Abstract
Through next generation sequencing, this study evaluated the circulating tumor DNA (ctDNA) of advanced breast cancer patients to prospectively explore the relationship between specific DNA mutations and prognosis as well as therapeutic decision making. The target region covered 1021 gene totally. Clinical characteristics, treatment and outcome data were collected. We analyzed progression-free survival (PFS) from first-line therapy and overall survival (OS), and found that their endpoints were correlated with observed gene mutations. We enrolled 54 patients, with a median follow-up time of 8 years. Mutations were found in TP53, PIK3CA, and ERBB family, at 40.7%, 35.2%, and 25.9%, respectively. PIK3CA more frequently occurred in the site of 3140 A>G (p.H1047R) for 20.4% and HER2+ diseases, and it was associated with shorter median PFS and worse OS among HER2+ patients [mutant vs. wild type: 4 (range 2-9) vs. 8 (range 2-22) months, P=0.006], and [mutant vs. wild type: 29 (range 12-74) vs. 64 (range 20-96) months, P=0.043], respectively. The patients with mutations in TP53 had shorter OS (median 64 vs. 121 months, P=0.006). Multivariate analysis for HER2+ disease demonstrated that the PIK3CA p.H1047R mutation was the only factor associated with shorter PFS (P=0.025); while the receiver operating characteristic (ROC) analysis produces an area under the curve (AUC) of 0.789. The ctDNA analysis, found PIK3CA p.H1047R mutation was more frequent in HER2+ disease and associated with worse OS. It was also the only mutation associated with shorter PFS through a multivariate analysis of HER2+ patients who were treated with trastuzumab, suggesting trastuzumab had lower activity in these patients. The presence of a TP53 mutation was associated with worse OS.
Keywords: Plasma ctDNA, next generation sequencing, PIK3CA mutation, clinical outcome, advanced breast cancer
Introduction
Breast cancer is the most common cancer in women worldwide, and it is the leading cause of cancer-related death in women. There are 272.4 thousands newly diagnosed breast cancer patients in China each year, among them about 10% are initially diagnosed with advanced disease, while most cases of the remaining 90% will progress to metastatic disease over time [1-3]. Therefore, it is of great significance to improve the management of metastatic disease.
According to current guidelines, the management of metastatic disease is chemotherapy, endocrine therapy and/or targeted therapy based on the specific molecular subtype, which is the subdivision determined by ER, PR, HER2 and Ki67 status. However, as we are entering the era of personalized medicine, much attention has been paid to identifying prognostic markers and cancer therapies that specifically target a tumor’s genetic background. In addition, the advent of new agents that target specific mutations are expected to increase the survival of patients substantially, especially those who are resistant to normal treatment. To apply these agents efficiently, better understandings of the relationships between specific mutations and prognosis are needed.
Although many studies have been conducted to address these relationships, controversial results still persist. One example is the study of PIK3CA. According to Berns et al., the PIK3CA pathway is associated with HER2 positive (HER2+) disease, and is resistant to trastuzumab [4]. In the NeoSphere study, Bianchini et al. also found that PIK3CA exon 9 mutations lacked sensitivity to HER2-targeted monoclonal antibody treatment [5]. However, Barbareschi et al. did not observe any relationship between PIK3CA mutations and trastuzumab-based therapy in neoadjuvant or metastatic breast cancer patients, although they did report PIK3CA hot-spot mutations in 25 breast cancer cases (19%): 12 (9%) in exon 9 and 13 (10%) in exon 20 [6]. Meanwhile, the CLEOPATRA study showed both exon 9 and 20 were resistant of antiHER2 therapy [7]. Therefore, further investigation on PIK3CA mutation is warranted. The contradictory findings might be related to the different patient populations of the studies. Since few studies have been conducted among the Chinese population, this study tries to assess this relationship using an original sample of Chinese advanced breast cancer patients. To our knowledge, this is one of the first studies to assess PIK3CA hotpot mutations in advance Chinese breast cancer patients using NGS, with a sufficiently large sample size.
The different findings might also be resulted from the limitations of conventional metrics. Conventional metrics are limited for studying metastatic cancer due to the lack of tissue and the potential for biological changes during treatment. This problem is addressed by using next-generation sequencing in liquid biopsy. The development of next-generation sequencing (NGS) platforms allows more comprehensive genetic screening. This high-throughput technology has revolutionized the sensitivity and specificity of detecting cell-free circulating tumor DNA (ctDNA), making liquid biopsy in breast cancer a more effective means to capture genomic mutations [8]. This technique allows us to identify actionable genomic alterations, monitor treatment responses, unravel therapeutic resistance, detect disease progression before clinical decision making, and characterize tumor heterogeneity and metastasis-specific mutations, all of which provide valuable information to adapt therapy decisions.
In this study, we evaluated ctDNA in advanced breast cancer patients using prospectively collected samples to explore the relationship between specific DNA mutations, prognosis and therapeutic decision making.
Material and methods
Patient selection
This was a formal-prospective/retrospective study. The patients who were enrolled matched the following criteria: having advanced (either metastatic or primary advanced) measurable disease with complete clinical follow-up data and a plasma sample collected prospectively at the time of diagnosis; some patients who had plasma samples during treatment were also enrolled in this study. All patients were confirmed to have ductal invasive breast cancer by immunohistochemical detection in our hospital. The study was approved by the Ethics Committee of Beijing Cancer Hospital (Beijing, China). Informed consents were obtained from all enrolled patients before sampling.
Study purpose
The primary purpose of the study is assessing the relationship between ctDNA-specific mutations and progression-free survival (PFS) in first line, especially trastuzumab-resistance in HER2+ patients. The tumor response rate was evaluated using the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 [9]; tumor measurements were performed by computed tomography scan or magnetic resonance imaging. Patients were evaluated once of two cycles of therapy, and an analysis was conducted to correlate ctDNA profiles in different patient subsets. Another purpose is assessing the relationship between ctDNA mutations and overall survival (OS), including both survival from diagnosed breast cancer (OS1) and from advanced disease (OS2).
