Abstract
The efficacy of most antibiotics is dependent on active bacterial growth, yet little is known about the growth dynamics during infection. Therefore, means to measure in-host bacterial growth rate is of importance. Here, we use chromosome replication as readout for in situ bacterial growth rate during infection; obtained from a single biological specimen. We have applied two independent methods: quantitative PCR (qPCR) and fluorescence microscopy, to quantify the level of chromosome replication present during Escherichia coli propagation in the mouse peritonitis model. We find that the methods complement each other and allow for quantification of growth rate, both on a population average and on a single-cell level. We demonstrate the presence of heterogeneous growth rates within bacterial populations propagating during infection. Also, no growth cessation was observed during the apparent stationary phase in vivo, and, by comparison of growth dynamics at different anatomical sites, we demonstrate that E. coli is unlikely to grow independently intravascularly. These findings provide novel insight into bacterial growth during host infection, and underscore the importance of pinpointing the primary site of infection in septicaemia of unknown origin and ensuring antibiotic availability at this site.
Introduction
The interplay between bacterial growth and chromosome replication has been studied extensively under laboratory conditions for decades1–6. Until recent years, the detection of chromosome replication status in bacteria propagating in their natural host (in vivo) had received scant attention. Bacterial growth in vivo has generally been assessed by viable bacterial counts (colony forming units (CFU)/ml)) plotted as a function of time. In a batch culture with a finite amount of nutrients, this curve is divided into well-defined growth phases7. However, bacterial growth dynamics in vivo are complex; growth rate is not solely a function of nutrient availability, but also of host immune-mediated clearance8–11. Applying bacterial count measurements to deduce an absolute bacterial growth rate in vivo is therefore not adequate, as this merely reflects the net change in bacterial population size. Neither does it reflect the level of growth rate heterogeneity within the population. To circumvent some of these limitations, recently published studies have explored the use of chromosome replication status as readout for bacterial growth rate in environmental and human microbiomes and found good correlation between the two12–14. However, this was achieved by whole genome sequencing, a method that besides being expensive and time-consuming, only provides a population average readout and fails to provide information on any possible differential growth rates within the population.
Escherichia coli, like many other bacteria, possesses one circular chromosome. DNA replication is initiated from a single origin of replication (oriC) and carried out bidirectionally, terminating in the opposite replication terminus region (terC)1,2,15,16. Coordination between chromosome replication and cell division ensures that the two identical daughter cells accommodate at least one fully replicated chromosome17,18. Initiation of replication takes place at a fixed cell mass per origin: the initiation mass3,19–22. Under favourable growth conditions multiple rounds of synchronously initiated replications per division cycle can occur4,23. This allows for faster growth rates, with daughter cells carrying chromosomes with overlapping rounds of replication, i.e. more than one number of oriC copies (2n, n = 1, 2, 3), at birth. Due to the consistency of the initiation mass, rapidly growing cells harbouring overlapping rounds of replication have larger cell mass proportional to the number of oriC copies. The growth rate of the population is reflected by the copy number ratio of oriC to terC (ori:ter), with an ori:ter of 1 representing non-replicating cells carrying a complete set of chromosome(s) and an ori:ter > 1 representing cells with ongoing chromosome replication (growing cells)1,24. Theoretically, this correlation can be expressed as ori:ter = 2 C/τ (1), where C is the chromosome replication time (C-period) and τ is the mass doubling time, and thus τ = C/log2(ori:ter) (2)24. However, such an unambiguous measure can only be inferred from a population of bacterial cells propagating at balanced growth rates, and only at τ < approximately 60 minutes, where C has been shown to be constant25.
Here, we applied quantitative PCR (qPCR), an inexpensive and easily accessible method, to provide population average measurements of bacterial growth rates during infection in vivo, combined with chromosomal fluorescent marker frequency analysis for quantification of bacterial growth at a single-cell level. The goal was to determine variation in in situ bacterial growth rates at various anatomical sites, which would add to our current understanding of growth dynamics during infection. We hypothesized that growth dynamics within and outside the primary site of infection would be different. Furthermore, the detection of an in situ bacterial growth rate obtained from one single specimen could form the basis for refining future antibacterial strategies, knowing that the efficacy of most antibiotics in clinical use is related to active bacterial growth26,27.
Results
Two methods for detection of ori:ter were tested in vivo in an experimental murine infection model; qPCR, where the ori:terqPCR was given as a population mean, and fluorescence microscopy, where the ori:termic was deduced from direct analysis of single fluorescently labelled live bacterial cells (exemplified in Fig. 1, where oriC foci are displayed in green (GFP labelling) and terC foci are displayed in red (mCherry labelling)). In ori:terqPCR the ‘oriC’ copy number was inferred from a site located immediately to the left of oriC, and the ‘terC’ copy number was inferred from a site located in close proximity to terC28. We applied these methods, in combination with bacterial counts, to reveal fundamental bacterial growth dynamics at various anatomical sites during infection. Furthermore, the correlation between cell length (µm) (measured by microscopy and used as a proxy for cell mass) and oriC/cell was measured to investigate initiation mass consistency during infection in vivo. As proof-of-concept, the same methods were tested and validated in vitro, using the same model infective organism; Escherichia coli ATCC® 25922™.
Figure 1.
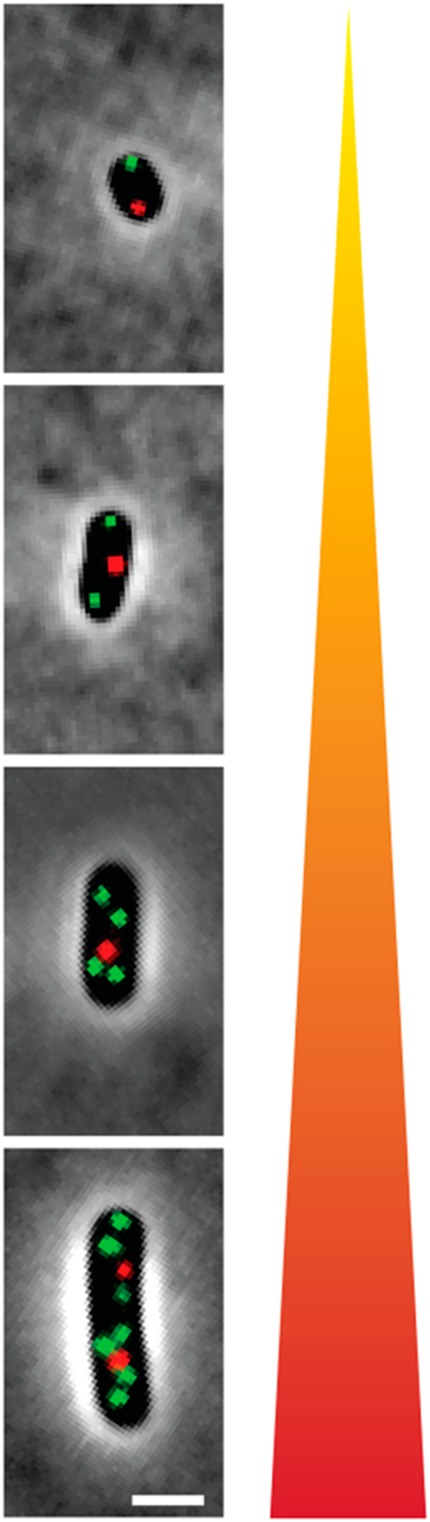
Visualisation of intracellular oriC foci (green) and terC foci (red) by fluorescence microscopy in ALO 4783 (shown in phase contrast) growing at various growth rates; from rapid (bottom) to slow (top) growth. Scale bar is 2 µm.
Bacterial growth in vitro
Bacteria grown in vitro in Lysogeny Broth (LB) batch cultures were used to validate the methods applied in the in vivo experiments. Here, stationary phase bacterial cells were diluted in fresh media and allowed to grow with repeated sample collections at time points 2, 4, 6, 8 and 10 hours of incubation. As a complete cessation of growth was observed at 10 hours of incubation, the in vitro experiments were not extended beyond this time point. We tested the wild-type strain (ATCC 25922) alongside a genetically modified derivative of the strain, with chromosomally incorporated fluorescent oriC and terC labels (ALO 4783). This was done to ensure that transgene insertion would not affect growth dynamics of the derivative strain, which was applied in the in vivo infection model.
