Abstract
Regulation of chromatin structure is critical for cell type-specific gene expression. Many chromatin regulatory complexes exist in several different forms, due to alternative splicing and differential incorporation of accessory subunits. However, in vivo studies often utilize mutations that eliminate multiple forms of complexes, preventing assessment of the specific roles of each. Here we examined the developmental roles of the TIP55 isoform of the KAT5 histone acetyltransferase. In contrast to the pre-implantation lethal phenotype of mice lacking all four Kat5 transcripts, mice specifically deficient for Tip55 die around embryonic day 11.5 (E11.5). Prior to developmental arrest, defects in heart and neural tube were evident in Tip55 mutant embryos. Specification of cardiac and neural cell fates appeared normal in Tip55 mutants. However, cell division and survival were impaired in heart and neural tube, respectively, revealing a role for TIP55 in cellular proliferation. Consistent with these findings, transcriptome profiling revealed perturbations in genes that function in multiple cell types and developmental pathways. These findings show that Tip55 is dispensable for the pre- and early post-implantation roles of Kat5, but is essential during organogenesis. Our results raise the possibility that isoform-specific functions of other chromatin regulatory proteins may play important roles in development.
Introduction
Regulation of chromatin structure is necessary to establish and maintain cell type-specific gene expression patterns during development. Generation of active or repressive chromatin architecture at gene enhancers and promoters is necessary for the binding and activities of lineage specific transcription factors1,2. On a broader scale, large chromosomal domains of active or repressive chromatin structure help to enforce gene expression patterns particular to each cell lineage3. Consequently, mutations in subunits of chromatin remodeling factors—multisubunit protein complexes that regulate chromatin architecture through a range of enzymatic activities—frequently result in developmental abnormalities4.
The lysine acetyltransferase KAT5 (originally named TIP60) is conserved throughout eukaryotes and activates gene expression through acetylation of histones H2A (and H2A variants), H4, and numerous transcription factors5–9. The KAT5 acetyltransferase activity is also necessary for the cellular response to DNA damage, in part by remodeling chromatin structure near sites of DNA damage7,8,10–12. Interestingly, KAT5 also has an essential non-catalytic role in regulation of chromatin accessibility and gene expression in embryonic stem cells and pre-implantation embryos13. KAT5 is a component of the 17-subunit TIP60-P400 complex, which remodels chromatin architecture not only through acetylation of histone tails, but also through incorporation of the H2A.Z histone variant into nucleosomes. Previously, individual knockdown of multiple subunits of TIP60-P400 resulted in loss of embryonic stem cell self-renewal, as well as a defect in differentiation14,15. Consistent with these phenotypes, mice with homozygous null mutations of the Kat5 gene arrest development at E3.5, prior to implantation, and Kat5−/− blastocysts fail to hatch from the zona pellucida when cultured in vitro16.
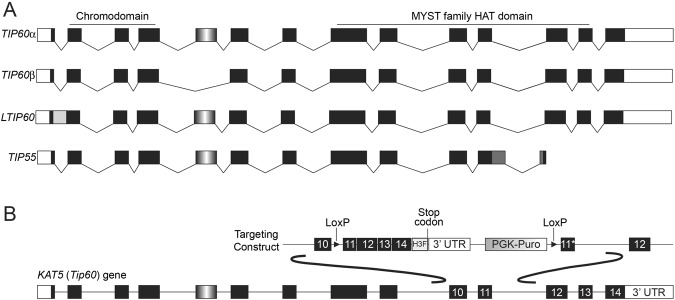
Although most studies of Kat5 functions have focused on a single isoform, denoted Tip60α, several isoforms are expressed in mouse and human by virtue of alternative splicing. Four isoforms are expressed in mouse, Tip60α, Tip60β, LTip60, and Tip55, which differ by alternative inclusion of intron 1, exon 5, or a portion of intron 11 combined with early termination of the reading frame (Fig. 1A)17–19. The amino acid sequences of LTIP60 and TIP55 differ from TIP60α and TIP60β near or within the critical chromodomain and MYST domain, respectively, raising the possibility that these sequence alterations affect their functions. The Kat5 gene is broadly expressed throughout development, although it is unclear which isoforms are expressed in each tissue, owing to a lack of reagents for distinguishing the different isoproteins. Therefore, the extent to which each Kat5 isoform functions specifically or redundantly with other isoforms is currently unknown.
Figure 1.