Sample processing and DNA extraction
Extraction of cell-free DNA and genomic DNA was performed as previously described [10]. Peripheral blood of advanced breast cancer patients were collected using EDTA vacutainer tubes, which was processed within 3 h after collection. Plasma was separated by centrifugation at 2,500 g for 10 min, transferred to microcentrifuge tubes and centrifuged at 16,000 g for 10 min to remove cell debris. The cell pellet from the initial spin was used for isolation of germline genomic DNA from Peripheral Blood Lymphocyte (PBL). PBL DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Cell-free circulating DNA was isolated from plasma using QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). Qubit fluorometer (Invitrogen, Carlsbad, CA, USA) dsDNA HS kit was adopted to measure DNA concentrations, after which size distributions of the cell-free DNA (cfDNA) were assessed using Agilent 2100 BioAnalyzer and the DNA HS kit (Agilent Technologies, Santa Clara, CA, USA).
Sequencing library construction and target enrichment
Microgramme PBL DNA was sheared to 300-bp fragments using a Covaris S2 instrument before library construction. Indexed Illumina NGS libraries were prepared from PBL germline and circulating DNA with KAPA Library Preparation Kit (Kapa Biosystems, Boston, MA, USA).
Target enrichment was then performed using a custom SeqCap EZ Library (Roche NimbleGen, Madison, WI, USA). In order to learn more about comprehensive tumor genetic character, the capture probe was designed based on genomic regions selected with 1021 genes in total, which covered the most frequently mutated genes and exons in solid tumors. Capture hybridization was carried out according to the manufacturer’s protocol. After hybrid selection, the captured DNA fragments were amplified and then pooled to several multiplexed libraries.
NGS Sequencing and data analysis
Sequencing was carried out using Illumina 2×75 bp paired-end sequencing on a Illumina HiSeq 3000 and NextSeq 500 instruments, using TruSeq PE Cluster Generation Kit v3 and the TruSeq SBS Kit v3 (Illumina, San Diego, CA, USA) according to the manufacturers’ recommendations. Terminal adaptor sequences and low-quality data were removed, after which reads were mapped to the reference human genome and aligned. GATK and MuTect were employed to call somatic small insert and delete (indel), and single nucleotide variants (SNV) by filtering PBL sequencing data [10-12].
Statistical analysis
According to clinically important baseline status, biomarker, metastatic status and treatment in first line, patient outcomes were analyzed using the Kaplan-Meier method and compared by the log-rank test. Relationships between mutational status and clinicopathological characteristics were detected using the Chi-square test. Using the Cox proportional hazards model, we also performed a multivariate analysis considering all the relevant variables, with forward stepwise variable selection. P-values less than 0.05 were considered statistically significant.
To assess the sensitivity and specificity of high frequency mutations, the area under the feature mutation curve (AUC) was calculated to measure the accuracy of predicting PFS. Statistical analyses were conducted using Statistical Package for the Social Sciences (SPSS) for Windows version 10.0 (SPSS Inc., Chicago, IL, USA).
Results
Patient characteristics
The study analyzed 72 samples from 61 patients in total. Every patient was tested at baseline, where no mutation was found for 7 patients, so they were excluded from the study. Among the 54 patients who had at least one mutation at baseline, 11 patients were also tested again after treatment, adding 11 samples to the study. Therefore, further analysis will be based on 65 samples (54 at baseline, 11 during treatment) from 54 patients. The 54 patients enrolled all had invasive ductal breast cancer, including 27 (50%) HER2+ patients (17 HER2+, 10 luminal B HER2+) and 27 HER2- patients [22 (41%) hormone receptor+ (HR+)/HER2- (luminal B/HER2- in 9, luminal A in 13), 5 (9%) triple negative breast cancer (TNBC)]. Median age was 48 years old (range 26-74 years old). The median follow-up was 96 months (range 12-180 months). Before being enrolled into the study, 50 (92.6%) patients received surgery, and 4 (7.4%) patients were diagnosed advanced disease initially. In adjuvant setting for patients after surgery, none of the them who were HER2+ had received trastuzumab in adjuvant setting due to economic reasons, 47 received chemotherapy; 30 estrogen/progesterone receptor positive (ER+/PR+) patients received hormonal therapy, and 15 patients received radiotherapy as individually indicated. Patient characteristics and disease-free survival (DFS) analysis are shown in (Table 1).
Table 1.
Patient’s characteristic and survival analysis of DFS by clinically important baseline status in the first diagnosed breast cancer and biomarker at the start of the study
| Total N=54 | No. of case (percentage %) | DFS (months) | χ2 | p-value | |
|---|---|---|---|---|---|
|
| |||||
| Median ± SD | Range | ||||
| Age (year) | 0.53 | 0.818 | |||
| ≤50 | 33 (61.1) | 38±7.46 | 0-145 | ||
| >50 | 21 (38.9) | 13±3.43 | 0-120 | ||
