mouth endowed with opposing and matching mandibles capable to support feeding and/or predation. These two levels of morphogenetic regulation must be coordinated to generate a functional oral organ capable of mastication. These morphogenetic processes occur in different cell types located in spatiotemporally separate embryonic territories suggesting that diverse Dlx5/6-depending pathways are involved. At least in the case of the jaws, the co-evolution of synergistic anatomical structures ostensibly involves the coordination of different regulations involving the same genes in distant embryonic territories.
These observations open very profound questions on how these different regulations might have evolved and be selected. It is remarkable however that both NCCs and the VCE derive from the same Dlx5-positive early progenitors at the very end of gastrulation suggesting interesting evolutionary scenarios that still need to be understood. Probing the origin of complementary, muscularized, jaws could open a window to understand more generally how different harmonious, but separate, parts of the body coordinate their morphogenesis.
Materials and Methods
Mice
Mice were housed in light, temperature (21 °C) and humidity controlled conditions; food and water were available ad libitum. WT animals were from Charles River France. Double Dlx5 and Dlx6 (Dlx5/6) null mice27,28 and the double conditional mutant Dlx5/6flox/flox 37 in which the DNA-binding region of both Dlx5 and Dlx6 is deleted by cre-recombinase were maintained and genotyped as reported. The inducible Cre driver strain Wnt1-creERT2 (Stock #008851) and Wnt1-cre (Stock #022501) were purchased from Jackson Laboratory Maine, USA through Charles River Laboratories (L’Arbresle, France) and maintained on a C57BL/6J genetic background.
To obtain mice carrying the R26RCAG-flox-DLX5/+ allele, an F3/FRT-flanked cassette containing the CAG promoter, a floxed stop sequence, Flag-tagged mouse Dlx5 cDNA and a poly(A) additional signal were inserted into the targeting vector pROSA26-1 (P. Soriano, Mount Sinai School of Medicine, New York, NY, USA) (Addgene, plasmid 21714). Homologous recombination was performed on the ROSA26 locus of B6129F1-derived ES cells. Targeted ES clones were injected into ICR blastocysts to generate chimeras. Chimeras were crossbred with ICR females. R26RCAG-flox-DLX5/+ mice were crossed with Wnt1-cre mice35 to induce NCC-specific expression of Dlx5 (NCCDlx5 mice).
Embryos were obtained from pregnant dams, washed and dissected in ice-cold phosphate buffered saline (pH7.4). Embryos selected for fixation were immersed in 4% paraformaldehyde from 15mn to 2 h, photographed and further processed17,37. Specimens selected for biochemical analyses were dissected in PBS and processed for RT-qPCR, Affymetrix, Western blot and Southern blot as described in Supplementary Text 1 and37.
Intraperitoneal tamoxifen (Sigma-Aldrich, France, #T5602) injections were performed as previously described34 using 3 to 5 mg/day/pregnant mouse. To cover most of the period of neural crest delamination and migration38 Dlx5/6Fl/Fl::Wnt1Cre-ERT2/+ pregnant dams received two IP injections of tamoxifen at E6 and E7, generating NCC∆Dlx5/6 embryos.
Histology and 3D reconstruction
Heads from E18.5 (mutant and wild type foetuses were fixed in Bouin’s solution (Sigma, France), embedded in paraffin and complete sets of frontal or parasagittal serial sections (12μm) were prepared. All sections were stained by Mallory’s trichrome31 and photographed (Nikon Digital Site DS-FI1). Pictures were aligned, piled and registered using the Fiji plug-in of NIH ImageJ “Register Virtual Stack Slices” (http://fiji.sc/wiki/index.php/Register_Virtual_Stack_Slices). 3D segmentation was performed with Mimics (Materialise, Belgium: http://biomedical.materialise.com/mimics) and visualized using Adobe Acrobat 9 pro.
Skeletal preparation and staining
Alizarin red/ alcian blue staining was performed, as previously described52. Samples were fixed in 95% ethanol for a week, permuted to acetone for three days and incubated with 0.015% alcian blue 8GS, 0.005% alizarin red S and 5% acetic acid in 70% ethanol for three days. After washing with distilled water, the samples were cleared in 1% KOH for several days and in 1% KOH glycerol series until the surrounding tissues turned transparent. The preparations were stored in glycerol.
