Abstract
Candida auris has recently emerged as a multi-drug resistant fungal pathogen that poses a serious global health threat, especially for patients in hospital intensive care units (ICUs). C. auris can colonize human skin and can spread by physical contact or contaminated surfaces and equipment. Here, we show that the mycoparasitic yeast Saccharomycopsis schoenii efficiently kills both sensitive and multi-drug resistant isolates of C. auris belonging to the same clade, as well as clinical isolates of other pathogenic species of the Candida genus suggesting novel approaches for biocontrol.
Introduction
Candida auris was first seen as an agent of human disease in 2009, when it had been isolated from the ear canal of a patient in Japan1. It has subsequently spread rapidly around the world and is now a major health threat on a global scale having been associated with potentially lethal infections in patients in ICUs in Eastern and South Asia, South Africa, Europe, the USA, and South America. Sequence-based analyses have grouped C. auris isolates from around the world into at least four different clades, represented by clonal populations2. More than 90% of isolates are fluconazole resistant and many isolates are cross-resistant to more than one of the three major classes of antifungals – azoles, echinocandins and polyenes2. A few strains of this fungus are resistant to all of the major classes of antifungals used by doctors to treat fungal infections. The species has a propensity to colonize skin, and it has proven to be difficult to eradicate from ICUs. Worldwide mortality rates for disease cases with C. auris infections of the bloodstream approach 50%. Concern about the emergence and spread of C. auris has resulted in alerts being posted by the CDC (Centers for Disease Control and Prevention, USA), the ECDC (European Centre for Disease Prevention and Control, Sweden), and PHE (Public Health England, UK)3–5.
C. auris colonizes the skin of patients and can be transmitted via contact with patients or contaminated hospital fixtures, which has already resulted in several health care associated outbreaks6. The horizontal transmission potential of C. auris demands strict decontamination methods and infection prevention protocols, since mortality rates in patients with systemic infections can be up to 50%2,7,8.
Necrotrophic mycoparasitism describes the ability of a fungal species to kill other fungi9. For example, filamentous fungi in the Trichoderma genus have been well characterized over the last decades and are used as biocontrol agents against fungal plant pathogens10. Mycoparasites typically exhibit wide host ranges and due to their general mode of action in killing their prey may use broad-acting lytic enzymes, such as proteases and chitinases. However, some species, e.g. Trichoderma, also generate specialized penetration structures or haustoria that allow them to efficiently target and kill prey cells11,12. Active mycoparasitism in yeast was discovered only in 1997, when species of the genus Saccharomycopsis were first described as necrotrophic predacious yeasts13. It has not been studied whether Saccharomycopsis species attack Candida species other than Candida albicans13,14. Predacious behaviour depends on solid structural support, presumably to allow for stable cell-cell contact and has been suggested to be starvation induced14. Specifically, a lack of organic sulfur-containing organic compounds such as methionine has been suggested as a trigger for predation, as all Saccharomycopsis yeasts share the, for microorganisms, rare feature of being unable to assimilate sulfate as their sole source of sulfur. Recently, we reported the lack of eight genes in the sulfate assimilation pathway in draft genomes of Saccharomycopsis fodiens and Saccharomycopsis fermentans15,16.
Here we show that S. schoenii efficiently attacks and kills a range of pathogenic Candida species, including the newly emerged human pathogenic fungus C. auris. We follow the predation process using time lapse microscopy in combination with fluorescent dyes. Efficient predation as shown here could be useful for biocontrol purposes in either clinical settings for skin clearance or in agricultural settings for combatting plant pathogens.