Four splice variants are expressed from the Kat5 locus. (A) Variants of Kat5 mRNA produced by alternative splicing. Black boxes: coding regions within exons; white boxes: non-coding sequences within exons; black lines: spliced introns; gray boxes: in frame intronic regions retained in specific Kat5 isoforms; gradient filled box: exon 5, which is spliced out in the Tip60β isoform. (B) Targeting construct used to generate both catalytic inactive Kat5 mutant mice13 and Tip55 knockout mice (described herein). The Tip55 allele was made by removing the intronic regions retained selectively in Tip55 and fusing the remaining exons (which are all included in all other Kat5 isoforms).
Here we test the hypothesis that Tip55 acts non-redundantly in development by specifically mutating this isoform within the mouse Kat5 gene. We show that Tip55 is necessary for embryonic development, with Tip55 homozygous mutant animals dying at or around E11.5. Prior to the appearance of overt developmental phenotypes, we observe defects in both heart and neural tube, which manifest as defects in cellular proliferation and increased cell death. Transcriptomic analyses of embryos prior to the appearance of developmental defects reveal alterations in developmental pathways that contribute to numerous tissues and organ systems. These data reveal a critical function of Tip55 in organogenesis that is distinct from the early, pre-implantation defect observed when all four Kat5 isoforms are lost and raise the possibility that additional isoforms of Kat5 have unique, non-redundant functions during development.
Results
Tip55 is essential for embryonic development
The TIP55 protein lacks the C-terminal 124 amino acids of the other three KAT5 isoforms, TIP60α, TIP60β, and LTip60. In its place, TIP55 has a unique 103 amino acid C-terminus encoded by a portion of intron 1117 (Fig. 1A). This unique C-terminal region lacks amino acid sequence features of any well-established protein domain. We previously generated a multifunctional Kat5 mutant mouse that specifically eliminates the Tip55 isoform and can be converted to a catalytically inactive Kat5 allele upon Cre-mediated recombination. To generate the Tip55 null mutation, we deleted introns 11, 12, and 13, fusing exons 11–14, and in the process removing the region encoding the unique 103 amino acids of TIP55 (Fig. 1B). This allele eliminates Tip55 expression, but does not alter the coding sequence or expression of the remaining isoforms (Fig. 1A, Fig. S1A–C), which are able to form normal TIP60-P400 complexes13. We previously examined the role of the Kat5 lysine acetyltransferase (KAT) activity after Cre-mediated recombination of the Tip55 KO cassette, and found that these KAT-deficient mutant mice progress beyond pre-implantation stages but exhibit defects at or before gastrulation13. However, the question of whether Tip55 is required at any stage of development has not been addressed.
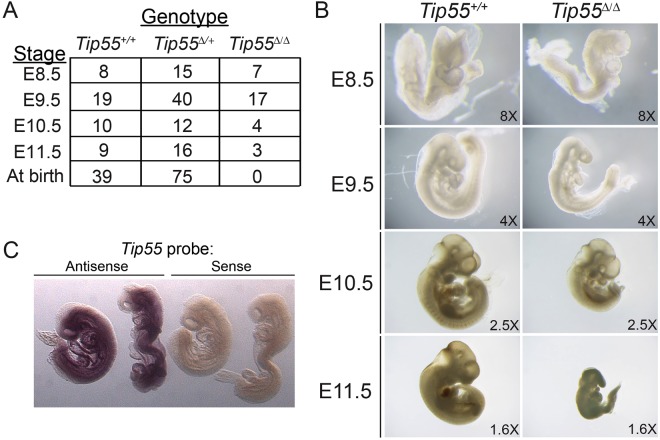
To test this possibility, we intercrossed mice heterozygous for the Tip55 mutant allele (hereafter, Tip55Δ/+) to generate Tip55Δ/Δ homozygotes. We recovered no Tip55Δ/Δ pups at birth (χ2 = 38.05; P < 0.0001), suggesting that the Tip55 isoform is essential for embryonic development (Fig. 2A). To determine the stage at which embryonic development of Tip55Δ/Δ mice was blocked, we dissected and genotyped embryos from E8.5 to E11.5. No overt phenotype was apparent at E8.5 (Fig. 2B). In contrast, Tip55Δ/Δ embryos were smaller than Tip55+/+ or Tip55Δ/+ at E9.5, although they appeared morphologically normal (Fig. 2B). Although Tip55Δ/Δ embryos could readily be recovered as late as E10.5 and occasionally as late as E11.5 (Fig. 2A), all E11.5 Tip55Δ/Δ embryos lacked beating hearts, suggesting a potential cause of death. The lack of Tip55 expression in Tip55Δ/Δ homozygotes was confirmed by RT-PCR (Fig. S1). These data reveal that, unlike null mutants lacking all Kat5 isoforms or Kat5 KAT-deficient mutants, Tip55Δ/Δ mice progress normally through early pre- and post-implantation developmental stages. However, Tip55Δ/Δ mice die during mid-gestation, potentially due to a fully penetrant defect in heart development.