In situ hybridization
Whole-mount in situ hybridization was performed, as described previously31. Embryos were fixed one overnight in 4% paraformaldehyde in PBT. After dehydration and rehydration with methanol, the embryos were bleached for 1 hour in 7.5% H2O2 in PBT and then washed in PBT 3 times. The samples were treated with 5 mg/ml proteinase K for 40 seconds at room temperature, treated with 2 mg/ml glycine in PBT to stop the enzyme reaction, and post-fixed in 4% paraformaldehyde and 0.2% glutaraldehyde for 20 minutes on ice. After the pretreatment, the samples were pre-hybridized for more than 1 hour at 70 °C in hybridization mix (50% formamide, 5 x SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1% SDS), 50 mg/ml heparin and 50 mg/ml yeast tRNA. With digoxygenin-labeled RNA probe in hybridization mix, the samples were hybridized overnight at 70 °C. The samples were then washed 3 times in hybridization mix at 70 °C, then in 0.2 M NaCl, 10 mM Tris-HCl (pH7.5), 0.1% Tween-20 for 5 minutes ant treated with 100 mg/ml RNase for 30 minutes at 37 °C. After a final wash in 50% formamide, 2 × SSC for 1 hour at 65 °C, the samples were pre-blocked with sheep serum, incubated with alkaline phosphatase-conjugated anti-digoxygenin antibody, and stained with nitro blue tetrazolium and 5-bromo-4-chloro-3-indoyl phosphate. Probes were prepared by RT-PCR and used in published17,53.
Quantitative real-time RT-PCR
The maxillary and mandibular processes were dissected from E10.5 control and NCCDlx5 mice. Total RNA was extracted from five sets of PAs by ISOGEN-II (Nippon Gene). One-μg samples were then reverse-transcribed using ReverTra Ace (TOYOBO) with RS19-15dT primer. Quantification of amount of each mRNA was performed by real-time PCR analysis using a LightCycler (Roche) and Real-Time PCR Premix with SYBR Green (RBC Bioscience) following the manufacturer’s protocol. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as internal control. PCR was performed using following primers, Dlx5 previously described54, 5′-AGACAGCCGCATCTTCTTGT-3′ and 5′-CTTGCCGTGGGTAGAGTCAT-3′ for Gapdh.
Gene expression profiling
The maxillary process, the mandibular process and the PA2 were collected from E10.5 control, Dlx5/6−/− and NCCDlx5 mice and subjected to Affymetrix GeneChip analysis. Each sample was a mixture from 3 littermates. Preparation of the cRNA and hybridization of probe array were performed on an Affymetrix GeneChip Mouse 430 2.0 array which contains 45,101 probe sets according to the manufacturer’s instructions (Affymetrix, Santa Clara, CA). The expression value for each mRNA was obtained by the Robust Multi-array Average (RMA) method. The gene set probes were filtered on an expression (20.0–100.0) percentile. Genes with the expression level lower than 20.0 percentile at least in one sample were eliminated from the analysis. After excluding the probes whose gene symbols were not identified, about 35,000 genes remained and used for further analysis. Annotation of the probe numbers and targeted sequences are shown on the Affymetrix web site.
Animal experimentation approvals
Procedures involving animals were conducted in accordance with the directives of the European Community (council directive 86/609), the French Agriculture Ministry (council directive 87–848, 19 October 1987, permissions 00782 to GL) and approved by the University of Tokyo Animal Care and Use Committee and by the “Cuvier” ethical committee (approval n° 68-028r1 to HK).
Electronic supplementary material
Acknowledgements
This research was partially supported by the EU Consortium HUMAN (EU-FP7-HEALTH-602757) and by the ANR grants TARGETBONE (ANR-17-CE14-0024) and METACOGNITION (ANR-17-CE37-0007) to GL, and an ATM grant (FORMs) to YG, CdL is supported by a grant of the French Ministry of Research. This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (15H02536) and Core Research for Evolutional Science and Technology (CREST) of the Japan Science and Technology Agency (JST), Japan (JPMJCR13W2). A particular thank goes to the team in charge of mouse animal care and in particular M Stéphane Sosinsky and M Fabien Uridat and to Pr. Amaury de Luze in charge of animal well-being. We thank the Stanford Exchange Program Scholars Mss. Shaheen Jeawoody, Andrea Fontaines and Heydi Malavé for the excellent help given to this work and Mss. Aicha Bennana and Lanto Courcelaud for administrative assistance.
Author Contributions
M.S. Performed experiments, prepared figures, wrote text and provided scientific discussion. N.N.-N. Performed and coordinated experimental work, wrote text and provided scientific input. Y.G. Performed experiments, wrote text and provided scientific discussion. C.d.L. Analyzed data. A.F. Performed experiments. G.A. Performed experiments. T.K. Performed experiments, analyzed data. Y.K. Performed experiments, analyzed data. E.H. Analyzed histological data, provided scientific input. L.M. Contributed 3D reconstructions. H.H. Analyzed data, prepared Figure 9 and S5. Y.W. Analyzed data. Y.K. Coordinated experimental work and mouse generation. H.K. Coordinated the work, analyzed data, wrote text. G.L. Coordinated the work, analyzed data, wrote the text, prepared figures, and took histological images.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Miki Shimizu, Nicolas Narboux-Nême and Yorick Gitton contributed equally.