Results and Discussion
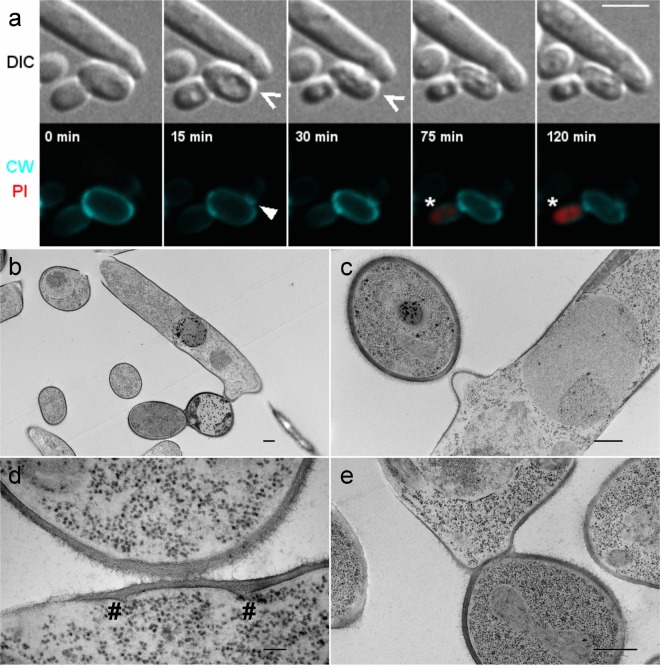
In this study, we prospected the use of a predatory yeast, Saccharomycopsis schoenii, as a potential biocontrol agent against human fungal pathogens of the Candida clade with a focus on C. auris. To this end we confronted multiple drug resistant strains including C. auris NCPF8985#20, a multi-drug resistant isolate from the South Asian clade (India), with S. schoenii (Supplementary Table 1). Equal numbers of dimorphic S. schoenii and ovoid C. auris NCPF8985#20 cells were seeded on minimal media agar on microscopy slides to offer solid support for a potential S. schoenii interaction. This minimal media lacked methionine and thus did not support proliferation of S. schoenii in pure culture. We found that S. schoenii attacked C. auris cells upon contact and killed prey cells using specialized penetration pegs (Fig. 1; Supplementary Movies 1–3). The chitin-staining fluorescent dye Calcofluor White, revealed septa at the bases of penetration pegs indicating the sites of entry (Fig. 1a; Supplementary Movies 1 and 2). Within minutes after S. schoenii cells generated penetration pegs, C. auris cells started to vacuolarize, take up dyes such as propidium iodide that are not permeable to living cells and then collapse, presumably because of feeding and material transfer to the predating S. schoenii cell (Fig. 1a; Supplementary Movies 1–3). We prepared Transmission Electron Microscopy (TEM) images of interactions between S. schoenii and C. auris after 1 h of co-culture on minimal media (Fig. 1b–e) and found that C. auris cells attacked by S. schoenii cells were necrotic (Fig. 1b). Penetration pegs were directed at prey cells (Fig. 1c) and cell wall interactions led to the formation of penetration peg start sites (# Fig. 1d). Ultimately this led to degradation of the prey cell wall (Fig. 1e). After killing of prey cells, penetration pegs did not grow further or develop into buds or daughter cells (Supplementary Movie 3). During their interaction with prey cells, S. schoenii cells did not show apical growth, however, S. schoenii cells resumed polar growth after successful attacks (Supplementary Movie 3).
Figure 1.
S. schoenii attacks and kills C. auris. (a) S. schoenii and C. auris NCPF8985#20 stained with Calcoflour White (CW, cyan, bottom panel), a fluorescent dye that stains chitin rich cell walls and septa, and ropidium iodide (PI, red, bottom panel), a fluorescent dye that stains nucleic acids of cells with a compromised cell membrane, i.e. dead or dying cells. We captured images twice per minute for two hours and found that at 15 min, a penetration peg [Δ] from S. schoenii is visualized by CW. The C. auris prey cell subsequently collapses in size between 15 and 30 min (Λ). Whereas the attacked C. auris cell was not stained by PI, its daughter cell accumulated PI between 75 min to 120 min (*). (b–e) TEM images of S. schoenii and C. auris that had been co-cultured for 1 h. Scale bar 500 nm in (b,c and e). Scale bar 100 nm in (d). (b) A dimorphic S. schoenii cell has formed a penetration peg to contact, attack and kill an ovoid C. auris cell. (c) A S. schoenii cell with a penetration peg protruding towards a prey cell. (d) Early interactions between A. schoenii and C. auris visualize potential penetration peg start sites (#). (e) Partial disintegration of the C. auris cell wall.
