Abstract
Airway epithelial cells (AECs) are the first point of contact with airborne antigens and are able to instruct resident immune cells to appropriate immune responses. Previous studies have shown that the abnormal expression of metastasis-associated lung adenocarcinoma transcript 1 (Malat1) was associated with tumorigenesis, progression, metastasis, and apoptosis in many cancer types. However, little is known about its functional involvement in the cross-talk of AECs with dendritic cells (DCs). The aim of the present study was to identify Malat1 as a novel epithelial cell-derived immune-modulating factor that contributes to the specific inflammatory-immune airway microenvironment. By using an in vitro co-culture model, where layers of AECs can interact with DCs, and transfecting Malat1 siRNA in AECs, AEC-conditioned DCs were harvested for further analysis of the celluar phenotype, secretion of inflammatory chemokines, and expression of apoptotic markers. The present study clearly demonstrated that Malat1 modulates the maturation process, pro-inflammatory cytokine secretion and apoptosis in AECs-conditioned DCs.
Keywords: lncRNA, Malat1, dendritic cell, airway epithelial cells, co-cultured
Introduction
The human lung exchanges thousands of liters of air daily and is often exposed to different stimuli, such as airborne allergens, viruses, and bacteria (1). The presence of certain special respiratory microenvironments controls and limits the inflammatory response (2–6). Airway epithelial cells (AECs) represent the mechanical immune barrier to the external environment. These cells cover the airway surface and serve as the frontline of defense against inhalable contamination. Respiratory epithelial cells, which is an essential part of the innate immune system, are also responsible for the initial immuno-inflammatory response. Among these different subtypes of AECs, type II AECs play a critical role in the induction of cellular innate immune response (7,8).
In recent years, epithelial cells have been established to play complex roles in organ-specific immunity. After antigen recognition, AECs determine the functional characteristics of innate immune cells through a soluble or contact-dependent mechanism. Several studies (9,10) demonstrated that respiratory epithelial cells could inhibit lipopolysaccharide (LPS)-stimulated dendritic cells (DCs) from secreting proinflammatory cytokine by inducing macrophage-derived chemokines and even immune-stimulatory DCs (11,12). Studies have also shown that via a constitutive release mediator, epithelial cells could regulate mucosal DCs as well as induce inflammatory response in DCs (13).
DCs as the strongest antigen-presenting cells, are the only cells that can activate naive immune cells and participate in recognizing and presenting antigen-initiated immune response. Epithelial modulation may also be involved in the induction or production of some special DCs, such induction can result in the dysfunction of T cells (14). Therefore, developing and validating the effective in vitro model system that establishes the crosstalk between different epithelial cells and DCs will contribute to respiratory and immunology research.
Long noncoding RNAs (lncRNAs) are a class of RNA molecules with transcript lengths greater than 200 nt. Initially, they were considered as the ‘noise’ of genomic transcription without biological functions. However, growing evidence suggests that lncRNAs are an important regulator of biological processes such as X-chromosome inactivation, modulation of protein activity and chromatin remodeling (15,16). Their potential importance in regulating immune responses, particularly, in the innate immune response, has only been discovered recently (17–20). Metastasis-associated lung adenocarcinoma transcript 1 (Malat1) is a 7 kb nuclear long non-coding RNA located at chromosome 11q13. Malat1 has been found to be a predictive biomarker of early metastasis of non-small cell lung cancer. Previous studies have shown that abnormal expression of Malat1 was associated with tumorigenesis, progression, metastasis, and apoptosis in many cancer types (21–32). Besides its oncogenic role, Malat1 is involved in many other diseases as well as in normal physiological processes, such as vascular growth, synaptogenesis, and muscle cell generation (33,34). However, knowledge about the direct crosstalk between this lncRNA and DCs remain largely unknown.
In the present study, by knocking down the expression of Malat1 in AECs and by using the in-vitro co-culture model, where cell layer of AECs can interact with DCs, we harvest AEC-conditioned DCs for further analysis of the cellular phenotype, secretion of inflammatory chemokines, and expression of apoptotic markers. Therefore, this study aims to explore the relationship between Malat1, AECs, and DCs, also intends to identify Malat1 as a novel epithelial cell-derived immunomodulatory factor for specific immuno-inflammatory microenvironment. Our results suggest that the Malat1-mediated crosstalk between epithelial cells and DCs can alter the phenotype and function of DCs.
Materials and methods
AECs culture and transfection
Murine type II alveolar epithelial cells (MLE-12) were isolated from female C57BL/6 mice (purchased from the Shanghai Institutes for Biological Sciences Shanghai, China) as described by Corti et al (35). siRNA control and siRNA Malat1 were obtained from Ribo (cat. no. stQ0019997-1; siR-Ribo, lnc., Guangzhou, China). Transfection reagent Lipofectamine 3000 reagent (cat. no. L3000-015; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and transfection media opti-MEM (cat. no. 31985062; Gibco; Thermo Fisher Scientific, Inc.) were purchased from Thermo Fisher Scientific, Inc. MLE-12 cells were seeded at 1×104/cm2 one day before transfection to reach 60–70% density the next day. For one well of 6-well plate, medium was changed to 1 ml empty medium without antibiotics or fetal bovine serum (FBS; cat. no. 10099141; Gibco; Thermo Fisher Scientific, Inc.) approximately 20 min before the transfection. In final experiments, mix A (50 µl opti-MEM/well+ 0.5 µl siRNA/well) and mix B (50 µl opti-MEM/well+2 µl RNAi Max/well) was prepared. The final concentration of siRNA was 50 nM.