| Total | 54 | 21±8.398 | 0-145 | ||
| Tumor grade | 2.235 | 0.327 | |||
| Grade I, II | 37 (68.5) | 21±5.052 | 0-145 | ||
| Grade III | 13 (24.1) | 30±11.234 | 2-73 | ||
| Unknown | 4 (7.4) | 65±28.000 | 12-120 | ||
| Tumor size | 2.737 | 0.254 | |||
| T1 ≤2 | 11 (20.4) | 40±15.276 | 3-107 | ||
| T2 2-≤5 | 26 (48.1) | 21±8.286 | 0-145 | ||
| T3-T4 >5 | 17 (31.5) | 13±6.174 | 0-72 | ||
| Nodal status | 56.533 | <0.001 | |||
| Negative | 15 (27.8) | 21±14.813 | 4-65 | ||
| 1-3 | 13 (24.1) | 42±17.375 | 2-145 | ||
| 4-9 | 13 (24.1) | 40±6.591 | 6-120 | ||
| ≥10 | 9 (16.7) | 12±2.981 | 3-107 | ||
| Primary IV | 4 (7.4) | 0 | 0 | ||
| ER | 0.006 | ||||
| Positive | 30 (55.6) | 41±6.162 | 0-145 | 7.653 | |
| Negative | 24 (44.4) | 12±2.449 | 0-24 | ||
| PR | 0.021 | ||||
| Positive | 27 (50) | 41±5.193 | 0-145 | 5.354 | |
| Negative | 27 (50) | 13±1.731 | 0-73 | ||
| HER2 | 6.486 | 0.011 | |||
| Negative | 27 (50.0) | 38±12.981 | 0-145 | ||
| Positive | 27 (50.0) | 13±1.731 | 0-73 | ||
| Ki67 activity | 4.386 | 0.223 | |||
| Negative | 1 (1.9) | 13 | 13 | ||
| ≤20% | 14 (25.9) | 21±7.951 | 0-145 | ||
| >20% | 33 (61.1) | 14±5.742 | 0-107 | ||
| Unknown | 6 (11.1) | 58±6.736 | 12-120 | ||
| PI3KCA | 0.023 | 0.878 | |||
| WT | 35 (64.8) | 27±10.560 | 0-145 | ||
| MT | 19 (35.2) | 14±4.897 | 0-120 | ||
| PI3KCA p.H1047R | 0.450 | 0.502 | |||
| WT | 43 (79.6) | 22±7.492 | 0-145 | ||
| MT | 11 (20.4) | 13±18.166 | 2-120 | ||
| ERBB1-4 | 2.111 | 0.146 | |||
| WT | 40 (74.1) | 21±5.053 | 0-120 | ||
| MT | 14 (25.9) | 38±28.062 | 2-145 | ||
| PI3KCA+ERBB2 | 2.551 | 0.110 | |||
| WT | 30 (55.6) | 21±6.148 | 0-73 | ||
| MT | 24 (44.4) | 36±17.759 | 0-145 | ||
| TP53 | 1.919 | 0.166 | |||
| WT | 32 (59.3) | 36±12.728 | 0-145 | ||
| MT | 22 (40.7) | 21±4.612 | 2-72 | ||
| Adjuvant CT | 1.881 | 0.597 | |||
| Anthracycline based | 9 (16.7) | 39±26.833 | 4-145 | ||
| Taxol based | 4 (7.4) | 12±21.000 | 0-65 | ||
| A+T | 34 (62.9) | 21±10.788 | 0-120 | ||
| No CT | 3 (5.6) | 14±6.532 | 6-59 | ||
| IV | 4 | 0 | |||
| Adjuvant ET | 17.710 | <0.001 | |||
| No | 24 (44.4) | 12±1.220 | 0-67 | ||
| Yes | 30 (55.6) | 43±6.847 | 2-145 | ||
| DFS (months) | |||||
| Total | 54 | 21±8.398 | 0-145 | ||
| <12 m | 19 (35.2) | ||||
| 12-24 | 9 (16.7) | ||||
| 24-36 | 3 (5.6) | ||||
| >36 | 23 (66.7) | ||||
Note: DFS: disease free survival; ER: estrogen receptor; PR: progesterone receptor; MT: mutation, WT: wild type; A: Anthracycline; T: Taxol; CT: chemotherapy; ET: endocrintherapy; IV: initially advanced disease; If one patients has more than one of same gene mutation but not same site will only count once.
Most patients (61.1%) had visceral metastases, and half of them (50%) had more than one metastasis site. After patients developed advanced disease, all HER2+ patients were treated with trastuzumab combined with chemotherapy; among them 20 patients were given combinations with taxane-based chemotherapy, while 7 were given combinations with vinorelbine-based chemotherapy agents; 17 HR+/HER2- patients were treated with chemotherapy sequential endocrine maintenance treatment and 5 with endocrine therapy alone; the 5 TNBC patients were treated with chemotherapy (Table 2).
Table 2.
Kaplan Meier survival analysis of PFS by clinically important baseline status, biomarker at the start of the study, metastatic status and treatment in first line
| N=54 | Number of case | PFS | χ2 | p-value | |
|---|---|---|---|---|---|
|
| |||||
| Median ± SD | Range | ||||
| Age (year) | 5.924 | 0.015 | |||
| ≤50 | 33 (61.1) | 10±1.136 | 2-25 | ||
| >50 | 21 (38.9) | 4±0.981 | 2-22 | ||
| Tumor grade | 3.069 | 0.216 | |||
| Grade I, II | 37 (68.5) | 8±1.299 | 2-24 | ||
| Grade III | 13 (24.1) | 8±1.348 | 4-22 | ||
| Unknown | 4 (7.4) | 8±9.500 | 3-25 | ||
| Tumor size | 0.934 | 0.627 | |||
| T1 ≤2 | 11 (20.4) | 9±4±2.890 | 3-25 | ||
| T2 2-≤5 | 26 (48.1) | 8±4±1.525 | 2-24 | ||
| T3-T4 >5 | 17 (31.5) | 84±4±0.676 | 2-22 | ||
| Nodal status | 1.358 | 0.851 | |||
| Negative | 15 (27.8) | 9±1.932 | 2-25 | ||
| 1-3 | 13 (24.1) | 10±1.797 | 2-22 | ||
| 4-9 | 13 (24.1) | 8±1.387 | 2-24 | ||
| ≥10 | 9 (16.7) | 8±1.414 | 3-20 | ||
| Primary IV | 4 (7.4) | 7±0.433 | 6-14 | ||
| ER | 0.167 | 0.683 | |||
| Positive | 30 (55.6) | 9±1.366 | 2-24 | ||
| Negative | 24 (44.4) | 8±0.949 | 2-25 | ||
| PR | 1.546 | 0.214 | |||
| Positive | 27 (50.0) | 10±1.277 | 2-25 | ||
| Negative | 27 (50.0) | 8±1.004 | 2-22 | ||
| HER2 | 0.778 | 0.378 | |||
| Negative | 27 (50.0) | 9±1.298 | 2-25 | ||
| Positive | 27 (50.0) | 8±0.418 | 2-22 | ||
| Ki67 activity | 3.326 | 0.344 | |||
| Negative | 1 (1.9) | 8 | 8 | ||
| <20% | 14 (25.9) | 6±2.806 | 2-18 | ||
| >20% | 33 (61.1) | 9±0.811 | 2-24 | ||
| Unknown | 6 (11.1) | 8±6.736 | 3-25 | ||
| PI3KCA | |||||
| WT | 35 (64.8) | 8±0.845 | 2-24 | 0.323 | 0.570 |
| MT | 19 (35.2) | 8±2.152 | 2-25 | ||
| PI3KCA p.H1047R | 0.220 | 0.639 | |||
| WT | 43 (79.6) | 9±0.892 | 2-24 | ||
| MT | 11 (20.4) | 4±0.826 | 2-25 | ||
| ERBB1-4 | 0.433 | 0.510 | |||
| WT | 40 (74.1) | 8±0.790 | 2-25 | ||
| MT | 14 (25.9) | 6±2.806 | 2-22 | ||
| ERBB2 | 1.080 | 0.299 | |||
| WT | 48 (88.9) | 8±0.767 | 2-25 | ||
| MT | 6 (11.1) | 8±3.062 | 4-12 | ||