Figure 2.
Tip55 homozygous knockout mice die at or before embryonic day 11.5. (A) Genotypes of embryos recovered at indicated stages of development or at birth. (B) Representative images of wild type (Tip55+/+) and knockout (Tip55Δ/Δ) embryos at stages indicated. Different magnifications as indicated (lower right) were required to image embryos at each stage, owing to differences in size. (C) Whole-mount in situ hybridization of Tip55 transcript. Shown are wild type E8.5-E9.0 mouse embryos hybridized with antisense or sense (as a negative control) in situ hybridization probes corresponding to the region of intron 11 retained in the Tip55 transcript but not found in all other Kat5 isoforms.
Tip55 is required for development of the heart and neural tube
Key early steps in organogenesis, including formation of the heart tube and closure of the neural tube, occur at approximately embryonic day 8 of mouse development, shortly before developmental defects are apparent in Tip55Δ/Δ embryos. To investigate the possibility that Tip55Δ/Δ embryos are defective in some aspects of organ formation, we first tested whether Tip55 was expressed in normal embryos at the initial stages of organogenesis. We performed RT-qPCR for Tip55, as well as for two additional Tip60 isoforms for which specific primers could be designed, LTip60 and Tip60β, using RNA isolated from E7.5-E11.5 embryos. We observed Tip55 expression at all stages, albeit to lower levels than observed for the other isoforms (Fig. S2). Furthermore, using whole-mount in situ hybridization and an intron 11 probe specific for the Tip55 isoform (dark gray boxes in Fig. 1A), we observed that expression of Tip55 was widespread in E8.5-E9.0 embryos (Fig. 2C).
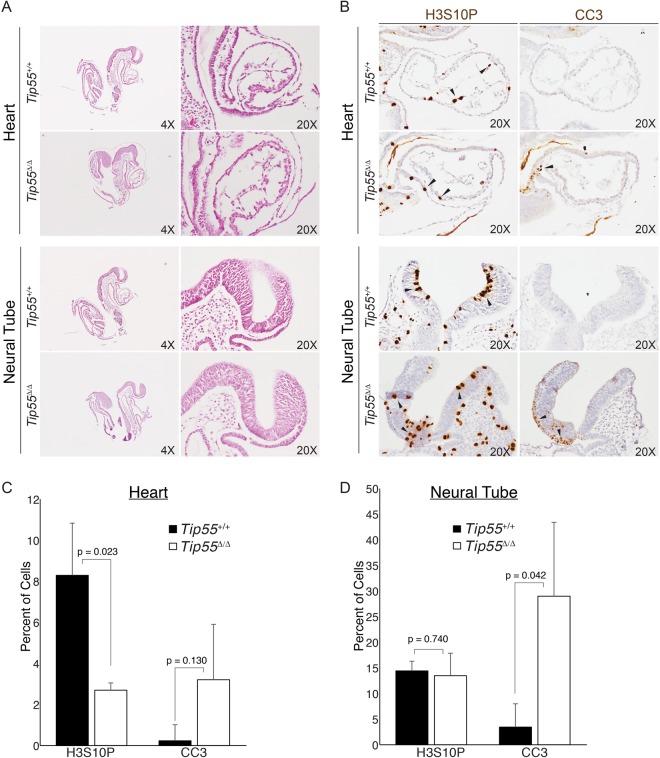
Next, we examined several features of Tip55+/+ and Tip55Δ/Δ embryos at E8.5 (prior to the appearance of developmental defects in Tip55Δ/Δ mutants) in embryo sections. We observed no obvious differences in organization of the body plan in sagittal sections of multiple embryos of each genotype (Fig. 3A). We performed immunohistochemistry with antibodies against Histone H3 phosphorylated at serine 10 (H3S10P) or cleaved caspase 3 (CC3) to test for proliferating or apoptotic cells, respectively. Although most regions of Tip55Δ/Δ embryos were normal, the proportion of H3S10P positive cells was significantly reduced in heart, whereas cleaved caspase 3 staining was significantly elevated in neural tube (Fig. 3B–D). (Cleaved caspase 3 staining also appeared to be higher in heart in Tip55Δ/Δ embryos, but the differences were not statistically significant.) These data suggest that loss of Tip55 results in defects in proliferation and/or increased apoptosis in multiple organs and further suggest that embryonic lethality caused by this mutation is due to defects in organ development.