Bone marrow-derived DCs (BMDCs)
BMDCs were generated from C57BL/6 mice as previously described (36). All animal experiments were performed according to protocols approved by the Institutional Committee for Use and Care of Laboratory Animals. In brief, femurs and tibia of mice were resected and washed with phosphate buffered saline (PBS; GNM-20012; GENOM., Ltd., Hangzhou, Zhejiang, China) to obtain bone marrow cells. After incubation with NH4Cl lysing buffer to remove red blood cell (RBC), bone marrow cells were placed in 6-well plates in RPMI-1640 (GNM-31800; GENOM., Ltd.). The cell layer was then gently washed by medium 2 h later to remove non-adhering cells. Adherent cells were incubated in the complete media (RPMI-1640 containing 2 mM L-glutamine (cat. no. 1294808-100MG; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with 10% FBS, 1% of nonessential amino acids (cat. no. 11140050; Gibco; Thermo Fisher Scientific, Inc.), 1% of sodium pyruvate, 20 ng/ml rmGM-CSF (cat. no. 415-ML-010/CF) and 10 ng/ml rmIL-4 (cat. no. 404-ML-010/CF; both R&D Systems, Inc., Minneapolis, MN, USA) for 6 days. Medium was changed every 2 days. On the sixth day, floating adherent cells were collected and used as BMDCs. To induce DCs mature, cells were stimulated with 100 ng/ml LPS (Escherichia coli 055B5; Sigma-Aldrich; Merck KGaA).
Co-culture model system
The AECs-DCs co-culture model was established as described before (37). In brief, the Malat1 treated MLE-12 cells were incubated for 48 h before cultured on the top of the inverted filter inserts, which were made of polyethylene terephthalate (cat. no. 353092; BD Pharmingen; Franklin Lakes, USA) with pore size of 3 µm, The filters were then reverted and placed into wells with RPMI-based culture media, and DCs suspended in RPMI-based culture media were added to the basal side within the insert well. DCs were harvest after 72 h.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol (cat. no. 12183555; Invitrogen; Thermo Fisher Scientific, Inc.) and then reversely transcribed into first-strand cDNA using PrimeScript RT reagent kit with gDNA Eraser (cat. no. RR047A; Takara Biomedical Technology Co., Ltd., Beijing, China) according to manufacturers' instructions. Real-time PCR was performed with Takara premix Ex Taq II (cat. no. DRR820A; Takara Biomedical Technology Co., Ltd.) and was run on CFX96TM Real-Time System (cat. no. 1855195.; Bio-Rad Laboratories, Inc., Hercules, CA, USA) in a total volume of 10 µl containing 5 ul Takara premix Ex Taq II and 5 ng cDNA template. The PCR thermal cycle parameters were as follows: 2 min at 50°C, 30 sec at 95°C and 40 cycles of 95°C for 5 sec and 60°C for 34 sec. Expression of mRNA was normalized to the expression of mouse GAPDH (mGAPDH) and quantified using the 2−ΔΔCq method (38). Primer pairs for qPCR are listed as follows: mmu F-malat1, 5′GGGCATTTCCATTCCTCTC 3′ and mmu R-malat1, 5′GTCTCTACGGGCACATTAC 3′; mmuF-CD80, 5′ACCCCCAACATAACTGAGTCT3′ and mmuR-CD80, 5′TTCCAACCAAGAGAAGCGAGG3′; mmuF-CD86, 5′TGTTTCCGTGGAGACGCAAG3′ and mmuR-CD86, 5′TTGAGCCTTTGTAAATGGGCA3′; mmuF-IL4, 5′GTTGTCATCCTGCTCTTC3′ and mmuR-IL4, 5′GTTTGGCACATCCATCTC3′; mmuF-IL6, 5′TGGAGCCCACCAAGAACGATAG3′ and mmuR-IL6, 5′TGTCACCAGCATCAGTCCCAAG3′; mmuF-INF-γ, 5′TCAAGTGGCATAGATGTG3′ and mmuR-INF-γ, 5′TGTTGCTGAAGAAGGTAG3′; mmuF-CCL2, 5′TTAAAAACCTGGATCGGAACCAA3′ and mmuR-CCL2, 5′GCATTAGCTTCAGATTTACGGGT3′; mmuF-CCL5, 5′GCTGCTTTGCCTACCTCTCC3′ and mmuR-CCL5, 5′TCGAGTGACAAACACGACTGC3′; mmuF-CXCR4, 5′GACTGGCATAGTCGGCAATG3′ and mmuR-CXCR4, 5′AGAAGGGGAGTGTGATGACAAA3′; mmuF-CXCR2, 5′ATGCCCTCTATTCTGCCAGAT3′ and mmuR-CXCR2, 5′GTGCTCCGGTTGTATAAGATGAC3′; mmuF-GAPDH, 5′CTGCCCAGAACATCATCC3′ and mmuR-GAPDH, 5′CTCAGATGCCTGCTTCAC3′.