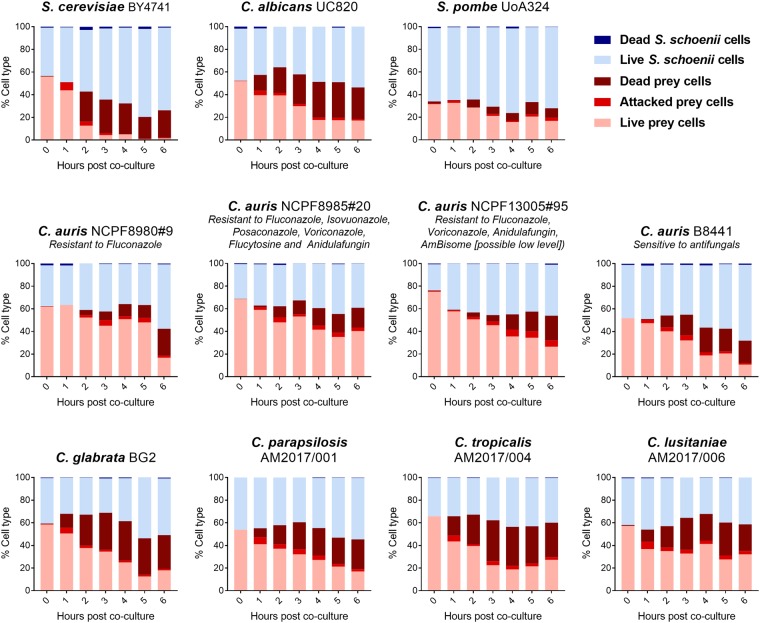
We determined rates of killing over a 6 h time-course using morphology and/or propidium iodide staining (PI) stain (Supplementary Figs 1 and 2). Several prey cells were found to accumulate PI staining upon predation, however, many prey cells were apparently killed without being stained by PI. In these cases, killed prey cells were “flattened” and or shrunken in size. This resulted in the death of around 34% of C. auris cells within a period of 6 h of co-culture with S. schoenii (Fig. 2, middle panel; Supplementary Table 2). As a control, almost none of the C. auris cells (0.6%) had died after 6 h when cultured alone under identical experimental conditions. To examine if predator-prey interactions differ with different C. auris isolates that exhibit variable drug resistance phenotypes, we analysed predator-prey interactions in three additional C. auris isolates (Supplementary Table 1). Furthermore, to elucidate host range of predator-prey interactions within Candida species we included clinical isolates of Candida albicans, Candida glabrata, Candida lusitaniae, Candida parapsilosis and Candida tropicalis in this analysis. For reference, we used Saccharomyces cerevisiae and Schizosaccharomyces pombe, two previously known prey species of S. schoenii14. All isolates of Candida species tested, including several drug resistant C. auris strains, were susceptible to predation by S. schoenii (Fig. 2 and Supplementary Table 2).
Figure 2.
Kill curves of prey species attacked and killed by S. schoenii. Cells were co-cultured on several slides with SD media with 1% agarose, for up to 6 h. Every hour, we took one slide and captured 3 representative frames. Cells were scored on viability based on morphology and PI stain as per Fig. S2.
Collectively, these results demonstrate that the predator yeast S. schoenii provides a novel opportunity to develop biocontrol methods for skin disinfection. Saccharomycopsis predator are unique within the Saccharomycetes in displaying predatory behaviour. Thus these yeasts may harbor potential as biocontrol agents of other fungi including human and plant pathogens. Based on genome survey sequencing, Saccharomycopsis yeasts, like the distantly related filamentous ascomycete Trichoderma, harbor multi-gene families of proteases and chitinases15,16 (our unpublished data). These multi-gene families probably represent a resource for the identification of lytic enzymes that have the potential to generate novel antifungal compounds.