| PI3KCA+ERBB1-4 | 0.118 | 0.731 | |||
| WT | 30 (55.6) | 8±0.781 | 2-24 | ||
| MT | 24 (44.4) | 8±1.831 | 2-25 | ||
| TP53 | 0.192 | 0.662 | |||
| WT | 32 (59.3) | 8±0.807 | 2-25 | ||
| MT | 22 (40.7) | 8±2.919 | 2-22 | ||
| Visceral metastasis | 1.035 | 0.309 | |||
| No | 21 (38.9) | 9±2.289 | 2-25 | ||
| Yes | 33 (61.1) | 8±0.568 | 2-22 | ||
| No. of metastasis | 0.428 | 0.934 | |||
| 1 | 27 (50.0) | 9±1.298 | 2-24 | ||
| 2 | 15 (27.8) | 8±0.949 | 2-25 | ||
| 3 | 10 (18.5) | 4±2.372 | 2-22 | ||
| 4 | 2 (3.7) | 8-10 | |||
| Liver primary metastasis | 0.295 | 0.587 | |||
| No | 28 (51.9) | 8±1.583 | 2-25 | ||
| Yes | 26 (48.1) | 8±1.017 | 2-22 | ||
| First line Treat after metastasis (for HER2-) | 1.395 | 0.498 | |||
| T based | 17 (62.9) | 7±2.572 | 2-24 | ||
| Other CT agents | 5 (18.5) | 10±2.191 | 3-22 | ||
| ET | 5 (18.5) | 11±2.191 | 3-25 | ||
| First line treat after metastasis (for HER2+) | 6.845 | 0.009 | |||
| H+T | 20 (74.1) | 8±0.445 | 2-22 | ||
| H+other CT agents | 7 (25.9) | 5±1.309 | 2-9 | ||
| PFS (month) | |||||
| Median (Range) | 8±0.733, 2-25 | ||||
| ≤12 m | 41 | ||||
| 12-25 | 13 | ||||
T: taxol; CT: chemotherapy; ET: endocrintherapy; MT: mutation, WT: wild type; H: trastuzumab.
DNA extraction and mutation analysis
In total, 72 samples got sufficient cell-free DNA (cfDNA), and were tested in mutation analysis, while 65 samples (54 at baseline, 11 during treatment) were detected to have at least one mutation per sample, and were enrolled in the subsequent study. The extraction quantity of cfDNA in these patients were between 9.2-530.4 ng (average 93.15 ng). No mutation was found for 7 samples, so they were excluded from further analysis, although the extracted quantity of cfDNA in these samples (around 14.04-65.73 ng, average 42.05 ng) was enough for mutation analysis.
Mutation analyses were performed by NGS and 255 mutations were found. Mutations were found in patients of TP53, PIK3CA, and ERBB (including ERBB1-4), at 40.7%, 35.2%, and 25.9% (Table 1), respectively. ERBB2 mutations more likely occurred in HER2+ disease than HER2- disease [5/27 (18.5%) vs. 1/27 (3.7%), P=0.192], but the difference was not significant; ESR1 (3 cases) mutations only occurred in HR+ patients in this study.
To compare mutation profiles in different situations, ctDNA examinations were extended from baseline to during treatment for 11 patients (shown in Table 3). We found that ERBB1-4, ERBB2, PIK3CA 3140 A>G (p.H1047R) and TP53 were present both at baseline and in some treatment samples. Among the 7 patients whose total number of mutations had decreased (including one who remained the same), 5 patients had responded to trastuzumab, while 2 patients had no response. These 2 patients had PIK3CA p.H1047R mutation in both baseline and progression disease (PD), which suggested that even when the total number of mutation had decreased, if PIK3CA p.H1047R mutation still existed, patients will respond poorly and have worse survival. Among the 5 patients who responded well, 1 patient had PIK3CA p.H1047R at baseline, but was free of it after trastuzumab treatment. This explained why her PFS was better than the 2 patients whose PIK3CA p.H1047R mutation did not disappear. The other 4 patients, 3 responded well to trastuzumab treatment, did not have PIK3CA p.H1047R mutation in both samples.
Table 3.
The relations of PFS and gene mutation changing in baseline and after trastuzumab (T) treatment
| Patient No | Time of Sampling | Disease status of Sampling | PFS on T treatment (m) | ERBB 1,3,4 mutation | ERBB2 | PIK3CA other mutation | PIK3CA p.H1047R | TP53 mutation | No. of all mutation (include other gene) | No.of mutation increase/decrease |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2014-11-05 | Baseline | 0 | 0 | 0 | 0 | 0 | 3 | ||
| 2015-06-24 | PD | 7 | 0 | 0 | 0 | 0 | 0 | 6 | ↑ | |
| 2 | 2014-07-12 | Baseline | 0 | 0 | 0 | 0 | 1 | 2 | ||
| 2015-08-12 | Duration | 22 | 0 | 0 | 0 | 0 | 0 | 1 | ↓ | |
| 3 | 2013-06-20 | Baseline | 0 | 0 | 1 | 0 | 1 | 4 | ||
| 2014-09-05 | PD | 15 | 0 | 0 | 1 | 0 | 1 | 4 | = | |
| 5 | 2013-09-09 | Baseline | 1 | 0 | 0 | 1 | 1 | 4 | ||
| 2014-04-09 | PD | 8 | 0 | 0 | 0 | 0 | 0 | 1 | ↓ | |
| 6 | 2014-03-25 | Baseline | 0 | 0 | 0 | 0 | 0 | 1 | ||
| 2014-07-28 | Duration | 8 | 0 | 0 | 0 | 0 | 1 | 3 | ↑ | |
| 8 | 2013-04-05 | Baseline | 0 | 0 | 0 | 0 | 0 | 2 | ||
| 2014-09-11 | PD | 14 | 0 | 0 | 0 | 1 | 1 | 4 | ↑ | |
| 11 | 2013-05-06 | Baseline | 0 | 0 | 1 | 0 | 1 | 5 | ||
| 2013-08-01 | Duration | 12 | 0 | 0 | 0 | 0 | 0 | 1 | ↓ | |
| 12 | 2015-05-16 | Baseline | 1 | 1 | 1 | 0 | 1 | 12 | ||
| 2015-09-28 | Duration | 10 | 0 | 0 | 0 | 0 | 0 | 1 | ↓ | |
| 14 | 2016-03-16 | Baseline | 1 | 0 | 0 | 1 | 1 | 6 | ||
| 2016-04-22 | PD | 2 | 0 | 0 | 0 | 1 | 0 | 1 | ↓ | |
| 20 | 2013-09-13 | Baseline | 0 | 0 | 0 | 0 | 1 | 3 | ||
| 2013-12-09 | PD | 4 | 0 | 0 | 0 | 0 | 1 | 5 | ↑ | |
| 21 | 2016-02-22 | Baseline | 0 | 1 | 0 | 1 | 1 | 4 | ||
| 2016-04-18 | PD | 2 | 0 | 1 | 0 | 1 | 1 | 3 | ↓ |
Note: PFS: Progression free survival in first line of trastuzumab; T: Trastuzumab; m: month; PD: progression disease; ↑: number of mutation increase; ↓: number of mutation decrease; =: number of mutation no change.