Transwell chemotaxis assay
Migration chemotaxis assay was performed by applying 24-well Boyden chambers with 8 µm pore size polycarbonate membranes (cat. no. 662638; Greiner Bio-One, Ltd., Frickenhausen, Germany) as described previously. After co-culturing with MEL-12 cells transfected with siRNA control or Malat1 siRNA, DC were seeded onto the upper chamber at 1×105 cells. The bottom chamber contained 600 µl 10% FBS medium with or without 200 ng/ml Chemokine ligand 19 (CCL19). After incubation for 6 h at 37°C, the cells remained on the upper side of the filters were removed by a cotton swab. The migrated cells on the underside of the membrane were fixed with 4% paraformaldehyde (PFA) prior to staining with 0.1% crystal violet solution for 15 min.
Flow cytometry
DCs were co-cultured for 6 h with MLE-12 cells interfered with siRNA control or Malat1 siRNA, followed by stimulation with LPS and further incubation for 72 h. DCs were detached from the flasks by trypsin (GNM-15050; GENOM., Ltd.), and cell numbers were counted. Cells were then fixed with 4% paraformaldehyde for 15 min at room temperature. After washing with PBS, cells were re-suspended in 5% swine serum and blocked with Fc Receptor Blocking Solution (cat. no. 422301; Biolegend, Inc., San Diego, CA, USA) for 5 min. Aliquots of about 2×105 cells were then incubated with specific antibodies (mouse CD80; cat. no. 561954), CD86 (cat. no. 561962; both BD Biosciences, Franklin Lakes, NJ, USA), MHC-II (cat. no. bs-4107R; Bioss Antibodies, Inc., Woburn, MA, USA), Annexin-V-FITC antibody (cat. no. 70-AP101-100; Multiscience, Hangzhou, Zhejiang, China) and IgG control for 1 h at 4°C. Cells were then washed three times and analyzed with the flow cytometer (BD FACS Canto™ II system). Further analysis was done with Flow Jo. Gating set as less than 1% of marker expression in IgG control group.
ELISA assays of cytokine
DCs were co-cultured with siRNA control or Malat1 siRNA interfered MLE-12 for 6 h, LPS was added and the cells were incubated for another 72 h. To detect the secretion of tumor necrosis factor-α (TNF-α), IL-4, INF-r and IL-6, the cell supernatant was harvest and analyzed by Sandwich Enzyme Immunoassay kits (R&D Systems Europe Ltd., Abingdon, UK) according to the manufacturer's protocol.
Methylthiazolyltetrazolium (MTT) assay
DCs were co-cultured with siRNA control or Malat1 siRNA interfered MLE-12 for 6 h, followed by stimulation with LPS and further incubation for 72 h. MTT (cat. no. c0009; Beyotime Institute of Biotechnology, Shanghai, China) of 0.5 mg/ml were then added to each well. After 4 h of incubation at 37°C, cells were treated with 1 ml of dimethyl sulfoxide (cat. no. D2650-5*5ML; Sigma-Aldrich; Merck KGaA) followed by incubation at 37°C overnight. Finally, optical densities were measured at 570 nm, and the growth inhibitory rate was calculated.
Immunoblotting
10–50 µg of the protein was mixed with a ¼ volume of the boiled 5X SDS loading buffer (cat. no. p0015) and then adjusted to the same volume with 1X SDS loading buffer (cat. no. P0015A; both Beyotime Institute of Biotechnology). The mix was then boiled at 95°C for 10 min before loaded to the NuPage 4–12% Tris-Glycine gel (cat. no. EC60249BOX) immersed in NuPage MOPS SDS running buffer (cat. no. NP0001; both Invitrogen; Thermo Fisher Scientific, Inc.) in a XCell SureLock Mini-Cell (cat. no. NP0335BOX; Thermo Fisher Scientific, Inc.). The gel was run at 160 V for about 75 min until the marker with lowest molecular weight was at the bottom of the gel. Protein was then transferred from the gel to the membrane in 1X transfer buffer for 45 min. The membrane was then blocked with 5% milk in PBS-Tween and incubated with primary antibody diluted in 5% milk at 4°C overnight. After being washed with PBS-Tween for three times with 10 min each, the membrane was incubated with secondary antibody for 1 h at room temperature. Secondary antibodies were HRP-conjugated and purchased from Dako (Polyclonal Rabbit anti-goat, cat. no. 103P0449; Polyclonal Rabbit anti-mouse, cat. no. P0260; Polyclonal Swine anti-rabbit, cat. no. P0217; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). Further 3 washes were performed before addition of ECL detection solutions. After incubation with the detection solution for 2 min, exposure of the films was carried out with the Compact X4 in the dark room. The bound primary antibodies: Rabbit-anti-mouse-caspase-3 (Ab44976), anti-mouse-Bax (Sc-493), anti-rabbit-Bcl2 (Sc-492), anti-mouse-GAPDH (CST-5174) diluted 1:1,000 in PBST.
Statistical analysis
Data between two groups with normal distribution were analyzed with unpaired and ungrouped t-tests, and data not normally distributed were analyzed with unpaired and nonparametric Mann-Whitney U test. Data across multiple groups were analyzed with one-way ANOVA test, followed by Bonferroni post-hoc analysis if P<0.05. Data were shown as the mean ± standard error of mean (SEM) and graphs were generated using GraphPad Prism 6 software. P<0.05 was considered to indicate a statistically significant difference (*P<0.05; **P<0.01 and ***P<0.001).