Growth curves from in vitro studies are presented as bacterial counts as a function of time in Fig. 2a, and as OD600 values as a function of time in Fig. 2b, respectively. Exponential growth was observed between the initial sampling time point and approximately 6 hours of incubation, after which growth progressively ceased towards stationary phase. The mass doubling time (τ) inferred from mean OD600 measurements during exponential growth was 25 minutes. No growth reduction due to transgene insertions in ALO 4783 was observed (Fig. 2b).
Figure 2.
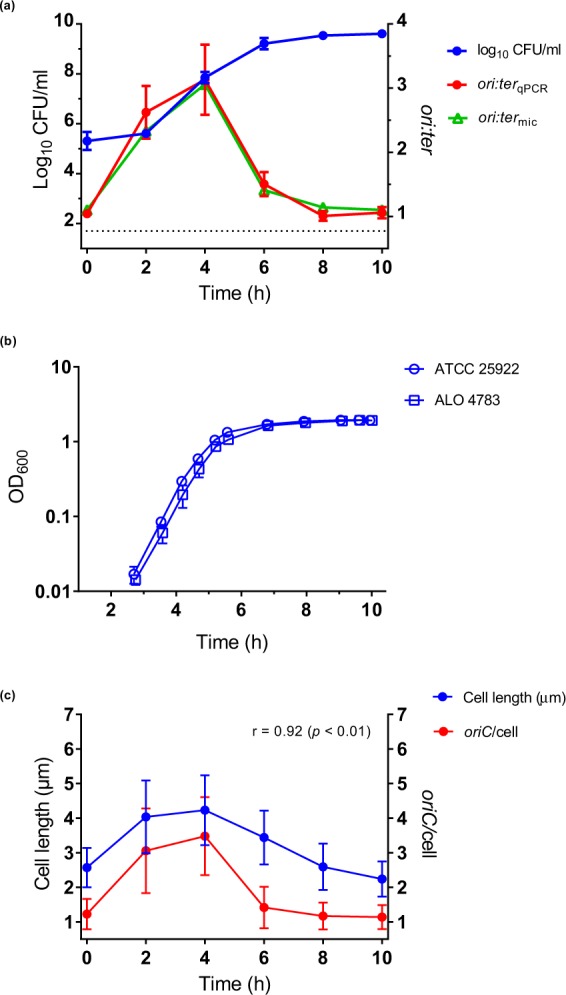
Bacterial growth in LB batch cultures (in vitro). (a) Bacterial growth measured as bacterial counts (log10 CFU/ml), ori:terqPCR and ori:termic. ATCC 25922 and ALO 4783 were tested in parallel and showed similar growth; hence, results from both strains are pooled in the data sets. Bland-Altman agreement between ori:terqPCR and ori:termic was good (Supplementary Fig. S1). 6 h, n = 12; all other time points, n = 6. (b) Comparisons of ATCC 25922 and ALO 4783 grown in LB batch cultures (in vitro) revealed no growth retardation due to transgene insertions. Growth measured as OD600. Both strains exerted doubling times of 25 minutes during exponential growth. (c) Bacterial cell length (µm) (used as proxy for cell mass) correlate with oriC/cell. Data represent pooled microscopically detected cells (ALO 4783). n = 500 at all time points, with the exception of 2 h, where n = 147. Data are presented as mean ± SD. Dotted line represents the limit of detection for bacterial count measurements. Time (h) represents hours of incubation; inoculum is presented as 0 h. r: Pearson’s correlation coefficient.
The two methods for detection of chromosome replication status (ori:terqPCR and ori:termic) were in good agreement (Supplementary Fig. S1). ori:terqPCR and ori:termic followed growth rate (Fig. 2a, Supplementary Table S1). We observed a rapid increase in ori:ter during the first 4 hours, followed by a gradual decline as the cells stopped growing by 8 hours; remaining unchanged at 10 hours of incubation. In accordance with these findings, single-cell fluorescence microscopy analysis of oriC/cell and cell size (µm) confirmed a shift from a homogenous bacterial population dominated by non-growing, small cells at 0 hours (i.e. the inoculum; oriC/cell ~1), to a heterogeneous population dominated by bacterial cells with ongoing chromosome replications at various levels (oriC/cell ≥ 2) and various cell sizes at 2 hours of incubation (Fig. 3). Starting from 4 hours of incubation, a tendency towards more growth homogeneity was observed. At this time point, an oriC/cell of 4 was observed in approximately 80 percent of the population, reflecting close to balanced growth (Fig. 3). Here, criteria for inferring mass doubling time (τ) from ori:ter were met, and τ was computed from equation (2) as (median (IQR)) 23.4 (21.5–29.9) minutes (assuming a constant C period of 40 minutes)25. Almost no overlapping rounds of replication were observed starting from 6 hours of incubation (oriC/cell ≤ 2). At 8 and 10 hours of incubation homogenous populations of small bacterial cells harbouring predominantly one copy of oriC/cell were observed (Fig. 3). The correlation between oriC/cell and cell size (µm) was very strong (r = 0.92, p < 0. 01) (Fig. 2c).
Figure 3.
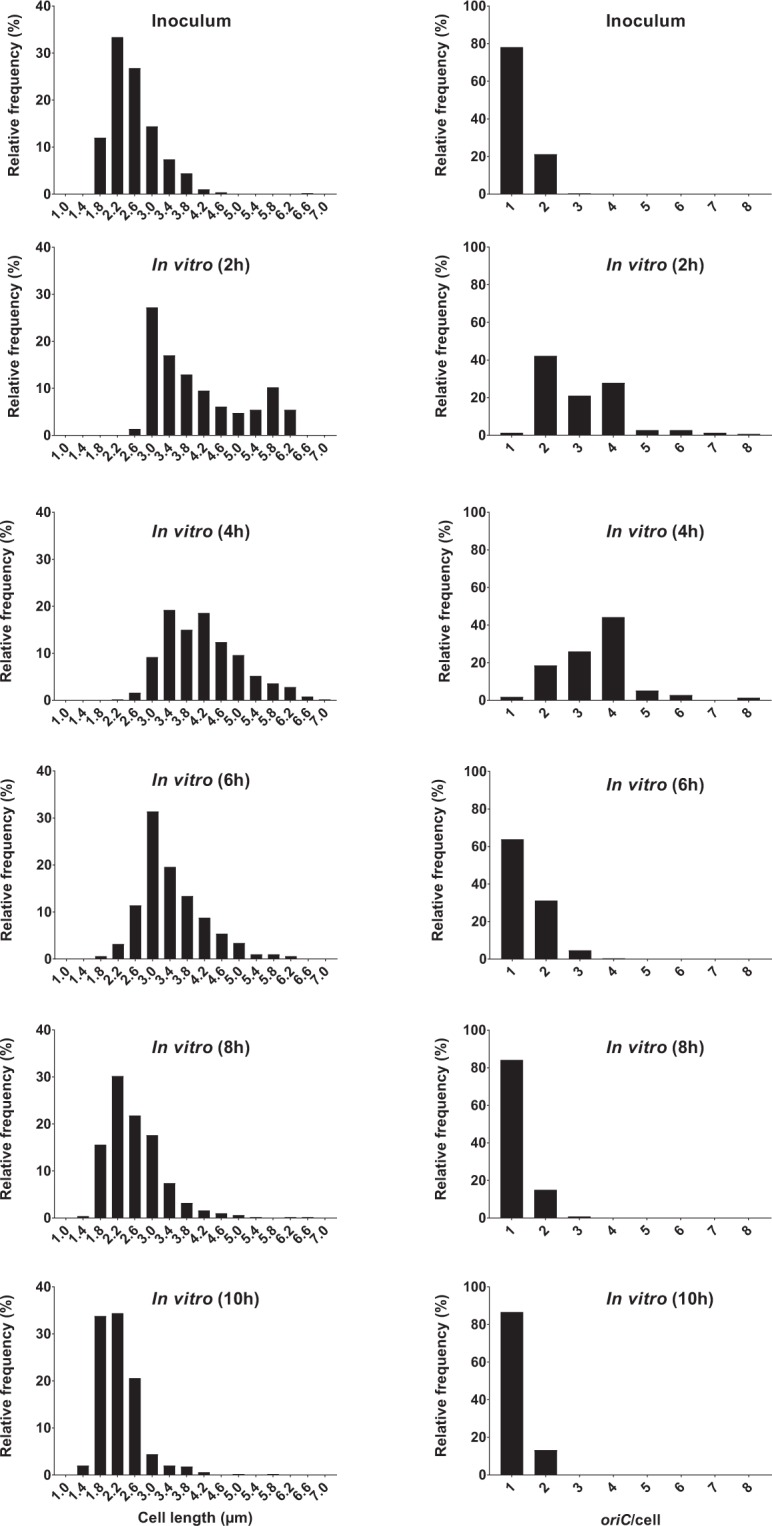
Relative frequency distributions (%) of pooled microscopically detected bacterial cells (ALO 4783) from growth in LB batch cultures (in vitro); cell length (µm) (left panel) and oriC/cell (right panel). n = 500 at each time point, with the exception of 2 h, where n = 147. Time (h) represents hours of incubation. For oriC/cell histograms, note that data are presented as detected by automated foci quantification, which is subject to the risk of underestimation due to co-localising oriCs48. Hence, it is likely that 3 detected foci truly represent 4 foci and that >4 detected foci truly represent 8 foci (see Methods).