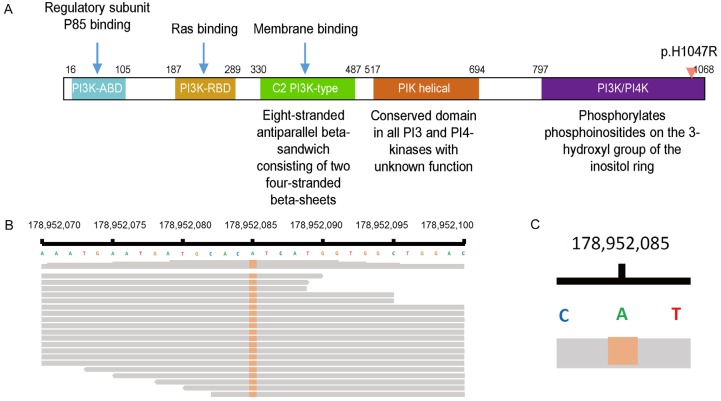
All mutations were missense, with either single nucleotide variation in ERBB1-4, PIK3CA, or other kinds of mutation in TP53, which were defined as insertion and deletion. PIK3CA mutations were most frequently observed in exon 20 (3140 A>G, p.H1047R) in 11 patients (20.3%) and exon 9 (1633 G>A, p.E545K) in 5 patients (9.3%). ERBB1-4, PIK3CA, PIK3CA p.H1047R and TP53 presented higher mutational frequencies and were considered in further survival analysis.
Survival analysis
Survival analysis by clinically important baseline status and biomarkers
The median DFS of the samples was 21±8.398 months (range 0-145 months). DFS was significantly longer for ER+, PR+, and HER2- patients, with p-values of 0.006, 0.021, and 0.011 respectively; while lymph node metastasis had a worse DFS. ERBB, PIK3CA and TP53 mutations were not correlated with DFS (Table 1).
Prognostic assessment of ERBB, PIK3CA and TP53 mutations for PFS
The frequency of ERBB mutations was the same in HER2+ 7/27 (25.9%) and HER2- 7/27 (25.9%) disease; and the mutation had no significant impact on PFS in any subset, P=0.707 and P=0.066, respectively. ERBB2 mutations more frequently occurred in HER2+ than HER2- disease [5/27 (18.5%) vs. 1/27 (3.7%), P=0.192], but the difference was not significant in this study. TP53 mutations were significantly higher in HER2+ (29.63%) than HER2- disease (11.11%, P=0.012); PIK3CA mutations were almost equally present in HER2+ (10 cases, 18.52%) and HER2- (9 cases, 16.67%) disease; none were related to PFS (Table 2).
PIK3CA p.H1047R mutation is an independent prognostic factor in HER2+ patients
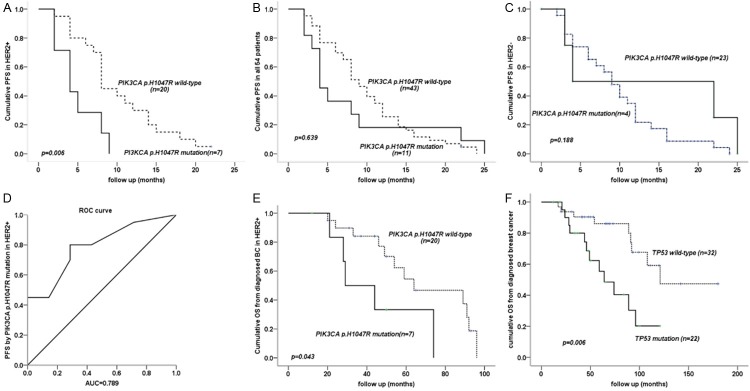
In total, the PIK3CA p.H1047R mutation was found in 11 cases (20%); it was more frequent in HER2+ (7/27, 25.9%) than HER2- (4/27, 14.8%) disease. PIK3CA p.H1047R was associated with shorter median PFS in HER2+ disease (mutant vs. wild type: 4 (range 2-9) vs. 8 (range 2-22) months, P=0.006) (Figure 1A); but not in HER2- disease or for all 54 patients, where no significant difference existed (P=0.188, 0.639, respectively) (Figure 1B, 1C).
Figure 1.
A. PIK3CA p.H1047R was associated with shorter median PFS in HER2+ disease; B, C. Total 54 patients and HER- patients, where no significant difference in PFS; D. Receiver operating characteristic (ROC) curve showed that the PIK3CA p.H1047R mutation had a large area uder the curve; E. PIK3CA p.H1047R mutation had shorter OS1 in HER2+ disease; F. ctDNA mutations in TP53 had a shorter OS1. Note: OS1: overall survival from diagnosed breast cancer.