Contributor Information
Hiroki Kurihara, Email: kuri-tky@umin.net.
Giovanni Levi, Email: glevi@mnhn.fr.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33207-2.
References
- 1.Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Compagnucci C, et al. Pattern and polarity in the development and evolution of the gnathostome jaw: both conservation and heterotopy in the branchial arches of the shark, Scyliorhinus canicula. Dev Biol. 2013;377:428–448. doi: 10.1016/j.ydbio.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 3.Tan SS, Morriss-Kay G. The development and distribution of the cranial neural crest in the rat embryo. Cell Tissue Res. 1985;240:403–416. doi: 10.1007/BF00222353. [DOI] [PubMed] [Google Scholar]
- 4.Tan SS, Morriss-Kay GM. Analysis of cranial neural crest cell migration and early fates in postimplantation rat chimaeras. J Embryol Exp Morphol. 1986;98:21–58. [PubMed] [Google Scholar]
- 5.Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- 6.Depew MJ, Olsson L. Symposium on the evolution and development of the vertebrate head. J Exp Zool B Mol Dev Evol. 2008;310:287–293. doi: 10.1002/jez.b.21219. [DOI] [PubMed] [Google Scholar]
- 7.Fish JL, Sklar RS, Woronowicz KC, Schneider RA. Multiple developmental mechanisms regulate species-specific jaw size. Development. 2014;141:674–684. doi: 10.1242/dev.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ealba EL, et al. Neural crest-mediated bone resorption is a determinant of species-specific jaw length. Dev Biol. 2015;408:151–163. doi: 10.1016/j.ydbio.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couly GF, Coltey PM, Le Douarin NM. The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- 10.Creuzet S, Couly G, Vincent C, Le Douarin NM. Negative effect of Hox gene expression on the development of the neural crest-derived facial skeleton. Development. 2002;129:4301–4313. doi: 10.1242/dev.129.18.4301. [DOI] [PubMed] [Google Scholar]
- 11.Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- 12.Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trainor PA, Tam PP. Cranial paraxial mesoderm and neural crest cells of the mouse embryo: co-distribution in the craniofacial mesenchyme but distinct segregation in branchial arches. Development. 1995;121:2569–2582. doi: 10.1242/dev.121.8.2569. [DOI] [PubMed] [Google Scholar]
- 14.Couly G, Creuzet S, Bennaceur S, Vincent C, Le Douarin NM. Interactions between Hox-negative cephalic neural crest cells and the foregut endoderm in patterning the facial skeleton in the vertebrate head. Development. 2002;129:1061–1073. doi: 10.1242/dev.129.4.1061. [DOI] [PubMed] [Google Scholar]
- 15.Barske L, et al. Essential Role of Nr2f Nuclear Receptors in Patterning the Vertebrate Upper Jaw. Dev Cell. 2018;44:337–347 e335. doi: 10.1016/j.devcel.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tucker AS, Lumsden A. Neural crest cells provide species-specific patterning information in the developing branchial skeleton. Evol Dev. 2004;6:32–40. doi: 10.1111/j.1525-142X.2004.04004.x. [DOI] [PubMed] [Google Scholar]
- 17.Gitton Y, et al. Evolving maps in craniofacial development. Semin Cell Dev Biol. 2010;21:301–308. doi: 10.1016/j.semcdb.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Ruhin B, et al. Patterning of the hyoid cartilage depends upon signals arising from the ventral foregut endoderm. Dev Dyn. 2003;228:239–246. doi: 10.1002/dvdy.10380. [DOI] [PubMed] [Google Scholar]
- 19.Depew MJ, Simpson CA. 21st century neontology and the comparative development of the vertebrate skull. Dev Dyn. 2006;235:1256–1291. doi: 10.1002/dvdy.20796. [DOI] [PubMed] [Google Scholar]
- 20.Clouthier DE, et al. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- 21.Kurihara Y, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- 22.Ozeki H, Kurihara Y, Tonami K, Watatani S, Kurihara H. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech Dev. 2004;121:387–395. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato T, et al. Recombinase-mediated cassette exchange reveals the selective use of Gq/G11-dependent and -independent endothelin 1/endothelin type A receptor signaling in pharyngeal arch development. Development. 2008;135:755–765. doi: 10.1242/dev.012708. [DOI] [PubMed] [Google Scholar]
- 25.Verzi MP, et al. The transcription factor MEF2C is required for craniofacial development. Dev Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J, et al. Endothelin signaling activates Mef2c expression in the neural crest through a MEF2C-dependent positive-feedback transcriptional pathway. Development. 2015;142:2775–2780. doi: 10.1242/dev.126391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merlo GR, et al. Multiple functions of Dlx genes. Int J Dev Biol. 2000;44:619–626. [PubMed] [Google Scholar]