A slow-down of growth was initiated between 4 and 6 hours of incubation, during which period we observed a decrease in both oriC/cell and ori:ter, manifesting cessation of chromosome replication (Fig. 2a,c). As expected1, cell division proceeded for some time longer, as demonstrated both by the bacterial count increase and cell size decrease (Fig. 2a,c). Eventually, no new initiations of chromosome replication took place during stationary phase (8–10 hours of incubation). Inferring τ from ori:ter for time points later than 4 hours of incubation would be deceptive, given that the growth was too slow (i.e. the criteria of τ < 60 minutes was not met).
We can conclude that, when these bacterial cells were growing fast the ori:ter and oriC/cell ratios were high and cell sizes were large, and when cells were growing slowly the ori:ter and oriC/cell ratios were low and cell sizes were small. Our findings are consistent with the existence of a fixed initiation mass at all in vitro growth rates, exemplified by the good correlation between oriC/cell and cell length (Fig. 2c). These data are consistent with previous studies that have shown correlation between ori:ter and initiation mass in E. coli K-12 strain MG1655 under various growth conditions in vitro29. At mid-exponential phase (4 hours of incubation; Fig. 2a) a mass doubling time (τ) could be inferred from ori:ter and was similar to that inferred from OD600 measurements; approximately 23 and 25 minutes, respectively.
Bacterial growth in vivo: mouse peritonitis model
Bacterial growth dynamics at separate anatomical sites during widespread bacterial infection were evaluated in the model of lethal peritonitis in mice, aiming both to validate the ori:ter method for use in vivo, and to determine any potential growth rate differences within and outside the primary site of infection.
An intraperitoneal (i.p.) dose of 1 × 106 CFU/ml stationary phase bacteria (ALO 4783) resulted in the establishment of septicaemia in a total of 51/51 animals within 2 hours of infection. Samples (peritoneal lavage fluid (PLF), blood, spleen and kidneys) were collected in biological triplicates at time points 2, 4, 6, 8 and 10 hours of infection. After 10 hours of infection animals demonstrated signs of distress to such an extent that criteria for euthanasia were met. Thus, this constituted the final possible sample collection time point in this model. Bacterial DNA and live bacterial cells were successfully isolated from PLF and blood for qPCR and microscopy analysis, respectively. Unfortunately, we were unable to adequately isolate bacterial DNA or live bacterial cells for the above mentioned analyses from spleen and kidneys. Consequently, the PLF and blood bacterial populations constituted representative populations of bacteria growing within and outside of the primary site of infection, respectively. It has previously been hypothesised that bloodstream bacterial populations represent a passive ‘spill-over’ of bacteria from the primary site of infection, rather than independently growing populations30. Using chromosome replication as readout for in situ bacterial growth rate, we aimed at improving insight into whether these populations truly differ or not.
Pooled bacterial count data from repeated in vivo experiments demonstrated that the net size of the bacterial populations found outside the primary site of infection (i.e. bloodstream, spleen or kidneys) increased in parallel with the bacterial population at the primary site of infection (i.e. peritoneum) (Fig. 4a). For all collected specimens (PLF, blood, spleen and kidneys) a pronounced increase in bacterial count was observed from 0 to 8 hours of infection, indicating bacterial growth. There was no apparent lag phase, however, this cannot be ruled out since no measurements were made prior to 2 hours of infection. Linear regression analysis derived slopes (β) for growth in blood, spleen and kidneys revealed no significant difference from growth in PLF (p > 0.05), whereas the blood, spleen and kidney bacterial counts differed significantly from the PLF bacterial count (p < 0.0001); meaning that bacterial population sizes outside the PLF were smaller, yet increasing in parallel with the PLF bacterial population (Fig. 4a). Also, attention should be drawn to the fact that the bacterial count found at the primary site of infection was under-estimated. The PLF bacterial count was in reality substantially higher than reported, taken into consideration the dilution factor from the required addition of lavage fluid during harvesting (see Methods). From 8 to 10 hours of infection the net bacterial count was stagnant (Fig. 4a).
Figure 4.
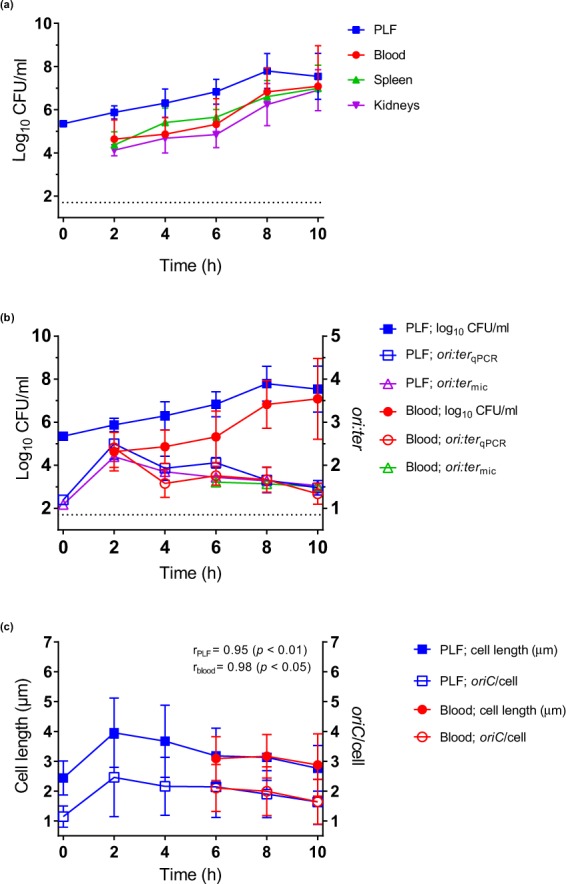
Bacterial growth (ALO 4783) in the mouse peritonitis model (in vivo). (a) Bacterial growth measured as bacterial counts (log10 CFU/ml) in peritoneal lavage fluid (PLF), blood, spleen and kidneys: 2 h, n = 15; 4 h, n = 9; 6 h, n = 6; 8 h, n = 12; 10 h, n = 9. Slopes (β) from linear regression analysis (0–8 h; blood, spleen and kidneys versus PLF) were not significantly different (p > 0.05), yet elevations (blood, spleen and kidneys versus PLF) were all significantly different (p < 0.0001) (b) Bacterial growth measured as bacterial counts, ori:terqPCR and ori:termic in PLF and blood. PLF bacterial count and ori:terqPCR: 2 h, n = 15, 4 h, n = 9; 6 h, n = 6; 8 h, n = 12; 10 h, n = 9. Blood bacterial count: 2 h, n = 15, 4 h, n = 9; 6 h, n = 6; 8 h, n = 12; 10 h, n = 9. Blood ori:terqPCR: 2 h, n = 12; 4 h, n = 8; 6 h, n = 5; 8 h, n = 12; 10 h, n = 9. ori:termic from 2 and 4 h (blood) are not presented due to insufficient number of microscopically detected cells (Supplementary Table S1). Bland-Altman agreement between ori:terqPCR and ori:termic was good (Supplementary Fig. S1). (c) Bacterial cell length (µm) (proxy for cell mass) correlate with oriC/cell, both in PLF and blood. Data represent pooled microscopically detected cells. Inoculum (0 h), n = 500. PLF: 2 h, n = 133; 4 h, n = 55; 6 h, n = 132; 8 h & 10 h, n = 500. Blood: 6 h, n = 164; 8 h, n = 157; 10 h, n = 500; 2 h and 4 h time points are not presented due to insufficient number of cells. Data are presented as mean ± SD. Dotted line represents the limit of detection for bacterial counts. Time (h) represents hours of infection; inoculum is presented as 0 h and extrapolated to PLF data. r: Pearson’s correlation coefficient.