Figure 3.
Tip55 loss leads to defects in heart and neural tube. (A) Hematoxylin and Eosin (H&E) stained E8.5 embryo sections of Tip55+/+ and Tip55Δ/Δ embryos, shown at multiple magnifications to reveal overall morphology (4X) or focus on heart (20X, above) and neural tube (20X, below). (B) Immunohistochemistry staining of cleaved caspase 3 (CC3) or Histone H3 phosphorylated on serine 10 (H3S10P) on E8.5 sections of Tip55+/+ and Tip55Δ/Δ embryos. Examples of positively staining cells denoted with arrowheads. (C,D) Quantification of cells staining positively for each H3S10P and CC3 in heart (C) and neural tube (D). N = 3 embryo sections were used. P-values were calculated using a two-sided t-test.
We next examined whether the observed phenotypes were associated with a failure to specify the heart and neural lineages. To test this possibility, we performed immunofluorescence staining of wild type and mutant embryo sections for markers of neural lineage (SOX2) and cardiac muscle (cardiac troponin T, cTNT). As above, we stained sections from E8.5 embryos to minimize indirect effects of developmental arrest. However, we observed no difference in staining of either marker in Tip55 mutant embryos relative to wild type (Fig. S3). These data suggest that Tip55 loss impairs proliferation and viability of cardiac and neural progenitor cells downstream of cell type specification in both organs.
Tip55 is required for proliferation of MEFs ex vivo
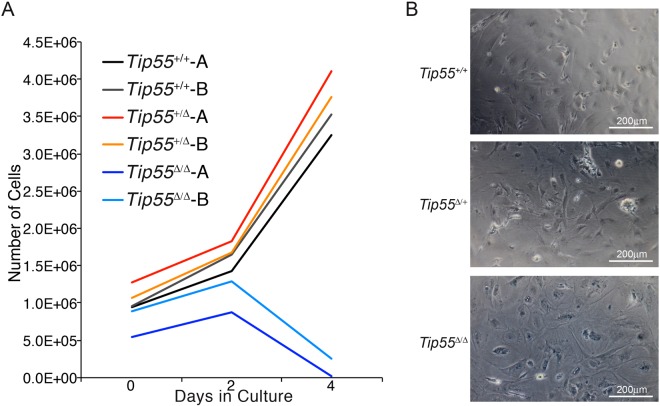
The reduced H3S10P staining of Tip55Δ/Δ embryos in E8.5 heart suggested that TIP55 may promote cellular proliferation. However, a reduction in the number of proliferating cells could also result indirectly from increased cell death or other defects. Therefore, to directly test whether TIP55 was necessary for proliferation, we measured the proliferation rate and morphology of cells isolated from embryos with and without Tip55 mutations. Fibroblasts (MEFs) were isolated from Tip55+/+, Tip55+/Δ, and Tip55Δ/Δ embryos and cultured during a four-day time course. We found that Tip55Δ/Δ MEFs proliferated minimally after isolation at E9.5, followed by growth arrest shortly thereafter (Fig. 4A). Furthermore, Tip55Δ/Δ MEFs exhibited flattened and elongated cell morphology reminiscent of cells undergoing senescence, in contrast to Tip55+/+ and Tip55Δ/+ cells (Fig. 4B). Indeed, Tip55Δ/Δ MEFs exhibited modest but reproducibly elevated staining for senescence-associated β-galactosidase activity compared to Tip55+/+ and Tip55Δ/+ MEFs (Fig. 4B). Together, these data suggest that Tip55 is required for the proliferation of some embryonic cell types and suppression of cellular senescence.
Figure 4.
Tip55 mutant MEFs exhibit premature proliferation arrest. (A) Growth curve indicating numbers of cells of each genotype after growth in culture for indicated lengths of time. Cells were seeded at approximately equivalent densities. (B) Bright field images of mouse embryonic fibroblasts (MEFs) isolated from mouse embryos with the indicated genotypes and stained for β-galactosidase activity (blue) after five days of culture. Scale bars are indicated.