We have done integrated analysis using log-rank test between multiple clinical factors including age, tumor stage, tumor size, nodal status, ER, HER2, ki67 and PIK3CA p.H1047R mutants, and found that PIK3CA p.H1047R was only significantly different between age ≤50 and >50 (P=0.009) (shown in Table S1).
Multivariate analysis for HER2+ disease considered age, ER, Ki67, ERBB, TP53, PIK3CA, and PIK3CA p.H1047R, and demonstrated that the PIK3CA p.H1047R mutation was the only factor associated with shorter PFS (P=0.025). Further analysis by receiver operating characteristic curve showed that the PIK3CA p.H1047R mutation had a large area under the curve, 0.789 (Figure 1D).
There were five TNBC cases among the HER2- patients. No ERBB2 or PIK3CA p.H1047R mutations were found in these patients (0/5), and ERBB mutation alone was not correlated with PFS. Based on the limited number of TNBC cases, it is hard to draw any useful conclusion.
OS analysis by clinically important baseline status and biomarkers
OS was calculated from diagnosis (OS1) and after advanced disease diagnosis (OS2); the median OS1 was 96±11.393 months (range 12-180 months). Longer OS1 was correlated with ER+, PR+, HER2-, Ki67 low, no visceral metastasis, and no primary liver metastasis; p-values were 0.008, 0.007, <0.001, 0.018, 0.006, 0.001, and 0.002, respectively. We found that PIK3CA p.H1047R mutation had shorter OS1 in HER2+ disease with P=0.043 (Figure 1E), while this relationship is not found for OS2. In an univariate analysis, patients with ctDNA mutations in TP53 had a shorter OS1 (median 64 vs. 121 months, P=0.006) (Figure 1F).
Median OS2 was 71±19.570 months (range 2-93 months). Longer OS2 was correlated with HER2-, Ki67 low, no visceral metastasis, fewer site of metastases, and no primary liver metastasis; p-values were <0.001, 0.001, 0.001, <0.001, and 0.006, respectively.
Discussion
In this study, we found that TP53, PIK3CA, and ERBB mutations occurred most frequently in advanced breast cancer (40.7%, 35.2%, and 25.9%, respectively) and were associated with different molecular subtypes. We also showed that some mutations found at baseline also occurred during the treatment.
The tumor suppressor gene TP53 is located on the short arm of chromosome 17 (17p13.1) [13], and is mutated in approximately 38.8% of breast cancer [12]. While mutation rates between intrinsic or immunophenotypical tumor subtypes vary, the TP53 mutation occurs in a high percentage (62% or 81%) [12,14] in TNBC and (67%) HER2+ patients [12]. However, patients with TP53 mutations have shown a tendency toward worse prognoses than those without it [13]. In this study, we also found that TP53 mutations were associated with shorter OS1 (P=0.006). We did not analyze TP53 mutation in TNBC, because of the limited number of samples available.
Few studies have addressed ERBB mutations [12,15]. In our study, ERBB mutations were higher, and the same in HER2- (25.9%) and HER2+ (25.9%) disease, while ERBB2 mutations were more frequent in HER2+ than HER2- disease (18.5% vs. 3.7%), but neither was associated with either OS or PFS.
The PIK3CA gene, encoding the p110α catalytic subunit, is the most frequently mutated gene in breast cancer, with mutations found in 25%-40% of all cases [16-20]. Two publications have reported PIK3CA mutations in 35.2% and 28.12% of Chinese breast cancer patients [21,22]. These findings are comparable to ours, in which the PIK3CA mutation was found in 35.2% of the cases. The frequency of PIK3A mutations is different in HER2+ positive breast cancer (31%) [23] and in HR+ (45%) [17], while low frequencies were found in TNBC (13.6%) [24]. In our study, the frequency of PIK3A mutation in different subgroups is HER2+ (37%), HR+ (40.9%) and TNBC (20%), which is also similar to existing literature. However, we did not find any significant survival differences correlated with PIK3CA mutations in any subgroup, while some previous studies reported an association between PIK3CA mutation and poor prognosis in HR+ [25,26] and HER2+ breast cancer [16]. Although the difference between studies in terms of sample size, sample selection, geographical distribution and the use of different techniques might partially explain why different results can occur, further studies should be carried out to address this problem.
Approximately 80% of PIK3CA mutations take place within the helical (E542K, E545K) and kinase (H1047R) domains (Figure 2A), as these lead to increased catalytic activity (i.e. production of phosphatidylinositol triphosphate) that enhances cell proliferation and survival. These mutations (exons 9: helical domain p.E542K and p.E545K, and exon 20: kinase domain p.H1047R) [27] were observed in over 90% of cases in our study, which was similar to previous reports. We found 11 (20%) PIK3CA exon 20 (p.H1047R) mutations (Figure 2B, 2C), all of which were missense mutations; while in studies of Western population, p.H1047L mutations were more frequent [4,20]. One explanation for this may be that Chinese breast cancer patients are more prone to p.H1047R than p.H1047L [22] mutations, but further study is needed to confirm this hypothesis. Since phosphorylation of PI3K kinase is an important downstream event of HER2 signaling pathway while p.H1047R may led to malfunction of PI3K/PI4K domain. Thus we sought to find out if the p.H1047R contributes to the disease progression in HER2 positive patients.
Figure 2.
The PIK3CA mutations take place frequently in kinase (H1047R) domains.
According to previous literature like the BOLERO-2 study [28], for HER2- cases, PIK3CA p.H1047R mutation was not associated with different PFS. For HER2+ cases, however, the prognostic implication of this activating mutation remains controversial. While some found that patients with exon 20 mutations had improved survival [29-31], some found no significant association [5,32], and some found significantly worse survival in patients with exon 20 mutations [7,26]. In this study, we examined 54 patients and found that PIK3CA p.H1047R exon 20 mutations, but not other PIK3CA mutations, were significantly associated with worse OS in HER2+ cases (P=0.043), but not in HER2- disease. More importantly, we found that PIK3CA p.H1047R was significantly associated with shorter PFS in HER2+ disease treated with trastuzumab (mutant vs. wild type: 4 vs. 8 months, P=0.006), but not for HER2-patients (P=0.188). Activating PIK3CA mutations alters cell proliferation and survival, and PIK3CA is essential in signaling cascades triggered by HER2. This may explain how PIK3CA p.H1047R mutations were resistant to trastuzumab treatment.