- 28.Beverdam A, et al. Jaw transformation with gain of symmetry after Dlx5/Dlx6 inactivation: mirror of the past? Genesis. 2002;34:221–227. doi: 10.1002/gene.10156. [DOI] [PubMed] [Google Scholar]
- 29.Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- 30.Fukuhara S, Kurihara Y, Arima Y, Yamada N, Kurihara H. Temporal requirement of signaling cascade involving endothelin-1/endothelin receptor type A in branchial arch development. Mech Dev. 2004;121:1223–1233. doi: 10.1016/j.mod.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Heude E, et al. Jaw muscularization requires Dlx expression by cranial neural crest cells. Proc Natl Acad Sci USA. 2010;107:11441–11446. doi: 10.1073/pnas.1001582107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fish JL, et al. Satb2, modularity, and the evolvability of the vertebrate jaw. Evol Dev. 2011;13:549–564. doi: 10.1111/j.1525-142X.2011.00511.x. [DOI] [PubMed] [Google Scholar]
- 33.Depew MJ, Compagnucci C. Tweaking the hinge and caps: testing a model of the organization of jaws. J Exp Zool B Mol Dev Evol. 2008;310:315–335. doi: 10.1002/jez.b.21205. [DOI] [PubMed] [Google Scholar]
- 34.Gitton Y, Narboux-Neme N, Levi G. Transitory expression of Dlx5 and Dlx6 in maxillary arch epithelial precursors is essential for upper jaw morphogenesis. F1000Res. 2014;2:261. doi: 10.12688/f1000research.2-261.v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chai Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 36.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 37.Bellessort B, et al. Dlx5 and Dlx6 control uterine adenogenesis during post-natal maturation: possible consequences for endometriosis. Hum Mol Genet. 2016;25:97–108. doi: 10.1093/hmg/ddv452. [DOI] [PubMed] [Google Scholar]
- 38.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/S0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 39.Jeong J, et al. Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development. 2008;135:2905–2916. doi: 10.1242/dev.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X, et al. Normal fate and altered function of the cardiac neural crest cell lineage in retinoic acid receptor mutant embryos. Mech Dev. 2002;117:115–122. doi: 10.1016/S0925-4773(02)00206-X. [DOI] [PubMed] [Google Scholar]
- 41.McBratney-Owen B, Iseki S, Bamforth SD, Olsen BR, Morriss-Kay GM. Development and tissue origins of the mammalian cranial base. Dev Biol. 2008;322:121–132. doi: 10.1016/j.ydbio.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugii H, et al. The Dlx5-FGF10 signaling cascade controls cranial neural crest and myoblast interaction during oropharyngeal patterning and development. Development. 2017;144:4037–4045. doi: 10.1242/dev.155176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romer A. Mirror image comparison of upper and lower jaws in primitive tetrapods. Anathomical Records. 1940;77:175–179. doi: 10.1002/ar.1090770206. [DOI] [Google Scholar]
- 44.Ruest LB, et al. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev Biol. 2003;257:263–277. doi: 10.1016/S0012-1606(03)00068-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charite J, et al. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15:3039–3049. doi: 10.1101/gad.931701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferguson CA, Tucker AS, Sharpe PT. Temporospatial cell interactions regulating mandibular and maxillary arch patterning. Development. 2000;127:403–412. doi: 10.1242/dev.127.2.403. [DOI] [PubMed] [Google Scholar]
- 48.Park BK, et al. Intergenic enhancers with distinct activities regulate Dlx gene expression in the mesenchyme of the branchial arches. Dev Biol. 2004;268:532–545. doi: 10.1016/j.ydbio.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 49.Yang L, et al. An early phase of embryonic Dlx5 expression defines the rostral boundary of the neural plate. J Neurosci. 1998;18:8322–8330. doi: 10.1523/JNEUROSCI.18-20-08322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Acampora D, et al. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 51.Vincentz JW, et al. Exclusion of Dlx5/6 expression from the distal-most mandibular arches enables BMP-mediated specification of the distal cap. Proc Natl Acad Sci USA. 2016;113:7563–7568. doi: 10.1073/pnas.1603930113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 53.Kitazawa T, et al. Distinct effects of Hoxa2 overexpression in cranial neural crest populations reveal that the mammalian hyomandibular-ceratohyal boundary maps within the styloid process. Dev Biol. 2015;402:162–174. doi: 10.1016/j.ydbio.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Vieux-Rochas M, et al. Spatio-temporal dynamics of gene expression of the Edn1-Dlx5/6 pathway during development of the lower jaw. Genesis. 2010;48:262–373. doi: 10.1002/dvg.20625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.