Drivers of organ and tissue development are misregulated in Tip55Δ/Δ mice
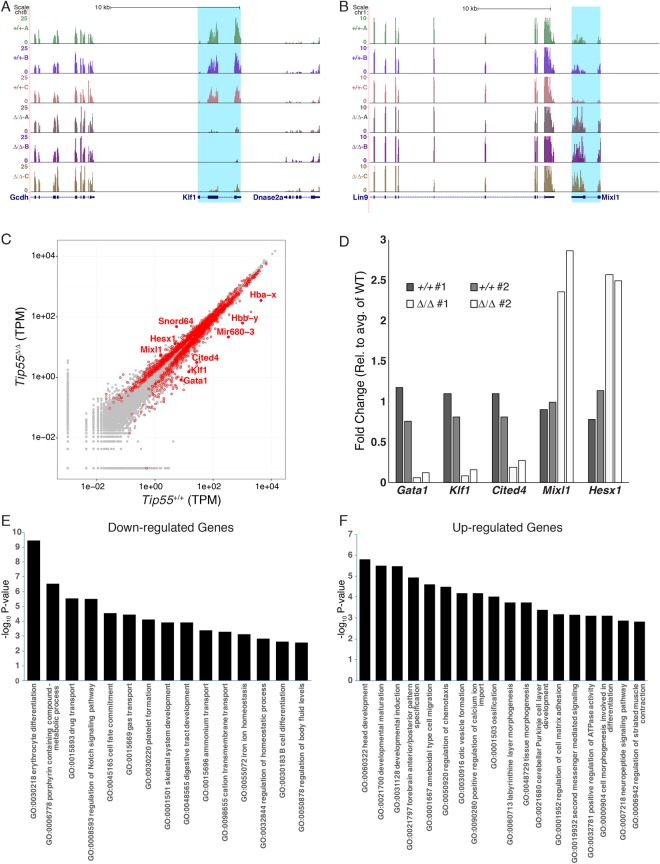
KAT5 isoforms act in part by acetylation of the N-terminal tails of histones H2A and H4 near gene regulatory regions, which promotes transcription10. To test whether genes required for development, cellular proliferation, or other processes were misregulated upon loss of the Tip55 isoform, we isolated RNA from Tip55+/+ or Tip55Δ/Δ embryos at E8.5, prior to the appearance of morphological defects in Tip55Δ/Δ embryos (Fig. 2B), and performed RNA-seq. We observed strong concordance among three biological replicates for Tip55+/+ and Tip55Δ/Δ (Fig. 5A,B). Next, we used EBseq analysis20 to identify genes that are significantly differentially expressed in Tip55Δ/Δ embryos. We identified 2507 genes that were significantly misregulated in Tip55Δ/Δ embryos (posterior probability of differential expression, PPDE > 0.95), with 278 of those genes misregulated more than two-fold (Fig. 5C and Table S1). We validated several differentially expressed genes by RT-qPCR and observed results consistent with the RNA-seq data (Fig. 5D).
Figure 5.
Genes necessary for organogenesis are misregulated in Tip55 mutant embryos. (A,B) Browser tracks of developmental regulators (highlighted in blue) downregulated (A) or upregulated (B) in Tip55Δ/Δ embryos (Δ/Δ) relative to Tip55+/+ embryos (+/+). Three biological replicate RNA-seq datasets (normalized for read number) for each geneotype were performed. (C) Average transcripts per million (TPM) for each genotype are shown on a log-log scale with genes significantly differentially expressed in Tip55Δ/Δ embryos (posterior probability of differential expression; PPDE > 0.95) highlighted with red circles. Several genes of interest labeled with solid red dots with gene names shown. (D) RT-qPCR validation of indicated genes from Tip55+/+ (+/+) or Tip55Δ/Δ (Δ/Δ) E8.5 embryos. Genes were selected based on differential expression (up or downregulation) in RNA-seq experiments. Expression levels in biological duplicate Tip55+/+ or Tip55Δ/Δ embryos are plotted individually, relative to the average of the Tip55+/+ (which is set to 1). (E,F) Significantly enriched gene ontology (GO) categories for genes down-regulated or up-regulated significantly (PPDE > 0.95) and |log2 (Fold Change)| >0.6 are depicted in (A) and (B), respectively.