Methods
Strains
Wild-type Saccharomycopsis schoenii (CBS 7425, CBS-KNAW collection, Utrecht, Netherlands) was provided by Marc-André Lachance. Schizosaccharomyces pombe UoA324, a derivative of FY15112 from NBRP yeast and SO2427 from Snezhana Oliferenko (King’s College London). Clinical isolates of the following C. auris and other Candida strains were used; C.auris (NCPF8980#9), C. auris (NCPF8985#20) and C. auris (NCPF13005#95) provided by Liz Johnson (PHE Bristol) and C. auris (B8441) provided by Shawn Lockhart (CDC Atlanta), Candida albicans (UC820) provided by Mihai Netea (Radboudumc, Nijmegen), Candida glabrata (BG2) provided by Brendan P. Cormack (Johns Hopkins University, Baltimore, MD), and Candida parapsilosis (AM2017/001), Candida lusitaniae (AM2017/006) and Candida tropicalis (AM2017/004) provided by Donna MacCallum (University of Aberdeen).
Media and Growth conditions
Cells were cultured to log phase in YPD media (10 g/L yeast extract, 20 g/L Bacto peptone, 20 g/L glucose), 30 °C, rotating. Cells were washed and stained with Calcofluor White (10 µg/mL) and propidium iodide (1 µg/mL). S. schoenii was mixed with prey cells at roughly 5*107 cells/mL each, in Synthetic Defined (SD) media (6.7 g/L YNB w/o amino acids with ammonium sulfate, 20 g/L glucose). Prior to imaging, cells were seeded on pads with SD media solidified with 1% agarose.
Microscopy
Imaging was performed using the PerkinElmer UltraVIEW VoX Spinning Disk Confocal Microscope controlled by Volocity software. Images for movies were captured 2–4 times/min for up to 2 h, using the Nikon Perfect Focus System to autofocus. For kill curve analyses, three frames were captured every hour per species and time point. FIJI/ImageJ17 was used for image processing and analysis. Drift in movies frames was corrected using the macro NMS fixTranslation v1 and the plugin Image Stabiliser. For kill curve analyses, individual cells were counted using the Cell counter plugin.
For TEM images, S. schoenii and C. auris cells were separately pre-cultured to log phase in YPD media, then washed and mixed together at equal ratios. A total 1*108 cells were seeded on SD media solidified with 2% agarose. After 1 h of co-culture, cells were scraped off, washed and pelleted. High Pressure Freezing was carried out using a Leica EM PACT 2 (Leica Microsystems, Milton Keynes, UK) and samples were freeze substituted in a Leica AFS 2. Freeze substitution was carried out using the following program: −95 °C to −90 °C for 30 h with 2% OsO4 in acetone, −90 °C for 10 h with 2% OsO4 in acetone, −90 °C to −30 °C for 8 h with 2% OsO4 in acetone, −30 °C to −10 °C for 1 h with acetone, −10 °C to 4 °C for 1 h in acetone, 4 °C to 20 °C for 1 h in acetone. Samples were then removed and placed in 10% Spurr’s (TAAB, UK): acetone for 72 h, followed by 30% Spurr’s overnight, 50% Spurr’s for 8 h, 70% Spurr’s overnight, 90% Spurr’s for 8 h and embedded in Spurr’s resin at 60 °C for at least 24 h. Ultrathin sections were cut to 90 µm using a diamond knife (Diatome Ltd, Switzerland) onto copper grids (TAAB, UK) using a Leica UC6 and were contrast stained with uranyl acetate and lead citrate in a Leica AC20. Samples were imaged on a JEM 1400 plus (JEOL UK) Transmission Electron Microscope and captured using an AMT UltraVUE camera (AMT, USA). All relevant data are available from the authors.