Results
Expression of Malat1 in AECs-DCs co-culture system
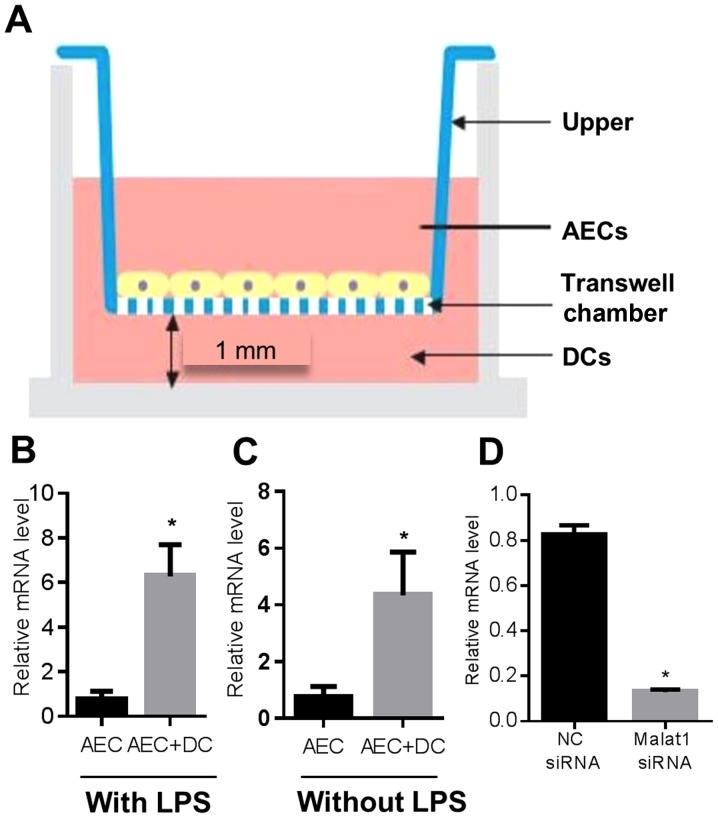
The tracheal epithelial cells have been found to regulate the gene expression in BMDCs (39), but whether these respiratory epithelial cell could modulate the long-non coding RNA expression is unclear. To contact the DCs with alveolar epithelial cells (MLE-12), MLE-12 cells were cultured on top of inverted filter inserts. The filters were then reverted and placed into wells containing RPMI-1640 media. The DCs stimulated with or without LPS were suspended in media and added to the basal side of the insert well (Fig. 1A), We term this system as AECs-conditioned DCs system. After 48 h, the expression of Malat1 in AECs was detected. The Malat1 expression was significantly enhanced in this co-culture system regardless of LPS addition (Fig. 1B and C). Furthermore, to elucidate the regulatory role of Malat1 in AECs-conditioned DCs, we used a chemically synthesized Malat1 inhibitor (Malat1 siRNA) to suppress the Malat1 expression. Fig. 1D revealed the significant diminution of Malat1 expression after the Malat1 knockdown in MLE-12 cells.
Figure 1.
Malat1 is implicated in AEC upon DC co-culture. (A) Simple summary of the co-culture system. The expression of Malat1 in AECs was detected by real-time PCR and found to be upregulated in the co-culture system in the presence (B) and absence (C) of LPS. (D) Malat1 knockdown AEC was generated by siRNA transfection, then the transfected AECs were harvested and subjected to q-PCR (n=5, *P<0.05 vs. AEC only group group and ‘NC siRNA’). AEC, airway epithelial cells; DC, dendritic cell; Malat1, metastasis-associated lung adenocarcinoma transcript 1; q-PCR, quantitative polymerase chain reaction; LPS, lipopolysaccharide.
Effects of Malat1 on the maturation of AECs-conditioned DCs
Immature DCs can assimilate antigens efficiently but express low levels of costimulatory molecules. Upon maturation, DCs dramatically augment their ability to stimulate naive T cells through the surface exposure of antigen-MHC complexes and increased expression of costimulatory molecule. Following co-culture for 48 h with AECs that had been treated with Malat1 silencing, the DCs were harvested to detect the specific maturity surface marker by real-time PCR and FACS. Our results showed that some surface molecules (CD80, CD86, and MHCII) of the AECs-conditioned DCs were upregulated in response to incubation with Malat1 inhibitor, both at the protein (Fig. 2A-F) and mRNA levels (Fig. 2G), implying Malat1 can partly induce the maturation of DCs cultured in AECs-conditioned system.
Figure 2.
Effects of Malat1 on the maturation of AEC-treated DCs. AECs were transfected with NC siRNA or Malat1 siRNA and then cocultured with DCs for 72 h. The costimulatory molecules (A and B) CD80, (C and D) CD86, and (E and F) MHCII in DCs were detected by FACS. (G) Relative mRNA levels of CD80, CD86, and MHCII in DCs were determined by real-time PCR (n=5, *P<0.05 vs. ‘NC siRNA’ group). AEC, airway epithelial cells; DC, dendritic cell; Malat1, metastasis-associated lung adenocarcinoma transcript 1; PCR, polymerase chain reaction.