The maximum ori:terqPCR (mean (SD)) measured in vivo (2.50 (0.36) in PLF) was lower than the maximum ori:terqPCR measured in vitro (3.13 (0.55)), accounting for an overall slower growth in vivo (Figs 4b and 2a, Supplementary Table S1). Nevertheless, ori:ter followed overall growth rate in vivo (Fig. 4b, Supplementary Table S1). In the PLF ori:terqPCR increased rapidly during the first 2 hours, followed by a slight decline towards unchanged levels at 4 and 6 hours of infection. After 6 hours of infection a gradual decline towards an ori:terqPCR approaching 1 was observed (Fig. 4b, Supplementary Table S1). In the blood, ori:terqPCR did not change between 4 and 8 hours. However, no significant difference (p > 0.05) in ori:terqPCR were found between PLF and blood, when compared both overall by linear regression analysis and time-point by time-point (2 to 10 hours of infection) (Fig. 4b).
There was no significant difference (p > 0.05) in oriC/cell or cell size (µm) between PLF and blood bacterial populations, compared at 6, 8 and 10 hours of infection (Fig. 4c). At 2 and 4 hours of infection, the total number of microscopically detected live bacterial cells from the blood was too low. Hence, these data are not included in statistical analyses or illustrations (Supplementary Table S1, Figs 4c and 5). oriC/cell and cell size (µm) data demonstrated a shift from a nearly homogenous bacterial population dominated by non-growing, small cells at 0 hours (i.e. the inoculum; oriC/cell ~1), to heterogeneous populations dominated by bacterial cells with ongoing chromosome replications at various levels (oriC/cell ≥ 2) and various cell sizes at 2 (PLF only), 4 (PLF only), 6 hours of infection (PLF and blood) (Figs 5 and 6). A minor decline in both oriC/cell and cell size was seen at 8 hours, and was even more pronounced at 10 hours of infection. However, unlike during stationary phase in vitro, we did not observe a complete cessation of chromosome replication during the length of infection. At 10 hours of infection (where minimal oriC/cell and cell size were observed) PLF and blood oriC/cell were significantly different from inoculum oriC/cell (p < 0.0001) (Figs 5 and 6). This indicated that fractions of the population were still growing, albeit slowly, at the terminal stage of infection. It has previously been reported, however, that bacteria may cease growth with 2 oriC/cell present31. Yet, under such conditions, ori:ter would go toward 1 (i.e. 2 fully replicated chromosomes), which we do not observe here.
Figure 5.
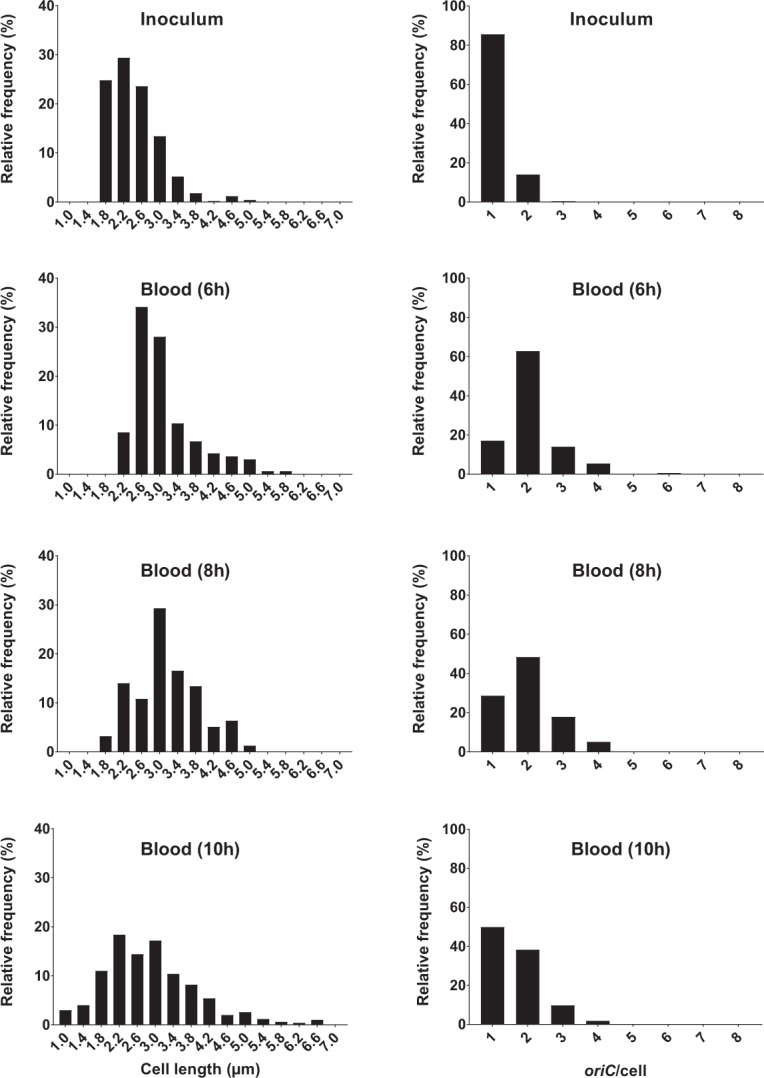
Relative frequency distributions (%) of pooled microscopically detected bacterial cells (ALO 4783) isolated from blood in the mouse peritonitis model; cell length (µm) (left panel) and oriC/cell (right panel). Inoculum, n = 500; 6 h, n = 164; 8 h; n = 157; 10 h, n = 500. Data from 2 and 4 h are not presented, due to insufficient number of microscopically detected cells (Supplementary Table S1). Time (h) represents hours of infection.
Figure 6.