To uncover classes of genes significantly misregulated in Tip55Δ/Δ embryos, we identified Gene Ontology (GO) terms significantly enriched among the differentially expressed genes. These analyses demonstrated that the differentially expressed genes were highly enriched for developmental regulators and genes required for development of multiple tissue types, including blood, muscle, and brain (Fig. 5E,F). Kat5 is necessary for proper silencing of a number of developmental regulators in embryonic stem cells13. Therefore, genes upregulated in Tip55Δ/Δ embryos may include some that are repressed by TIP55. In addition, developmental delays in Tip55Δ/Δ embryos may cause expression of genes that peak early in wild type embryos (and normally exhibit reduced expression by E8.5) to be more highly expressed in Tip55Δ/Δ mutants at E8.5 as a result of their delayed or extended window of expression.
Among numerous developmentally regulated genes that were downregulated in Tip55Δ/Δ embryos were factors that promote blood cell development – including key erythroid factors Gata1 and Klf1 – suggesting Tip55 loss leads to a failure to initiate erythropoiesis. However, these data do not distinguish whether the effect of Tip55Δ/Δ on erythroid development is due to a direct role for TIP55 at the regulatory regions of key erythroid regulators or an indirect effect on gene expression due to other developmental abnormalities (such as in heart). In sum, we conclude that Tip55 is necessary in multiple cell and tissue types for normal expression of differentiation genes that are critical for organ development.
Discussion
Here we have shown an essential, non-redundant role of the Tip55 isoform of Kat5 during post-implantation embryonic development. These findings contrast with the phenotype of a mouse mutant that lacks all four Kat5 transcripts, which causes pre-implantation lethality at the blastocyst stage16, as well as a KAT-deficient mutant that exhibits defects in gastrulation13. The defects of Tip55Δ/Δ embryos in formation of heart and neural tube suggest this mutation impairs development of multiple cell lineages, a finding that is verified by our transcriptomic data indicating mis-expression of developmental regulators of multiple tissue types. On a cellular level, Tip55Δ/Δ results in a combination of reduced proliferation and increased apoptosis in several different tissues and cell types. Therefore, widespread developmental defects may result from poor proliferation or survival of cells that comprise multiple tissues. Alternatively, these cellular phenotypes may result from more complex developmental impairments owing to defects in the gene regulatory networks specific to each lineage.
The finding that Tip55, a poorly-studied splice variant of Kat5, plays specific roles in development raises the possibility that each of the other Kat5 isoforms play non-redundant roles in development that have yet to be discovered. This raises a larger question regarding regulation of developmental gene expression in mammals – do additional chromatin regulatory enzymes that are expressed in multiple isoforms have distinct functions that are specific for each isoform? Although the functions of splice variants of a few mammalian chromatin regulatory enzymes have been addressed in vitro21,22, the generation of isoform specific mutant animals will be necessary to assess their potential functions in vivo. Such studies have been performed in Drosophila for the SWI/SNF family ATPase, domino, in which two splice isoforms were found to function in distinct cell types23. Although multiple isoforms of one of two mammalian homologs of domino, Ep400, have been identified, it remains to be tested whether they exhibit similar cell type specificity. Therefore, an effort to make and characterize isoform-specific alleles of numerous mammalian chromatin remodeling enzymes will likely provide important insights into developmental gene regulation.
Methods
Antibodies
Antibodies used in this study were as follows: H3S10P (9701, Cell Signaling Technologies), and Cleaved caspase 3 (9661, Cell Signaling Technologies).
Generation of Tip55 knockout mice
All animal experiments were approved by the Institutional Animal Care and Use Committee of University of Massachusetts Medical School (approval number A-2165). All animal procedures were performed in accordance with UMMS and NIH guidelines on animal care. The mutant mouse line with the Tip55 deletion is equivalent to the parental line in which the Tip60ci allele was generated by Cre mediated recombination of fused exons 11–14, placing a catalytically inactive version of exon 11 into the Kat5 gene. This parental allele (prior to Cre mediated recombination) specifically lacks the Tip55 isoform13,14. Mice were genotyped by PCR with primers listed in Table S2. Tip55Δ/+ mice were maintained as heterozygotes on an inbred FVB/N background and intercrossed to generate Tip55+/+, Tip55Δ/+ and Tip55Δ/Δ embryos.
RNA in situ hybridization
Whole mount in situ hybridization of wild type embryos were performed as previously described24. A unique region of Tip55 cDNA (293 bases) was used to generate Tip55 sense and anti-sense probes.