AECs-conditioned DCs cytokine secretion in response to Malat1
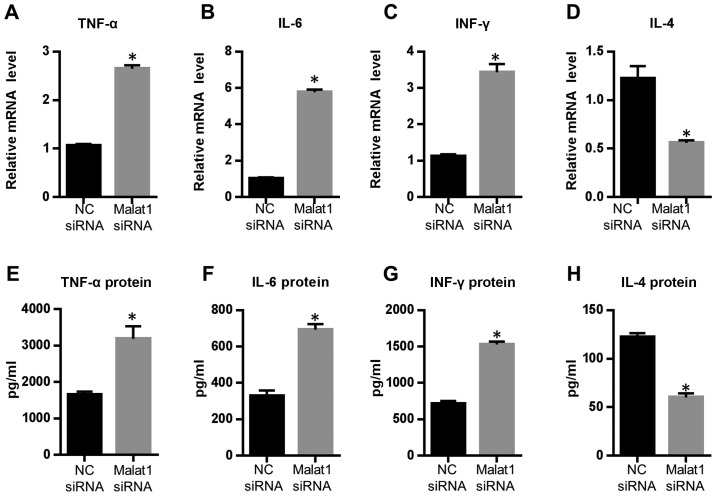
The cytokine secreted by DCs plays a vital role during T cell polarization. To investigate the functional role of Malat1 in AECs-conditioned DCs, levels of certain inflammatory cytokines were analyzed. The Malat1 inhibitor increased the secretion of some inflammatory cytokines by DCs such as TNF-alpha, IL-6, and IFN-γ significantly (more than 3–5 fold at the RNA level and approximately 2 fold at the protein level) (Fig. 3). By contrast, IL-4 expression was slightly downregulated by Malat1 inhibitor (50% off) (Fig. 3). These results indicated that Malat1 could affect the inflammatory response of AECs-conditioned DCs.
Figure 3.
Effects of Malat1 on the inflammatory response of AEC-treated DCs. AECs were incubated in the presence of NC siRNA or Malat1 siRNA and then cocultured with DCs for further 72 h. The expression of TNF-α, IL-4, IL-6, and IFN-r in DCs were regulated after the suppression of Malat1. Relative mRNA levels of (A) TNF-α, (B) IL-4, (C) IL-6, and (D) IFN-r detected by RT-qPCR. The protein levels of (E) TNF-α, (F) IL-4, (G) IL-6, and (H) IFN-r were detected by ELISA. Mean ± SD was obtained from five different experiments (*P<0.05 vs. ‘NC siRNA’ group). AEC, airway epithelial cells; DC, dendritic cell; Malat1, metastasis-associated lung adenocarcinoma transcript 1; RT-qPCR, real-time quantitive polymerase chain reaction; TNF-α, tumor necrosis factor-α.
Chemokine production of AECs-conditioned DCs in response to Malat1
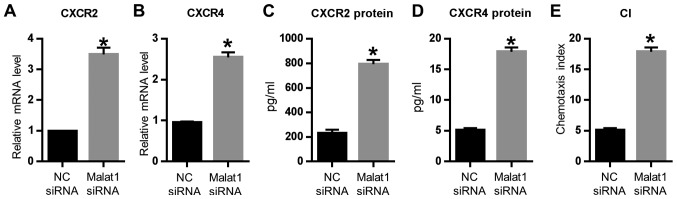
We also detected the expression of chemokines in the AECs-conditioned DCs. The Malat1 siRNA dramatically increased the secretion of chemokines, such as CXCR2 and CXCR4, in AECs-conditioned DCs, both at the mRNA and protein levels (Fig. 4A-D). Given that Malat1 siRNA-treated MLE-12 significantly increased the chemokine expression of DCs, we further performed Transwell assay to confirm the effect of Malat1 on the chemotaxis of co-cultured DCs. As expected, the Malat1 siRNA-treated MLE-12 also remarkably induced DC chemotaxis (Fig. 4E). All these results suggested that Malat1 could affect the chemokine secretion and chemotaxis of AECs-conditioned DCs.
Figure 4.
Effects of Malat1 on the chemokine expression of AEC-treated DCs. AECs were incubated in the presence of the NC siRNA or Malat1 siRNA and then cocultured with DCs for 72 h. Relative mRNA level of (A) CXCR2 and (B) CXCR4 as detected by RT-qPCR. Relative protein level of (C) CXCR2 and (D) CXCR4 as detected by ELISA. (E) CI of the Transwell results. Mean ± SD calculated from five different experiments (*P<0.05 vs. ‘NC siRNA’ group). CI, chemotaxis index; AEC, airway epithelial cells; DC, dendritic cell; Malat1, metastasis-associated lung adenocarcinoma transcript 1; RT-qPCR, real-time quantitive polymerase chain reaction.