More evidence is found from cell line and animal model studies. Using transfected MDA361 cells that contained E545K PIK3CA mutation, a previous study found that trastuzumab and lapatinib were both effective alone and in combination under normal PTEN conditions. Under low PTEN conditions, however, these cells were resistant to trastuzumab, while lapatinib was still effective. Another cell line study also found that mutant PIK3CA enhanced HER2-mediated transformation of MCF10A breast epithelial cells in vitro [33,34]. Similar results were found in animal model studies. Using a mouse model, a study found that mice expressing both human HER2 and mutant PIK3CA in the mammary epithelium developed tumors with shorter latencies compared with mice expressing either oncogene alone. In addition, HER2+/PIK3CA tumors were resistant to trastuzumab alone and in combination with lapatinib. They found that PIK3CA H1047R accelerates HER2-mediated breast epithelial transformation and metastatic progression, alters the intrinsic phenotype of HER2-overexpressing cancers, and generates resistance to approved combinations of anti-HER2 therapies [35].
Finally, in a multivariate Cox survival model that included age, ER, Ki67, TP53, PIK3CA, ERBB, and PIK3CA p.H1047R, we found that PIK3CA p.H1047R was the only biomarker that served as an independent risk factor (P=0.025) for PFS in HER2+ patients, with relatively high sensitivity (AUC=0.789). This specific mutation site in PIK3CA p.H1047R suggests that each cancer sample could present a specific pattern of molecular alteration, leading to the concept of individualized medicine in cancer treatment.
Conclusions
Using NGS on ctDNA, we found that the PIK3CA p.H1047R mutation was more frequent in HER2+ disease and associated with worse OS. This was the only mutation associated with shorter PFS on multivariate analysis in HER2+ patients who were treated with trastuzumab, suggesting trastuzumab had lower activity in these patients. The presence of a TP53 mutation was also associated with worse OS from diagnosed. Overall, evaluating ctDNA is feasible in a general breast cancer population and has prognostic impact.
Acknowledgements
We thank James P. Mahaffey, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text review of this manuscript. This study was funded by China Medical Women’s Association.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Sun S, Yuan JP, Wang YH, Cao TZ, Zheng HM, Jiang XQ, Gong YP, Tu Y, Yao F, Hu MB, Li JJ, Sun SR, Wei W. Characteristics of breast cancer in Central China, literature review and comparison with USA. Breast. 2016;30:208–213. doi: 10.1016/j.breast.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Shao B, Yan Y, Song G, Liu X, Wang J, Liang X. Efficacy and safety of trastuzumab combined with chemotherapy for first-line treatment and beyond progression of HER2-overexpressing advanced breast cancer. Chin J Cancer Res. 2016;28:330–338. doi: 10.21147/j.issn.1000-9604.2016.03.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, Beijersbergen RL, Mills GB, van de Vijver MJ, Bernards R. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 5.Bianchini G, Kiermaier A, Bianchi GV, Im YH, Pienkowski T, Liu MC, Tseng LM, Dowsett M, Zabaglo L, Kirk S, Szado T, Eng-Wong J, Amler LC, Valagussa P, Gianni L. Biomarker analysis of the NeoSphere study: pertuzumab, trastuzumab, and docetaxel versus trastuzumab plus docetaxel, pertuzumab plus trastuzumab, or pertuzumab plus docetaxel for the neoadjuvant treatment of HER2-positive breast cancer. Breast Cancer Res. 2017;19:16. doi: 10.1186/s13058-017-0806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbareschi M, Cuorvo LV, Girlando S, Bragantini E, Eccher C, Leonardi E, Ferro A, Caldara A, Triolo R, Cantaloni C, Decarli N, Galligioni E, Dalla Palma P. PI3KCA mutations and/or PTEN loss in Her2-positive breast carcinomas treated with trastuzumab are not related to resistance to anti-Her2 therapy. Virchows Arch. 2012;461:129–139. doi: 10.1007/s00428-012-1267-2. [DOI] [PubMed] [Google Scholar]
- 7.Baselga J, Cortes J, Im SA, Clark E, Ross G, Kiermaier A, Swain SM. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J. Clin. Oncol. 2014;32:3753–3761. doi: 10.1200/JCO.2013.54.5384. [DOI] [PubMed] [Google Scholar]
- 8.De Mattos-Arruda L, Caldas C. Cell-free circulating tumour DNA as a liquid biopsy in breast cancer. Mol Oncol. 2016;10:464–474. doi: 10.1016/j.molonc.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Zhou J, Chang L, Guan Y, Yang L, Xia X, Cui L, Yi X, Lin G. Application of circulating tumor DNA as a non-invasive tool for monitoring the progression of colorectal cancer. PLoS One. 2016;11:e0159708. doi: 10.1371/journal.pone.0159708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fei M, Jianghua W, Rujuan M, Wei Z, Qian W. The relationship of plasma Abeta levels to dementia in aging individuals with mild cognitive impairment. J Neurol Sci. 2011;305:92–96. doi: 10.1016/j.jns.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Roy-Chowdhuri S, de Melo Gagliato D, Routbort MJ, Patel KP, Singh RR, Broaddus R, Lazar AJ, Sahin A, Alvarez RH, Moulder S, Wheler JJ, Janku F, Gonzalez-Angulo AM, Chavez-MacGregor M, Valero V, Ueno NT, Mills G, Mendelsohn J, Yao H, Aldape K, Luthra R, Meric-Bernstam F. Multigene clinical mutational profiling of breast carcinoma using next-generation sequencing. Am J Clin Pathol. 2015;144:713–721. doi: 10.1309/AJCPWDEQYCYC92JQ. [DOI] [PubMed] [Google Scholar]