Derivation of MEFs
MEFs were generated from E9.5 embryos as previously described25. Briefly, E9.5 embryos were dissected, and trypsin (0.05%) digested for 12 minutes at 37 °C. Embryos were pipetted to obtain single cells and cultured in a 12 well plate in DMEM with 10% FBS.
Cell proliferation assays
MEFs from two independent embryos from each genotype (Tip55+/+, Tip55Δ/+ and Tip55Δ/Δ) were seeded at approximately 50% confluency and cultured in wells of a 12-well plate. The total number of cells were counted and re-plated into new 12-well plates every 48 hours for a total of four days to measure proliferation rate.
β-galactosidase staining
MEFs were isolated from E9.5 embryos and cultured in a 12-well plate. Three days after plating, cells were sub-cultured for an additional two days followed by staining for β-galactosidase activity using a kit (Millipore KAA002) according to manufacturer’s protocol.
Hematoxylin and Eosin (H&E) Staining
E8.5 embryos were collected and sectioned at 8μm thickness for morphological analysis as previously described26. Hematoxylin- and eosin- staining was performed by de-paraffinizing sections in xylene, rehydrating slides through an ethanol gradient, staining for 30 s with 30% Harris modified hematoxylin and a 30 s counterstain with eosin Y. Slides were rinsed and dehydrated with ethanol, cleared with xylene, and mounted using Vectashield mounting media.
Immunohistochemistry and Immunofluorescence
Sections from E8.5 embryos were examined for proliferation and apoptosis defects by immunohistochemistry, following protocols described previously26. Briefly, sections were rehydrated through an ethanol gradient, followed by heat antigen retrieval (Buffer A, Electron Microscopy Sciences). Immunostaining was conducted using the Vectastain Elite ABC and DAB Peroxidase Substrate kit according to manufacturer guidelines. Sections were incubated with H3S10P (1:100; Cell signaling, CSG 9706) or cleaved caspase 3 (1:100; Cell signaling, CSG 9661) biotinylated primary antibodies overnight at 4 °C26. For counterstaining, slides were rinsed and then incubated with 30% hematoxylin for 30 s after developing staining with 3,3′ diaminobenzimidine. All slides were ethanol-dehydrated, cleared with xylene, and mounted with Vectashield mounting medium.
Expression of cardiac and neural markers in embryonic sections was examined by immunofluorescence. E8.5 embryo sections were rehydrated, then subjected to heat antigen retrieval as described above. Sections were incubated with Sox2 antibody (1:200; R&D research, AF2018) from goat and cTNT antibody (1:50; DSHB, RV-C2) from mouse overnight at 4 °C. The following secondary antibodies were used: Alexa 488 anti-goat (1:200; Life Technologies, A-11055) and Alexa 564 anti-mouse (1:500; Life Technologies, A-11037). Slides were mounted with Vectashield mounting media. Images were taken with a Nikon Eclipse 80i microscope and NIS-Elements 4.00.03 software.
Proliferation and Apoptosis Quantification
For quantification of H3S10P or cleaved caspase 3 immunostaining, digital images were taken using a Nikon Eclipse 80i microscope and the NIS-Elements 4.00.03 software. Positively stained cells were counted manually using ImageJ (v 1.6.0_65). The percentage of H3S10P positive or cleaved caspase-3 positive cells relative to the total number of nuclei was calculated for a minimum of three embryos per genotype.
RNA sequencing
RNA was isolated from E8.5 embryos using a Direct-zol RNA MicroPrep kit (Zymo research). Enrichment of mRNA, library preparation, and sequencing were performed at BGI, using the BGI-seq format. Reads were mapped to the mm10 genome and quantified using RSEM27. Identification of differentially expressed genes was performed using a combined RSEM-EBseq pipeline20. Gene ontology enrichment was performed using Metascape (http://metascape.org)28.
Electronic supplementary material
Acknowledgements
We thank K. McCannell, T. Wu, and J. Benanti for critical comments on the manuscript. This work was supported by the National Institutes of Health [R01HD072122 to T.G.F.; R01HL118100 to C.M.T.; and R01HD083311 to J.A.R.-P.] and the American Cancer Society [RSG-14-220-01] to T.G.F. B.N. was supported by a Translationtional Cancer Biology Postdoctoral training grant from the NIH (T32CA130807). T.G.F. is a Leukemia and Lymphoma Society Scholar.