Malat1 knockdown attenuates the apoptosis of AECs-conditioned DCs
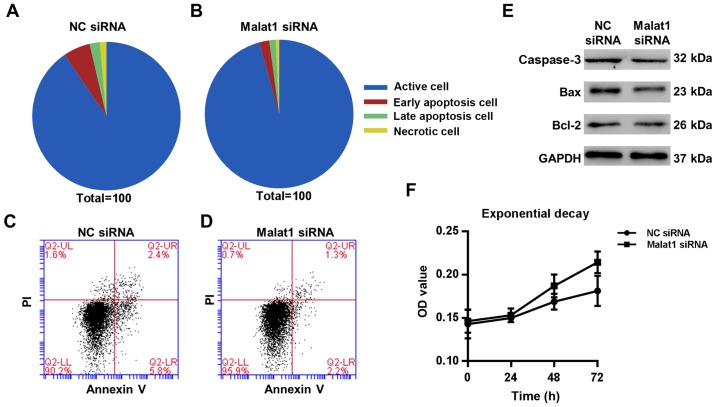
To explore the effect of Malat1 knockdown on the apoptosis of AECs-conditioned DCs, the apoptosis marker Annexin V in the DCs was detected by flow cytometry. Compared with siRNA control group, the mean apoptotic cell fractions (early apoptotic + late apoptotic) were significantly decreased, and viable cell populations were significantly decreased upon Malat1 inhibition (P=0.04) (Fig. 5A-D). Expression of apoptotic markers, such as caspase-3, Bax, and Bcl2, in the co-cultured DCs was decreased after transfection with Malat1 siRNA (Fig. 5E). Meanwhile, we detected the effect of Malat1 on the viability of AECs-conditioned DCs by using MTT assay, Our results found that the Malat1 siRNA slightly induced cell viability (18.2±1.3%) as compared to the control group at 72 h (Fig. 5F), indicating an apoptotic-inducing role of Malat1 in AECs-conditioned DCs.
Figure 5.
Effects of Malat1 on the apoptosis and proliferation of AEC-treated DCs. AECs were incubated with NC siRNA or Malat1 siRNA and then cocultured with DCs for 72 h. The percentage of apoptosis of (A) NC siRNA and (B) Malat1 siRNA groups. FITC-Annexin-V of DCs was detected by FASC in (C) NC siRNA and (D) Malat1 siRNA groups. (E) Protein levels of caspase-3, Bax, and Bcl-2 in DCs were detected by WB. (F) DC cell viability was determined by MTT assay. Mean ± SD calculated from five different experiments (P<0.05 vs. ‘NC siRNA’ group). AEC, airway epithelial cells; DC, dendritic cell; Malat1, metastasis-associated lung adenocarcinoma transcript 1.
Discussion
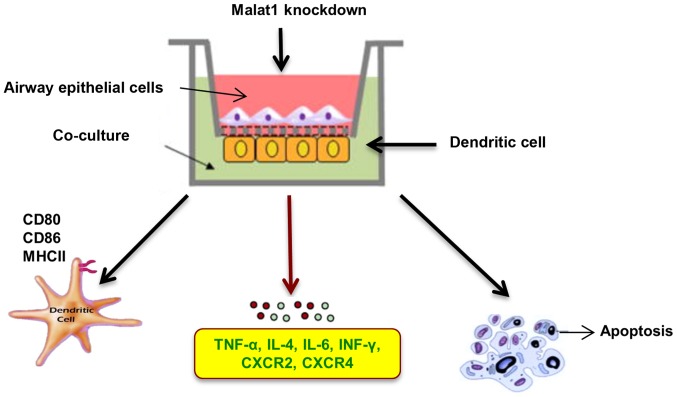
Mucosal epithelial cells usually induce a specific microenvironment to regulate the intensity of classical immune responses (3). Given their frequent contact with inhaled antigens or microbial substances, immunomodulatory functions appear particularly important in the trachea (40). Previous study (39) have revealed the interrelationship between epithelial cells and DCs at the transcriptional level. In this study, we provide the first evidence that Malat1 can modulate the maturation process, pro-inflammatory cytokine secretion and apoptosis in AECs-conditioned DCs as Schematic representation in Fig. 6.
Figure 6.
Schematic of the functional role of Malat1 in regulating AECs-treated DCs. AEC, airway epithelial cells; DC, dendritic cell; Malat1, metastasis-associated lung adenocarcinoma transcript 1.
Recent studies (41–46) showed that lncRNAs were expressed in a highly lineage-specific manner with the role of regulating differentiation and physiological functions of innate and acquired cell types (47). Specific lncRNAs have been identified to regulate the differentiation of DCs and to inhibit their ability to stimulate T cells by activating the transcription factor STAT3. However, lncRNAs that could potentially interact with DCs and AECs have not been found. In our study, significant upregulation of Malat1 in AECs was observed when co-cultured with DCs, suggesting potential involvement of Malat1 in the interaction between AECs and DCs.
We also observed that CD80, CD86, and MHCII were upregulated in DCs after co-culturing with the Malat1-inhibited AECs, while the expression of membrane receptors CD40, CD209, and HLA-DR (data not shown) were unchanged. CD40, CD80, CD86, and HLA-DR have been reported to be downregulated by a specific lncRNA in DCs (8). Our results, at least, partly indicated that Malat1 can inhibit the maturation of AECs-conditioned DCs.
It was demonstrated that AECs could induce a tolerogenic phenotype of immune cells, including reduced T cell responsiveness (2,3) and decreased pro-inflammatory cytokine production (3). In our study, we observed that the Malat1 knockdown could induce the secretion of the pro-inflammatory cytokines TNF-α, IL-6, and IFN-r in AECs-conditioned DCs. However, the Malat1 siRNA suppressed the secretion of IL-4, which is a canonical type 2 cytokine. IL-4 can reduce the expression of B1 and B2 receptors, which are induced by either IL-1β or TNF-α (48). These evidences support the negative effect between IL-4 and other pro-inflammatory cytokines. Besides the effect of Malat1 on inflammatory cytokines, we also explored the role of this lncRNA in chemokine production in the AECs-DCs co-cultured system. Our finding confirmed that the Malat1 suppression can augment chemotaxis by inducing the secretion of CXCR2 and CXCR4. All the results implied that Malat1 plays a vital role in cytokine production in AECs-conditioned DCs.