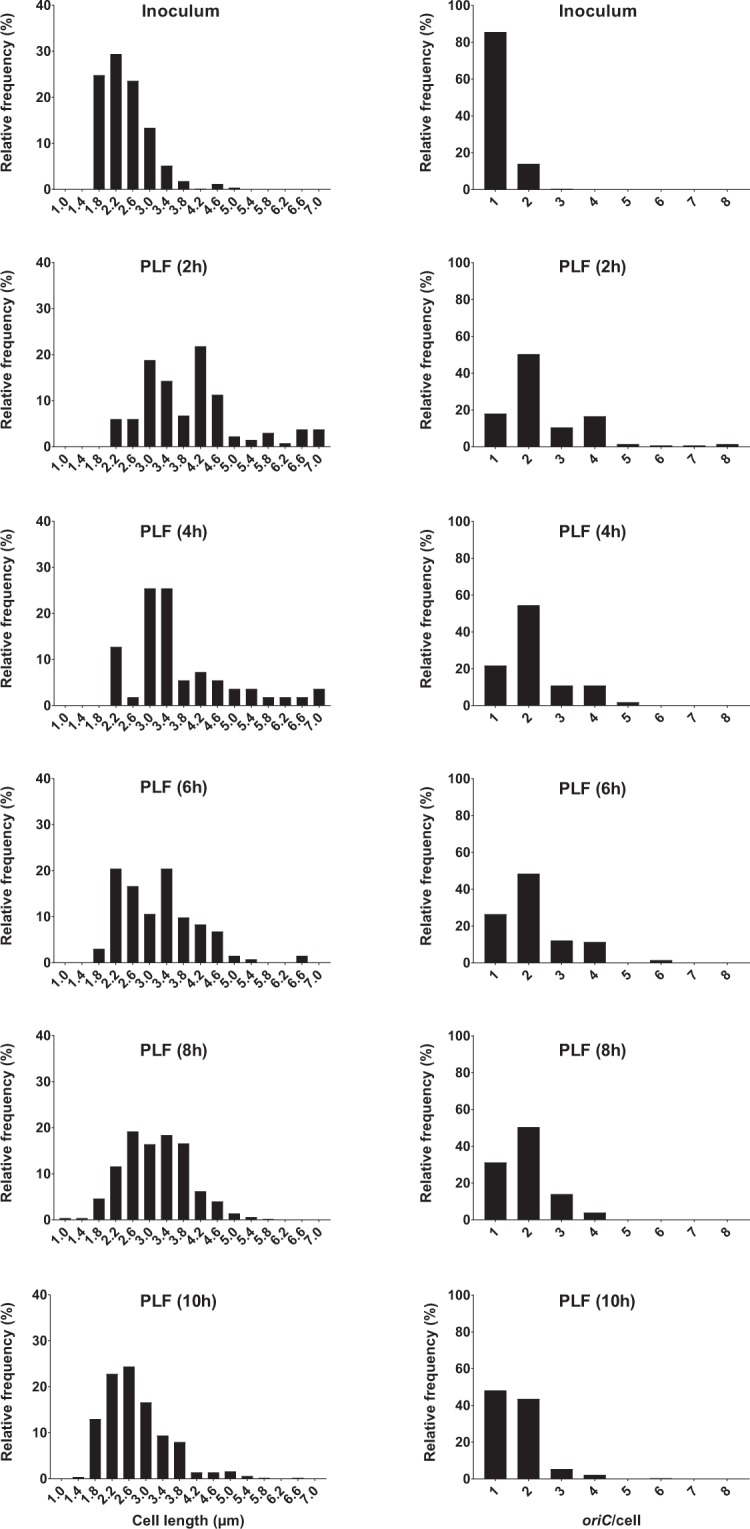
Relative frequency distributions (%) of pooled microscopically detected bacterial cells (ALO 4783) isolated from peritoneal lavage fluid (PLF) in the mouse peritonitis model; cell length (µm) (left panel) and oriC/cell (right panel). Inoculum, n = 500. PLF: 2 h, n = 133; 4 h, n = 55; 6 h, n = 132; 8 h, n = 500; 10 h, n = 500. We emphasize that data representing 4 hours of infection are subject to uncertainty, due to low number of detected cells (n < 100). Time (h) represents hours of infection.
The correlation between oriC/cell and cell size (µm) was very strong, both in PLF (r = 0.95, p < 0.01) and blood (r = 0.98, p < 0.05) bacterial populations (Fig. 4c).
In summary, when net population size was increasing, indicating growth, oriC/cell and ori:ter were high, and cell sizes were large (0–6 hours of infection) (Fig. 4b,c). After 6 hours of infection growth was gradually beginning to slow down, demonstrated by a slight decrease in both oriC/cell, ori:ter and cell size (µm). The data indicate the presence of a fixed initiation mass, also during infection in vivo. Unlike during growth in vitro, we did not observe a complete cessation of chromosome replication in vivo (oriC/cell and ori:ter remained >1 at all times during the course of the experiment) (Figs 4b, 5 and 6 and Supplementary Table S1). As demonstrated by the persistent growth heterogeneity (i.e. unbalanced growth), inferring τ from ori:ter during growth in vivo would be deceptive (Figs 5 and 6). However, ori:ter, as demonstrated, may be used as an independent measure of bacterial growth rate during infection in vivo.
Bacterial growth in vivo: mouse intravenous (i.v.) septicaemia model
To determine whether the bacterial population found in the bloodstream during peritonitis represented cells growing independently of that at the primary site of infection or not, we attempted to induce primary septicaemia directly via i.v. inoculation of 1 × 106 CFU/ml bacteria (ALO 4783). We were unable to establish an infection, as a complete clearance of bacteria from the bloodstream had occurred prior to the initial sample collection time point; 2 hours post bacterial challenge (Supplementary Fig. S2). No viable bacterial counts were found in blood, PLF or kidneys at any time. However, in the spleen we observed the presence of a stable bacterial population of (mean (SD)) 3.41 (0.03) log10 CFU/ml of all animals at all times (2–8 hours post challenge), likely as a result of bacterial bloodstream clearance.
None of the animals presented any signs of distress during the observation period, in contrary to the mouse peritonitis model, where signs of distress were starting to appear from 2 hours of infection.
Discussion
We have applied and validated two well-established methods for detection of bacterial chromosome replication to shed light on fundamental growth dynamics of E. coli during infection in vivo. ori:ter was determined by qPCR and fluorescence microscopy, respectively, overall with good agreement between methods. The microscopy method has the advantage that it allows for direct observation of in situ ori:ter at a single-cell level, which can be used to observe the level of replication heterogeneity within a bacterial population. Also, microscopy can be used to determine cell size, which demonstrated good correlation with the number of replication origins (oriC/cell), in agreement with initiation of chromosome replication taking place at a fixed initiation mass under all growth conditions. However, the fluorescence microscopy method requires precedent fluorescent labelling at relevant chromosome loci, along with a certain bacterial quantity for cells to be detected, which is a limitation to the method. The qPCR method, by contrast, reports only the mean ori:ter of a population, without regard to any possible heterogeneity. However, this method is less time consuming, uncomplicated and inexpensive, compared to whole genome sequencing. We conclude that both qPCR and fluorescence microscopy are valid methods for measuring in situ bacterial growth during infection, and that they complement each other favourably.
Bacterial growth in a closed batch-culture in vitro was shown to be directly related to nutrient availability, as anticipated32,33. Ori:ter levels were high, with a high level of replication heterogeneity, during the first hours of incubation. Subsequently, as the bacterial count increased, we observed a complete cessation of growth within a homogenous population of predominantly non-replicating cells, due to starvation after prolonged propagation33. A similar pattern of growth was observed in vivo during infection in the mouse peritonitis model. However, unlike during growth in vitro, we observed no complete cessation of growth in neither PLF nor blood as the bacterial densities reached their maximum in the terminal stage of infection. At this stage (8 to 10 hours of infection), the stagnant development in bacterial count is deceptive, as it gives the perception of growth cessation. Indeed, a complete cessation of growth might never occur during infection in vivo, where bacterial life-sustaining resources presumably are still available as long as the animal is alive. We show in this model that the apparent stationary phase during infection represents fractions of both slowly growing and non-growing cells, conceivably balanced by cells taken out by the host immune system. Hence, it is evident that one cannot extrapolate findings from in vitro growth studies to explain pharmacodynamics in relation to bacterial growth during infection in vivo, particularly not in regard to the apparent stationary phase. The in vitro effect of most commonly used antibiotics, including β-lactams, fluoroquinolones and aminoglycosides, has been shown to correlate to bacterial growth rate26,27. Chromosome replication as a readout for bacterial growth rate could form the basis for refined future studies on antibiotic treatment effect as a function of bacterial growth rate during infection in vivo.
In the mouse peritonitis model, significantly lower bacterial count densities could be found in the blood, relative to the PLF, yet ori:ter and oriC/cell remained similar in blood and PLF. These findings, taken together with the observed inability of the same strain to establish an infection upon direct introduction into the bloodstream (as demonstrated by the i.v. septicaemia model), suggest that E. coli found outside the primary site of infection (e.g. in the bloodstream) is not constituted by an independently growing population, but rather mirrors the population found at the primary site of infection. This is in consistence with previous in vivo studies of Streptococcus pneumoniae in the same experimental model, describing the phenomenon as ‘spill-over’30. Conceivably, bacterial cells grow at a given rate at the primary site of infection (e.g. the peritoneal cavity), followed by a translocation through lymphatic drainage into the systemic venous system via the thoracic duct and onward to relevant tissues, such as the spleen and kidneys where bacterial phagocytic clearance and filtration, respectively, take place11,34. Our findings are in accordance with previous studies indicating that E. coli, unlike bacteria such as Haemophilus influenzae, do not grow independently intravascularly8,35,36. Our data suggest that bacterial cells surviving in the bloodstream remain within the same state of growth as that of bacterial cells at the primary site of infection, without acceleration or significant deceleration; likely as the net result of constant influx- and kill-rate of bacteria. These findings underscore the importance of source control in septicaemia, i.e. locating and ensuring antibiotic availability at the primary site of infection, as this is where active bacterial growth is taking place.