Electronic supplementary material
Acknowledgements
This research was supported by the European Union Marie Curie Initial Training Network Fungibrain (607963), the Wellcome Trust (101873, 204815, 086827, 075470, 099215, and 097377), and the MRC Centre for Medical Mycology at the University of Aberdeen (MR/N006364/1). We thank Gillian Milne of the Microscopy and Histology facility at the University of Aberdeen for expert assistance with TEM. Fungal strains were kindly provided by Brendan P. Cormack, Elizabeth Johnson, Marc-André Lachance Shawn Lockhart, Donna MacCallum, Mihai Netea, Snezhana Oliferenko, and the National BioResource Project (NBRP) Japan.
Author Contributions
K.J., J.W., A.L. and N.A.R.G. conceived this study. K.J., G.B.R., A.L., and L.W. conducted the experiments. K.J. and A.L. designed and conducted the predation assays and its analyses. J.W., K.J. and N.A.R.G. drafted the manuscript. All authors read, revised and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Neil A. R. Gow, Email: n.gow@abdn.ac.uk
Jürgen Wendland, Email: jurgen.wendland@vub.be.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33199-z.
References
- 1.Satoh K, et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41–4. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart SR, et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin Infect Dis. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeffery-Smith, A. et al. Candida auris: a Review of the Literature. Clin Microbiol Rev31, e00029-17 (2018). [DOI] [PMC free article] [PubMed]
- 4.Chowdhary Anuradha, Sharma Cheshta, Meis Jacques F. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLOS Pathogens. 2017;13(5):e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lockhart SR, Berkow EL, Chow N, Welsh RM. Candida auris for the Clinical MicrobiologyLaboratory: Not Your Grandfather’s Candida Species. Clin Microbiol Newsl. 2017;39:99–103. doi: 10.1016/j.clinmicnews.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welsh RM, et al. Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J Clin Microbiol. 2017;55:2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schelenz S, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kean R., Sherry L., Townsend E., McKloud E., Short B., Akinbobola A., Mackay W.G., Williams C., Jones B.L., Ramage G. Surface disinfection challenges for Candida auris : an in-vitro study. Journal of Hospital Infection. 2018;98(4):433–436. doi: 10.1016/j.jhin.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson, M., Atanasova, L., Jensen, D. F. & Zeilinger, S. Necrotrophic Mycoparasites and Their Genomes. Microbiol Spectr5 (2017). [DOI] [PMC free article] [PubMed]
- 10.Hermosa R, Viterbo A, Chet I, Monte E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology. 2012;158:17–25. doi: 10.1099/mic.0.052274-0. [DOI] [PubMed] [Google Scholar]
- 11.Druzhinina IS, et al. Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol. 2011;9:749–59. doi: 10.1038/nrmicro2637. [DOI] [PubMed] [Google Scholar]
- 12.Schmoll M, et al. The Genomes of Three Uneven Siblings: Footprints of the Lifestyles of Three Trichoderma Species. Microbiol Mol Biol Rev. 2016;80:205–327. doi: 10.1128/MMBR.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lachance MA, Pang WM. Predacious yeasts. Yeast. 1997;13:225–32. doi: 10.1002/(SICI)1097-0061(19970315)13:3<225::AID-YEA87>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 14.Lachance MA, Pupovac-Velikonja A, Natarajan S, Schlag-Edler B. Nutrition and phylogeny of predacious yeasts. Can J Microbiol. 2000;46:495–505. doi: 10.1139/w00-021. [DOI] [PubMed] [Google Scholar]
- 15.Junker, K., Hesselbart, A. & Wendland, J. Draft Genome Sequence of Saccharomycopsis fodiens CBS 8332, a Necrotrophic Mycoparasite with Biocontrol Potential. Genome Announc5, e01278-17 (2017). [DOI] [PMC free article] [PubMed]
- 16.Hesselbart, A., Junker, K. & Wendland, J. Draft Genome Sequence of Saccharomycopsis fermentans CBS 7830, a Predacious Yeast Belonging to the Saccharomycetales. Genome Announc6, e01445-17 (2018). [DOI] [PMC free article] [PubMed]
- 17.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.