The impact of Malat1 on cellular apoptosis seems to be disease-dependent. While one study showed that Malat1 promoted neuronal apoptosis by sponging miR-124 in Parkinson's disease (49), another study found that it suppressed apoptosis of retinal ganglion cells in glaucoma through the PI3K/Akt signaling pathway (50). Another study (51) demonstrated that Malat1 shRNA plasmids enhanced apoptosis, weakened proliferation, and decreased migration and invasion of gallbladder cancer cells. We thus assessed the impact of Malat1 on the apoptosis and proliferation of AECs-conditioned DCs. As expected, Malat1 siRNA inhibited apoptosis and induced proliferation of AECs-conditioned DCs, confirming the pro-apototic role of Malat1 in the co-culture system.
Taken together, this is the first time to demonstrate the functional role of Malat1 in AECs-DCs co-culture system. That is, Malat1 induced the anti-inflammatory, alternatively inactivated phenotype and suppressed apoptosis in the microenvironment. We recognize a few limitations of our study, we did not explore the mechanism involved the Malat1 in AECs and DCs crosstalk, the future studies investigating the mechanism of Malat1 in the current model are still pending, such as microRNA sponge (49) and the potential signaling pathway as suggested by some previously studies (Stat3 and PI3K/Akt) (47,50). Through the emerging use of in vitro co-culture models, our study may help the understanding and uncovery of the complicated interplay between the respiratory epithelium and immune cells in airway disorders.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- lncRNA
long non-coding RNA
- DCs
dendritic cells
- AECs
airway epithelial cells
- miRNA
microRNA
- siRNA
small interfering RNA
- Malat1
metastasis-associated lung adenocarcinoma transcript 1
- MLE-12
murine type II alveolar epithelial cells
- BMDCs
bone marrow-derived dendritic cells
Funding
The present study was supported by the Zhejiang Provincial Natural Science Foundation of China (grant no. LY17H010002), National Natural Science Foundation of China (grant nos. 81770435, 81700238 and 81400224), China Cardiovascular Association-Cardiac Rehabilitation and Metabolic Therapy Research Fund (grant no. CCA-CRMT-1614) and the Xinxin-Merck Cardiovascular Scientific Research Fund (grant no. 2017-CCA- Xinxin-Merck fund-004).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WW and TC designed the research; ZL, QZ, YW, FH and LG performed the research; ZL analyzed the data; and TC wrote the manuscript.
Ethics approval and consent to participate
Ethical approval for the study was granted from The First Affiliated Hospital of Zhejiang University (Hangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Folkerts G, Nijkamp FP. Airway epithelium: More than just a barrier! Trends Pharmacol Sci. 1998;19:334–341. doi: 10.1016/S0165-6147(98)01232-2. [DOI] [PubMed] [Google Scholar]
- 2.Mayer AK, Bartz H, Fey F, Schmidt LM, Dalpke AH. Airway epithelial cells modify immune responses by inducing an anti-inflammatory microenvironment. Eur J Immunol. 2008;38:1689–1699. doi: 10.1002/eji.200737936. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt LM, Belvisi MG, Bode KA, Bauer J, Schmidt C, Suchy MT, Tsikas D, Scheuerer J, Lasitschka F, Gröne HJ, Dalpke AH. Bronchial epithelial cell-derived prostaglandin E2 dampens the reactivity of dendritic cells. J Immunol. 2011;186:2095–2105. doi: 10.4049/jimmunol.1002414. [DOI] [PubMed] [Google Scholar]
- 4.Wissinger E, Goulding J, Hussell T. Immune homeostasis in the respiratory tract and its impact on heterologous infection. Semin Immunol. 2009;21:147–155. doi: 10.1016/j.smim.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Holt PG, Strickland DH, Wikström ME, Jahnsen FL. Regulation of immunological homeostasis in the respiratory tract. Nat Rev Immunol. 2008;8:142–152. doi: 10.1038/nri2236. [DOI] [PubMed] [Google Scholar]
- 6.Mayer AK, Dalpke AH. Regulation of local immunity by airway epithelial cells. Arch Immunol Ther Exp (Warsz) 2007;55:353–362. doi: 10.1007/s00005-007-0041-7. [DOI] [PubMed] [Google Scholar]
- 7.Castro-Garza J, King CH, Swords WE, Quinn FD. Demonstration of spread by Mycobacterium tuberculosis bacilli in A549 epithelial cell monolayers. FEMS Microbiol Lett. 2002;212:145–149. doi: 10.1111/j.1574-6968.2002.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 8.Sato K, Tomioka H, Shimizu T, Gonda T, Ota F, Sano C. Type II alveolar cells play roles in macrophage-mediated host innate resistance to pulmonary mycobacterial infections by producing proinflammatory cytokines. J Infect Dis. 2002;185:1139–1147. doi: 10.1086/340040. [DOI] [PubMed] [Google Scholar]
- 9.Sekiya T, Miyamasu M, Imanishi M, Yamada H, Nakajima T, Yamaguchi M, Fujisawa T, Pawankar R, Sano Y, Ohta K, et al. Inducible expression of a Th2-type CC chemokine thymus- and activation-regulated chemokine by human bronchial epithelial cells. J Immunol. 2000;165:2205–2213. doi: 10.4049/jimmunol.165.4.2205. [DOI] [PubMed] [Google Scholar]
- 10.Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B, Gilliet M, Ho S, Antonenko S, Lauerma A, et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol. 2002;3:673–680. doi: 10.1038/ni805. [DOI] [PubMed] [Google Scholar]