Author Contributions
D.A. and B.N. performed initial characterization of the Tip55 mutant and most experiments. Z.J.M. and L.B. performed IHC experiments with assistance from C.M.T. Y.Y. and B.N. performed in situ hybridizations with assistance from J.A.R.-P. D.A. and T.G.F. analyzed RNA-seq data. D.A., B.N., and T.G.F. wrote the manuscript with input from all authors.
Data Availability
Sequence reads for all RNA-seq libraries have been deposited at Gene Expression Omnibus (GEO) and are available with accession number: GSE111691.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Diwash Acharya and Bernadette Nera contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33213-4.
References
- 1.Guertin MJ, Lis JT. Mechanisms by which transcription factors gain access to target sequence elements in chromatin. Curr Opin Genet Dev. 2013;23:116–123. doi: 10.1016/j.gde.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villar D, Flicek P, Odom DT. Evolution of transcription factor binding in metazoans — mechanisms and functional implications. Nat Rev Genet. 2014;15:221–233. doi: 10.1038/nrg3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez-Sandoval A, Gasser SM. On TADs and LADs: Spatial Control Over Gene Expression. Trends Genet. 2016;32:485–495. doi: 10.1016/j.tig.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Mirabella AC, Foster BM, Bartke T. Chromatin deregulation in disease. Chromosoma. 2015;125:75–93. doi: 10.1007/s00412-015-0530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 6.Sykes SM, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/S0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 10.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends in Cell Biology. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Kusch T, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 12.Legube G, et al. Role of the histone acetyl transferase Tip60 in the p53 pathway. J Biol Chem. 2004;279:44825–44833. doi: 10.1074/jbc.M407478200. [DOI] [PubMed] [Google Scholar]
- 13.Acharya D, et al. KAT-Independent Gene Regulation by Tip60 Promotes ESC Self-Renewal but Not Pluripotency. Cell Reports. 2017;19:671–679. doi: 10.1016/j.celrep.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen PB, et al. Hdac6 regulates Tip60-p400 function in stem cells. Elife. 2013;2:e01557. doi: 10.7554/eLife.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, et al. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev Dyn. 2009;238:2912–2921. doi: 10.1002/dvdy.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M-S, Merlo X, Wilson C, Lough J. Co-activation of atrial natriuretic factor promoter by Tip60 and serum response factor. J Biol Chem. 2006;281:15082–15089. doi: 10.1074/jbc.M513593200. [DOI] [PubMed] [Google Scholar]
- 18.Legube G, Trouche D. Identification of a larger form of the histone acetyl transferase Tip60. Gene. 2003;310:161–168. doi: 10.1016/S0378-1119(03)00547-X. [DOI] [PubMed] [Google Scholar]
- 19.Ran Q, Pereira-Smith OM. Identification of an alternatively spliced form of the Tat interactive protein (Tip60), Tip60(beta) Gene. 2000;258:141–146. doi: 10.1016/S0378-1119(00)00410-8. [DOI] [PubMed] [Google Scholar]
- 20.Leng N, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zibetti C, et al. Alternative Splicing of the Histone Demethylase LSD1/KDM1 Contributes to the Modulation of Neurite Morphogenesis in the Mammalian Nervous System. Journal of Neuroscience. 2010;30:2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barak O, Lazzaro MA, Cooch NS, Picketts DJ, Shiekhattar R. A Tissue-specific, Naturally Occurring Human SNF2L Variant Inactivates Chromatin Remodeling. J Biol Chem. 2004;279:45130–45138. doi: 10.1074/jbc.M406212200. [DOI] [PubMed] [Google Scholar]
- 23.Börner K, Becker PB. Splice variants of the SWR1-type nucleosome remodeling factor Domino have distinct functions during Drosophila melanogaster oogenesis. Development. 2016;143:3154–3167. doi: 10.1242/dev.139634. [DOI] [PubMed] [Google Scholar]
- 24.Rivera-Pérez JA, Magnuson T. Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev Biol. 2005;288:363–371. doi: 10.1016/j.ydbio.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milstone ZJ, Lawson G, Trivedi CM. Histone deacetylase 1 and 2 are essential for murine neural crest proliferation, pharyngeal arch development, and craniofacial morphogenesis. Dev Dyn. 2017;246:1015–1026. doi: 10.1002/dvdy.24563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathi S, et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host and Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence reads for all RNA-seq libraries have been deposited at Gene Expression Omnibus (GEO) and are available with accession number: GSE111691.