In conclusion, we present data supporting the applicability of both qPCR and fluorescence microscopy as valid methods for determination of bacterial growth rate in vivo. The methods could be extrapolated to other infection models and other pathogenic bacteria. Ultimately, it would be evident to pursue the inexpensive and easily accessible qPCR method in a clinical setting to examine bacterial growth rates in infected body fluids, which could prove helpful in evaluating future antibacterial strategies.
Methods
Bacterial strains and inoculum preparations
Escherichia coli ATCC® 25922™, a clinical isolate from the American Type Culture Collection (Manassas (VA), USA) and CLSI and EUCAST control strain for antibiotic susceptibility testing, was used throughout the study. This strain was applied in vitro both as a wild-type and as a genetically modified version expressing fluorescent fusion-proteins at chromosomal sites corresponding to oriC and terC, respectively (ALO 4783). Only ALO 4783 was applied in the in vivo models.
For in vitro experiments, inoculum from frozen stock cultures were grown overnight in Lysogeny Broth (LB) broth, shaking 140 rpm, at 37 °C in ambient air. Bacterial counts were quantified by spotting 10 µl of 10-fold serial dilutions made in sterile physiological saline in duplicate on LB agar plates.
For in vivo experiments, bacteria were grown from frozen stock cultures overnight at 35 °C in ambient air on 5% blood agar plates (SSI Diagnostica, Copenhagen, Denmark), after which inocula were suspended in sterile physiological saline and quantified by measurement of optical density at 546 nm (OD546). Porcine mucin (M-2378, Sigma-Aldrich, Munich, Germany) was added as adjuvant to a final concentration of 5% (wt/vol). The mucin stock solution was prepared in physiological saline, sterilised and adjusted to physiological pH before application. The final inoculum (106 CFU/ml) was quantified by spotting 20 µl of serial dilutions in duplicate on selective bromothymol lactose blue agar plates (SSI Diagnostica, Copenhagen, Denmark).
Construction of double-loci fluorescent-labelled strain
For microscopic visualisation of cellular origins and termini, transgenes were inserted into the ATCC 25922 chromosome, by use of the lambda red recombineering technique previously described37,38. P1parS::KAN was PCR amplified from plasmid pMS2438 and inserted into the chromosome in the oriC region by use of lambda red recombineering plasmid pTP22339, using primers 5′-CATCATTAAAGTGCTGTACCGTAACTAACAGAAAGGCCTTAAAGCCGAAGCCTTAAACTT-3′ and 5′-GGGTTAATAGCGATTCAGAGTTCAAGGCCTTCTCCCGGAGGGAACTTCAAGATCCCCTTA-3′. Also, pMT1parS::CHL was PCR amplified from plasmid pGBkD3-parSpMT138 and inserted, by the same method, into the chromosome in the terC region using primers 5′- ATATAAATTCTATAATTAGATGTATCTTTCCATTTACGGCGTGTAGGCTGGAGCTGCTTC-3′ and 5′- TCGGTGTGAGATGCTTTACGTCTTCCAAGCCCCCTTCCTTCGTCTTACTGTCGGGAATTC-3′.
Furthermore, the pTrc-mCherry-pMTparB-GFP-P1parB cassette was PCR amplified from pJFM440 and inserted into the chromosomal attTN7 site at the 3′ end of the glmS gene, using the protocol for Tn7 transgene insertion as previously described41, resulting in strain ALO 4783. Co-expression of GFP-labelled P1-ParB proteins and mCherry-labelled pMT-ParB proteins29 in live cells allowed for visualisation of origins as GFP foci (green) and termini as mCherry foci (red), respectively. Correct insertion of all transgenes was confirmed by PCR.
It should be noted that a substantial part of the LacI operator on pRN01042, from which the pTrc-mCherry-pMTparB-GFP-P1parB cassette on pJFM4 originated, had unintentionally been lost during plasmid construction. Thus, the use of IPTG (isopropyl β-D-1-thiogalactopyranoside) for induction of the fluorescence fusion proteins proved redundant.
In vitro experiments
Overnight liquid cultures of both wild-type and genetically modified version of ATCC 25922 with a bacterial density of 109 CFU/ml were diluted 1:10,000 into fresh media and grown with shaking 140 rpm, at 37 °C.
Growth was observed by repeated measurements of optical density at 600 nm (OD600). Samples for quantification of bacterial count, qPCR analysis and fluorescence microscopy were withdrawn at time points 2, 4 and 6 hours of incubation in the exponential growth phase study, and at times 6, 8 and 10 hours of incubation for the stationary growth phase study, respectively. All samples were immediately set on ice after withdrawal. Both studies were performed in duplicate (i.e. including both wild-type ATCC 25922 as control of growth, and the genetically modified version, ALO 4783) in three independent experiments. As both growth curves and ori:ter were the same for both versions of the strain, these results were pooled for statistical analyses. For fluorescence microscopy, only ALO 4783 was applied.
Experimental animal models
Mouse peritonitis model
The mouse peritonitis model has been previously described and studied extensively43,44. Here, outbred female NMRI mice (BomTac: NMRI; weight 26–30 g; Taconic, Denmark) were applied throughout the study. The animals were kept in Macrolon type III cages in groups of three and allowed free access to feed and water. Experiments were initiated after an acclimatisation period of 5 days.
Inoculation was performed by intraperitoneal (i.p.) injection of 0.5 ml bacterial suspension containing 106 CFU/ml and 5% (wt/vol) mucin. At various time points of infection, the mice were anaesthetized with a subcutaneous (s.c.) injection of pre-mixed Zolazepam/Tiletamin (Zoletil, Virbac, Kolding, Denmark), Xylasin (Xysol Vet., ScanVet, Fredensborg, Denmark) and Butorphanol (Torbugesic Vet inj., Orion Pharma, Copenhagen, Denmark) prior to blood collection from total cardiocentesis, performed with a 30-gauge needle via subxiphoid access. Blood was stored in EDTA covered micro tubes (Sarstedt, Nümbrecht, Germany). After euthanasia by cervical dislocation, a peritoneal lavage was performed by injecting 2.0 ml of sterile physiological saline i.p. After 1 minute of abdominal massage, the peritoneum was opened and peritoneal lavage fluid (PLF) withdrawn with a pipette. Spleen and both kidneys were surgically removed using sterile procedures and placed in Eppendorf tubes.
All specimens were immediately placed in an insulated 4 °C cooling box for transportation and kept on ice at 4 °C until application in subsequent tests. Standard bacterial quantification and DNA purification were performed within two hours; fluorescence microscopy the subsequent day. To ensure that bacterial cells in the blood and PLF specimens did not undergo any alterations in any of the growth parameters measured (ori:ter, oriC/cell or cell size) while kept on ice at a non-permissive growth temperature45 post harvesting, we performed a confirmatory in vitro experiment where chromosome replication analyses (qPCR and fluorescence microscopy) were performed on exponentially growing cells left on ice for up to 24 hours (Supplementary Materials). The results combined confirmed that storage up to 24 hours on ice at non-permissive growth temperatures did not alter these parameters (Supplementary Fig. S3).
The mouse peritonitis model was repeated in a total of 6 independent experiments, where groups of three cohabitant animals were sacrificed at various combinations of time points; 2 (n = 15), 4 (n = 9), 6 (n = 6) 8 (n = 12) or 10 (n = 9) hours of infection.
Data from repeated experiments were pooled for statistical analyses.
Mouse intravenous septicaemia model
To examine the possibility of inducing primary bacteraemia without an established infection outside of the bloodstream, we applied a direct intravenous (i.v.) septicaemia mouse model. Mice, housing, acclimatisation and feeding procedures were identical to that of the mouse peritonitis model. However, here, the mice were inoculated via i.v. injection of 0.2 ml bacterial suspension containing 106 CFU/ml, without adjuvant, into the lateral tail vein. Animals were subsequently handled as above mentioned, with the extraction of blood, peritoneal wash fluid, spleen and kidneys, which were all kept at 4 °C after harvesting. The animals were sacrificed in groups of three biological replicates at 2 (n = 3), 4 (n = 3) and 8 (n = 3) hours of infection.