- 11.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Willart MA, Deswarte K, Pouliot P, Braun H, Beyaert R, Lambrecht BN, Hammad H. Interleukin-1α controls allergic sensitization to inhaled house dust mite via the epithelial release of GM-CSF and IL-33. J Exp Med. 2012;209:1505–1517. doi: 10.1084/jem.20112691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rimoldi M, Chieppa M, Salucci V, Avogadri F, Sonzogni A, Sampietro GM, Nespoli A, Viale G, Allavena P, Rescigno M. Intestinal immune homeostasis is regulated by the crosstalk between epithelial cells and dendritic cells. Nat Immunol. 2005;6:507–514. doi: 10.1038/ni1192. [DOI] [PubMed] [Google Scholar]
- 14.Jonuleit H, Schmitt E, Schuler G, Knop J, Enk AH. Induction of interleukin 10-producing, nonproliferating CD4(+) T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J Exp Med. 2000;192:1213–1222. doi: 10.1084/jem.192.9.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Cao X. Long noncoding RNAs in innate immunity. Cell Mol Immunol. 2016;13:138–147. doi: 10.1038/cmi.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berindan-Neagoe I, Pdel Monroig C, Pasculli B, Calin GA. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 22.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neuro-Oncol. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 23.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 24.Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 25.Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 26.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 29.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36:2403–2407. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 30.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 31.Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, Noguchi M. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 2006;97:106–112. doi: 10.1111/j.1349-7006.2006.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, Wu JW. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36:3355–3359. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]
- 33.Gutschner T, Hämmerle M, Diederichs S. MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 34.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corti M, Brody AR, Harrison JH. Isolation and primary culture of murine alveolar type II cells. Am J Respir Cell Mol Biol. 1996;14:309–315. doi: 10.1165/ajrcmb.14.4.8600933. [DOI] [PubMed] [Google Scholar]
- 36.Nie W, Yan H, Li S, Zhang Y, Yu F, Zhu W, Fan F, Zhu J. Angiotensin-(1–7) enhances angiotensin II induced phosphorylation of ERK1/2 in mouse bone marrow-derived dendritic cells. Mol Immunol. 2009;46:355–361. doi: 10.1016/j.molimm.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 37.Papazian D, Chhoden T, Arge M, Vorup-Jensen T, Nielsen CH, Lund K, Würtzen PA, Hansen S. Effect of polarization on airway epithelial conditioning of monocyte-derived dendritic cells. Am J Respir Cell Mol Biol. 2015;53:368–377. doi: 10.1165/rcmb.2014-0183OC. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Weitnauer M, Schmidt L, Leong Ng Kuet N, Muenchau S, Lasitschka F, Eckstein V, Hübner S, Tuckermann J, Dalpke AH. Bronchial epithelial cells induce alternatively activated dendritic cells dependent on glucocorticoid receptor signaling. J Immunol. 2014;193:1475–1484. doi: 10.4049/jimmunol.1400446. [DOI] [PubMed] [Google Scholar]
- 40.Diamond G, Legarda D, Ryan LK. The innate immune response of the respiratory epithelium. Immunol Rev. 2000;173:27–38. doi: 10.1034/j.1600-065X.2000.917304.x. [DOI] [PubMed] [Google Scholar]
- 41.Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol. 2015;16:318–325. doi: 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dijkstra JM, Ballingall KT. Non-human lnc-DC orthologs encode Wdnm1-like protein. Version 2. F1000Res. 2014;3:160. doi: 10.12688/f1000research.4711.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo F, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517–522. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Spurlock CF, III, Tossberg JT, Guo Y, Collier SP, Crooke PS, III, Aune TM. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nature Commun. 2015;6:6932. doi: 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: Influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. 2012;189:2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L, Zhang H, Chen ZW, Zeng G. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci USA. 2015;112:E3883–E3892. doi: 10.1073/pnas.1501662112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 48.Souza PP, Brechter AB, Reis RI, Costa CA, Lundberg P, Lerner UH. IL-4 and IL-13 inhibit IL-1β and TNF-α induced kinin B1 and B2 receptors through a STAT6-dependent mechanism. Br J Pharmacol. 2013;169:400–412. doi: 10.1111/bph.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W, Zhang Q, Zhang J, Pan W, Zhao J, Xu Y. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 2017;7:19. doi: 10.1186/s13578-017-0147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li HB, You QS, Xu LX, Sun LX, Majid Abdul AS, Xia XB, Ji D. Long non-coding RNA-MALAT1 mediates retinal ganglion cell apoptosis through the PI3K/Akt signaling pathway in rats with glaucoma. Cell Physiol Biochem. 2017;43:2117–2132. doi: 10.1159/000484231. [DOI] [PubMed] [Google Scholar]
- 51.Sun KK, Hu PP, Xu F. Prognostic significance of long non-coding RNA MALAT1 for predicting the recurrence and metastasis of gallbladder cancer and its effect on cell proliferation, migration, invasion, and apoptosis. J Cell Biochem. 2018;119:3099–3110. doi: 10.1002/jcb.26451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.