Ethics statement
Mice in both experimental models were regularly observed and scored for signs of distress. Humane end points were constituted by signs of irreversible sickness. The mice would be euthanized upon presentation of any of these signs. All animal experiments were approved by the Danish Animal Experimentation Inspectorate (Licence No. 2014-15-0201-00171) and performed according to institutional guidelines.
Quantification of bacterial growth
Samples from in vitro experiments were spotted (10 µl) in duplicate of serial dilutions on LB agar plates, and the bacterial count was recorded as the mean of two plates after overnight incubation at 37 °C in ambient air. The detection limit was 100 CFU/ml.
To the tubes containing spleen or kidneys from the in vivo experiments, sterile physiological saline was added to a total volume of 1 ml, after which they were homogenised for 2 minutes at 30 oscillations/sec using a TissueLyser II (Qiagen, US), before being spotted (20 µl) in serial dilutions in duplicate on bromothymol lactose blue agar plates. The required addition of saline before homogenisation of the tissues entailed a dilution factor of approximately 1:1. Blood and peritoneal wash fluid from in vivo experiments were spotted directly, without further dilution. Colony counts were performed after overnight incubation at 35 °C in ambient air and recorded as the mean of two plates. The detection limit for blood specimens was 50 CFU/ml. The other materials were recorded as CFU/ml of solution.
Quantitative PCR (qPCR)
ori:terqPCR was calculated as the population mean level of qPCR amplified oriC to terC, respectively.
Samples from in vitro experiments were prepared by pelleting 1 ml of culture by 5 min centrifugation at 15,0000 × g, after which bacterial cells were re-suspended in 100 µl 10 mM Tris pH 7.4 and 900 µl 77% Ethanol and kept at 4 °C until qPCR was performed. Here, cells were spun down and re-suspended in serial dilutions of sterile DNA/RNA free water prior to analysis.
Bacterial DNA from blood and peritoneal lavage fluid from in vivo experiments were purified for qPCR using QIAamp DNA Mini Kit (51304, Qiagen, Hilden, Germany), according to the manufacturer’s instructions. We were unable to purify adequate quantities of bacterial DNA from spleen and kidney tissue for use in this analysis, possibly due to co-purification of murine DNA and/or low bacterial counts in the tissues.
qPCR was performed as previously described, using primers 5′-CGCAACAGCATGGCGATAAC-3′ and 5′-TTCGATCACCCCTGCGTACA-3′ for partial amplification of the highly conserved gidA gene located immediately leftwards of oriC (representing the oriC region), and primers 5′-TCAACGTGCGAGCGATGAAT-3′ and 5′-TTGAGCTGCGCTTCATCGAG-3′ for partial amplification of the dcp gene, located in close proximity to terC, opposite the oriC region (representing the terC region), respectively28.
The analysis was performed using Takara SYBR Premix Ex Taq II (RR820A, Takara Bio, Saint-Germain-en-Laye, France) in a BioRAD CFX96 (95 °C 30 s, 39 × (95 °C 5 s + 58 °C 30 s), 95 °C 15 s, 60 °C 60 s, 95 °C −15 s, 65 °C −5 s, 95 °C − ∞), as previously reported40. The ori:terqPCR was calculated using comparative cycle threshold (Ct) analysis; the 2∆∆Ct method46. A fixed sample of the same strain grown into late stationary phase, where the population would be expected to have an ori:ter corresponding to 1, was used for normalisation in every cycling run.
Each biological replicate was run as a minimum as three technical replicates in each cycling run, and the mean Ct value of the technical triplicates (or more) was used to calculate the ori:terqPCR. DNA/RNA-free water was used as negative control template in each run. Furthermore, QIAamp PCR purified blood and peritoneal lavage fluid specimens without viable bacterial growth were regarded as negative controls for blood and peritoneal lavage fluid derived specimens. There was a qPCR detection limit corresponding to approximately 103 CFU/ml for purified blood specimens. Hence, not all purified blood specimens yielded adequate qPCR results. No detection limit was observed in samples from in vitro experiments.
Fluorescence microscopy
ori:termic was calculated from automated photomicrographic detection of pooled single bacterial cells carrying intracellular fluorescent markers corresponding to oriC and terC, respectively.
For phase contrast and fluorescence microscopy, bacterial cells were spun down for 2 minutes at 6,000 × g, re-suspended in a small volume of AB minimal medium47 and mounted on microscope slides covered with a thin 2% (wt/vol) AB minimal medium agarose pad.
Blood specimens from in vivo studies underwent murine cell lysis prior to pelleting, by the addition of cold, sterile water mixed with the sample at a ratio of approximately 1:1 for 1 minute. Harvested tissues were not observed in the microscope.
A total of 500 live bacterial cells were pooled and analysed from each time point, with the exception of certain specimens, where this was not possible due to the quantity of live cells available for microscopy detected being too low (Supplementary Table S1). The ori:termic was deduced from the mean of oriC foci detected divided by the mean of terC foci detected.
It should be noted that fractions of the bacterial cells were recorded with a number oriC copies ≠ 2n (n = 1, 2, 3). The risk of underestimation of co-localising oriCs is a known microscopy resolution limitation in photomicrographic foci quantification48. It is unlikely that the bacterial cells propagating in vitro underwent asynchronous replication initiation4. Accordingly, cells reported with an oriC/cell of 3 in all likelihood represent 4 oriC/cell, and cells reported with an oriC/cell > 4 in all likelihood represent 8 oriC/cell (Fig. 3). Since less is known about chromosome replication during infection in vivo, we cannot rule out any possible replication initiation asynchrony in bacterial populations from the in vivo experiments (Figs 5 and 6).
Medial axis cell length (µm) was recorded by automated detection and used as surrogate measure for cell mass.
Images were acquired with a Zyla 5.5 sCMOS camera attached to a Nikon Eclipse Ti-E inverted microscope and analysed with NIS-ELEMENTS and MicrobeJ software for automated bacterial cell and fluorescent foci detection and quantification49.
Statistical analyses
Bacterial quantification data were log10 transformed prior to analysis. D’Agostino and Pearson omnibus normality test was applied to all data sets. In general, the bacterial count and the qPCR data sets represented a normal distribution. Cell length and numbers of oriC foci detected by microscopy did not all meet the assumptions for normal distribution. Linear regression analysis was used to compare bacterial counts or ori:ter between different materials harvested in the in vivo experiments. Additionally, two-way ANOVA followed by Sidak’s multiple comparisons test was applied within the same data sets to test difference between blood and peritoneal ori:ter at each time point. Pearson’s correlation coefficient was applied to estimate correlation between cell length (µm) and oriC/cell. The Kolmogorov-Smirnov test was applied to test differences in oriC/cell distributions between bacterial populations in the blood and PLF, respectively, at different time points. Bland-Altman analysis was performed to evaluate agreement between the qPCR and the microscopy method for detection of ori:ter. A two-tailed p value < 0.05 was considered significant.
GraphPad Prism version 7 (GraphPad Software, CA, USA) was applied for statistical analysis and illustration.
Electronic supplementary material
Acknowledgements
Plasmids pMS24 and pGBkD3-parSpMT1 were generously provided by Francois Cornet, and plasmid pTP223 likewise by Martin Marinus. We thank Jytte Mark Andersen and colleagues at Statens Serum Institut for technical assistance with the animal experiments. The study was partly funded by an EU-IMI Joint Undertaking, grant agreement No. 115583 (ENABLE), and by the Danish National Research Foundation (DNRF120) through the Centre for Bacterial Stress Response and Persistence (BASP).
Author Contributions
N.F.-M., A.L.-O., G.C. and M.S.H. conceptualised the study. N.F.-M. and A.L.-O. secured funding. M.S.H. carried out experiments, performed statistical analyses and drafted the manuscript. A.L.-O., N.F.-M. and G.C. provided critical analysis and discussions. All of the authors reviewed, approved and contributed to the final version of the manuscript.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Niels Frimodt-Møller and Anders Løbner-Olesen jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33264-7.
References
- 1.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli Br. J. Mol. Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 2.Wang JD, Levin PA. Metabolism, cell growth and the bacterial cell cycle. Nat. Rev. Microbiol. 2009;7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donachie WD. Relationship between Cell Size and Time of Initiation of DNA Replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 4.Skarstad K, Boye E, Steen HB. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 1986;5:1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stracy M, Uphoff S, Garza de Leon F, Kapanidis AN. In vivo single-molecule imaging of bacterial DNA replication, transcription, and repair. FEBS Lett. 2014;588:3585–3594. doi: 10.1016/j.febslet.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Haeusser DP, Levin PA. The Great Divide: Coordinating cell cycle events during bacterial growth and division. Curr. Opin. Microbiol. 2008;11:94–99. doi: 10.1016/j.mib.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peleg M, Corradini MG. Microbial Growth Curves: What the Models Tell Us and What They Cannot. Crit. Rev. Food Sci. Nutr. 2011;51:917–945. doi: 10.1080/10408398.2011.570463. [DOI] [PubMed] [Google Scholar]
- 8.Rubin LG. Comparison of in vivo and in vitro multiplication rates of Haemophilus influenzae type b. Infect. Immun. 1986;52:911–913. doi: 10.1128/iai.52.3.911-913.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith H. What Happens in Vivo to Bacterial Pathogens? Ann. N. Y. Acad. Sci. 1996;797:77–92. doi: 10.1111/j.1749-6632.1996.tb52951.x. [DOI] [PubMed] [Google Scholar]
- 10.Brock TD. Microbial growth rates in nature. Bacteriol. Rev. 1971;35:39–58. doi: 10.1128/br.35.1.39-58.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith H. Host factors that influence the behaviour of bacterial pathogens in vivo. Int. J. Med. Microbiol. 2000;290:207–213. doi: 10.1016/S1438-4221(00)80117-4. [DOI] [PubMed] [Google Scholar]
- 12.Korem T, et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015;349:1101–1106. doi: 10.1126/science.aac4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown CT, Olm MR, Thomas BC, Banfield JF. Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 2016;34:1256–1263. doi: 10.1038/nbt.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olm MR, et al. Identical bacterial populations colonize premature infant gut, skin, and oral microbiomes and exhibit different in situ growth rates. Genome Res. 2017;27:601–612. doi: 10.1101/gr.213256.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewar JM, Walter JC. Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 2017;18:507. doi: 10.1038/nrm.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leonard, A. C. & Grimwade, J. E. Initiation of DNA replication. EcoSal Plus4 (2010). [DOI] [PMC free article] [PubMed]
- 17.Wallden M, Fange D, Lundius EG, Baltekin Ö, Elf J. The Synchronization of Replication and Division Cycles in Individual E. coli Cells. Cell. 2016;166:729–739. doi: 10.1016/j.cell.2016.06.052. [DOI] [PubMed] [Google Scholar]
- 18.Huisman O, D’Ari R. An inducible DNA replication–cell division coupling mechanism in E. coli. Nature. 1981;290:797. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- 19.Hill Norbert S., Kadoya Ryosuke, Chattoraj Dhruba K., Levin Petra Anne. Cell Size and the Initiation of DNA Replication in Bacteria. PLoS Genetics. 2012;8(3):e1002549. doi: 10.1371/journal.pgen.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Løbner-Olesen A, Skarstad K, Hansen FG, von Meyenburg K, Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989;57:881–889. doi: 10.1016/0092-8674(89)90802-7. [DOI] [PubMed] [Google Scholar]
- 21.Skarstad K, Katayama T. Regulating DNA Replication in Bacteria. Cold Spring Harb. Perspect. Biol. 2013;5:a012922–a012922. doi: 10.1101/cshperspect.a012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Si F, et al. Invariance of Initiation Mass and Predictability of Cell Size in Escherichia coli. Curr. Biol. 2017;27:1278–1287. doi: 10.1016/j.cub.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmstetter CE, Cooper S. DNA synthesis during the division cycle of rapidly growing Escherichia coli Br. J. Mol. Biol. 1968;31:507–518. doi: 10.1016/0022-2836(68)90424-5. [DOI] [PubMed] [Google Scholar]
- 24.Bremer H, Churchward G. An examination of the Cooper-Helmstetter theory of DNA replication in bacteria and its underlying assumptions. J. Theor. Biol. 1977;69:645–654. doi: 10.1016/0022-5193(77)90373-3. [DOI] [PubMed] [Google Scholar]
- 25.Michelsen O. Precise determinations of C and D periods by flow cytometry in Escherichia coli K-12 and B/r. Microbiology. 2003;149:1001–1010. doi: 10.1099/mic.0.26058-0. [DOI] [PubMed] [Google Scholar]
- 26.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. The rate of killing of Escherichia coli byβ-lactam antibiotics is strictly proportional to the rate of bacterial growth. Microbiology. 1986;132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 27.Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 1991;35:1824–1828. doi: 10.1128/AAC.35.9.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riber L, et al. Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev. 2006;20:2121–2134. doi: 10.1101/gad.379506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 2006;61:383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- 30.Frimodt-Møller N, Sebbesen O, Frølund Thomsen V. The pneumococcus and the mouse protection test: importance of the lag phase in vivo. Chemotherapy. 1983;29:128–134. doi: 10.1159/000238186. [DOI] [PubMed] [Google Scholar]
- 31.Boye E, Løbner-Olesen A. Bacterial growth control studied by flow cytometry. Res. Microbiol. 1991;142:131–135. doi: 10.1016/0923-2508(91)90020-B. [DOI] [PubMed] [Google Scholar]
- 32.Bren A, Hart Y, Dekel E, Koster D, Alon U. The last generation of bacterial growth in limiting nutrient. BMC Syst. Biol. 2013;7:27. doi: 10.1186/1752-0509-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli Physiology in Luria-Bertani Broth. J. Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohnsack JF, Brown EJ. The Role of the Spleen in Resistance to Infection. Annu. Rev. Med. 1986;37:49–59. doi: 10.1146/annurev.me.37.020186.000405. [DOI] [PubMed] [Google Scholar]
- 35.Rogers DE. Host mechanisms which act to remove bacteria from the blood stream. Bacteriol. Rev. 1960;24:50–66. doi: 10.1128/br.24.1.50-66.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander WJ, Alexander LS, Curtiss R. Bactericidal activity of human serum against Escherichia coli chi1776. Infect. Immun. 1980;28:837–841. doi: 10.1128/iai.28.3.837-841.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stouf M, Meile J-C, Cornet F. FtsK actively segregates sister chromosomes in Escherichia coli. Proc. Natl. Acad. Sci. 2013;110:11157–11162. doi: 10.1073/pnas.1304080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charbon G, Bjørn L, Mendoza-Chamizo B, Frimodt-Møller J, Løbner-Olesen A. Oxidative DNA damage is instrumental in hyperreplication stress-induced inviability of Escherichia coli. Nucleic Acids Res. 2014;42:13228–13241. doi: 10.1093/nar/gku1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenzie GJ, Craig NL. Fast, easy and efficient: site-specific insertion of transgenes into Enterobacterial chromosomes using Tn 7 without need for selection of the insertion event. Bmc Microbiol. 2006;6:39. doi: 10.1186/1471-2180-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadoya R, Chattoraj DK. Insensitivity of Chromosome I and the Cell Cycle to Blockage of Replication and Segregation of Vibrio cholerae Chromosome II. mBio. 2012;3:e00067-12–e00067-12. doi: 10.1128/mBio.00067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knudsen, J. D. & Frimodt-Moller, N. Animal models in bacteriology. In Animal Testing in Infectiology9, 1–14 (Karger Publishers, 2001). [DOI] [PubMed]
- 44.Frimodt-Møller N. The mouse peritonitis model: present and future use. J. Antimicrob. Chemother. 1993;31:55–60. doi: 10.1093/jac/31.suppl_D.55. [DOI] [PubMed] [Google Scholar]
- 45.Jones T, Gill CO, McMullen LM. The behaviour of log phase Escherichia coli at temperatures that fluctuate about the minimum for growth. Lett. Appl. Microbiol. 2004;39:296–300. doi: 10.1111/j.1472-765X.2004.01593.x. [DOI] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.Clark DJ, Maaløe O. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 1967;23:99–112. doi: 10.1016/S0022-2836(67)80070-6. [DOI] [Google Scholar]
- 48.Nielsen HJ, Hansen FG. An automated and highly efficient method for counting and measuring fluorescent foci in rod-shaped bacteria. J. Microsc. 2010;239:194–199. doi: 10.1111/j.1365-2818.2010.03374.x. [DOI] [PubMed] [Google Scholar]
- 49.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.