- 13.Fountzilas G, Giannoulatou E, Alexopoulou Z, Zagouri F, Timotheadou E, Papadopoulou K, Lakis S, Bobos M, Poulios C, Sotiropoulou M, Lyberopoulou A, Gogas H, Pentheroudakis G, Pectasides D, Koutras A, Christodoulou C, Papandreou C, Samantas E, Papakostas P, Kosmidis P, Bafaloukos D, Karanikiotis C, Dimopoulos MA, Kotoula V. TP53 mutations and protein immunopositivity may predict for poor outcome but also for trastuzumab benefit in patients with early breast cancer treated in the adjuvant setting. Oncotarget. 2016;7:32731–53. doi: 10.18632/oncotarget.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madic J, Kiialainen A, Bidard FC, Birzele F, Ramey G, Leroy Q, Rio Frio T, Vaucher I, Raynal V, Bernard V, Lermine A, Clausen I, Giroud N, Schmucki R, Milder M, Horn C, Spleiss O, Lantz O, Stern MH, Pierga JY, Weisser M, Lebofsky R. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer. 2015;136:2158–2165. doi: 10.1002/ijc.29265. [DOI] [PubMed] [Google Scholar]
- 15.Ross JS, Gay LM, Wang K, Ali SM, Chumsri S, Elvin JA, Bose R, Vergilio JA, Suh J, Yelensky R, Lipson D, Chmielecki J, Waintraub S, Leyland-Jones B, Miller VA, Stephens PJ. Nonamplification ERBB2 genomic alterations in 5605 cases of recurrent and metastatic breast cancer: An emerging opportunity for anti-HER2 targeted therapies. Cancer. 2016;122:2654–2662. doi: 10.1002/cncr.30102. [DOI] [PubMed] [Google Scholar]
- 16.Xu B, Guan Z, Shen Z, Tong Z, Jiang Z, Yang J, DeSilvio M, Russo M, Leigh M, Ellis C. Association of phosphatase and tensin homolog low and phosphatidylinositol 3-kinase catalytic subunit alpha gene mutations on outcome in human epidermal growth factor receptor 2-positive metastatic breast cancer patients treated with first-line lapatinib plus paclitaxel or paclitaxel alone. Breast Cancer Res. 2014;16:405. doi: 10.1186/s13058-014-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Saenz JA, Ayllon P, Laig M, Acosta-Eyzaguirre D, Garcia-Esquinas M, Montes M, Sanz J, Barquin M, Moreno F, Garcia-Barberan V, Diaz-Rubio E, Caldes T, Romero A. Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer. 2017;17:210. doi: 10.1186/s12885-017-3185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Top Microbiol Immunol. 2011;347:21–41. doi: 10.1007/82_2010_68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cejalvo JM, Perez-Fidalgo JA, Ribas G, Burgues O, Mongort C, Alonso E, Ibarrola-Villava M, Bermejo B, Martinez MT, Cervantes A, Lluch A. Clinical implications of routine genomic mutation sequencing in PIK3CA/AKT1 and KRAS/NRAS/BRAF in metastatic breast cancer. Breast Cancer Res Treat. 2016;160:69–77. doi: 10.1007/s10549-016-3980-z. [DOI] [PubMed] [Google Scholar]
- 21.Bai X, Zhang E, Ye H, Nandakumar V, Wang Z, Chen L, Tang C, Li J, Li H, Zhang W, Han W, Lou F, Zhang D, Sun H, Dong H, Zhang G, Liu Z, Dong Z, Guo B, Yan H, Yan C, Wang L, Su Z, Li Y, Jones L, Huang XF, Chen SY, Gao J. PIK3CA and TP53 gene mutations in human breast cancer tumors frequently detected by ion torrent DNA sequencing. PLoS One. 2014;9:e99306. doi: 10.1371/journal.pone.0099306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng J, Fu S, Wei C, Tania M, Khan MA, Imani S, Zhou B, Chen H, Xiao X, Wu J, Fu J. Evaluation of PIK3CA mutations as a biomarker in Chinese breast carcinomas from Western China. Cancer Biomark. 2017;19:85–92. doi: 10.3233/CBM-160380. [DOI] [PubMed] [Google Scholar]
- 23.Xu B, Guan Z, Shen Z, Tong Z, Jiang Z, Yang J, DeSilvio M, Russo M, Leigh M, Ellis C. Association of phosphatase and tensin homolog low and phosphatidylinositol 3-kinase catalytic subunit alpha gene mutations on outcome in human epidermal growth factor receptor 2-positive metastatic breast cancer patients treated with first-line lapatinib plus paclitaxel or paclitaxel alone. Breast Cancer Res. 2014;16:405. doi: 10.1186/s13058-014-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad F, Badwe A, Verma G, Bhatia S, Das BR. Molecular evaluation of PIK3CA gene mutation in breast cancer: determination of frequency, distribution pattern and its association with clinicopathological findings in Indian patients. Med Oncol. 2016;33:74. doi: 10.1007/s12032-016-0788-y. [DOI] [PubMed] [Google Scholar]
- 25.Deng L, Chen J, Zhong XR, Luo T, Wang YP, Huang HF, Yin LJ, Qiu Y, Bu H, Lv Q, Zheng H. Correlation between activation of PI3K/AKT/mTOR pathway and prognosis of breast cancer in Chinese women. PLoS One. 2015;10:e0120511. doi: 10.1371/journal.pone.0120511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai YL, Mau BL, Cheng WH, Chen HM, Chiu HH, Tzen CY. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann Surg Oncol. 2008;15:1064–1069. doi: 10.1245/s10434-007-9751-7. [DOI] [PubMed] [Google Scholar]
- 27.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 28.Moynahan ME, Chen D, He W, Sung P, Samoila A, You D, Bhatt T, Patel P, Ringeisen F, Hortobagyi GN, Baselga J, Chandarlapaty S. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR+, HER2- advanced breast cancer: results from BOLERO-2. Br J Cancer. 2017;116:726–730. doi: 10.1038/bjc.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbareschi M, Buttitta F, Felicioni L, Cotrupi S, Barassi F, Del Grammastro M, Ferro A, Dalla Palma P, Galligioni E, Marchetti A. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064–9. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
- 30.Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, Hedvat CV, Traina TA, Solit D, Gerald W, Moynahan ME. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–59. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 31.Dupont Jensen J, Laenkholm AV, Knoop A, Ewertz M, Bandaru R, Liu W, Hackl W, Barrett JC, Gardner H. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res. 2011;17:667–677. doi: 10.1158/1078-0432.CCR-10-1133. [DOI] [PubMed] [Google Scholar]
- 32.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 33.Dave B, Migliaccio I, Gutierrez MC, Wu MF, Chamness GC, Wong H, Narasanna A, Chakrabarty A, Hilsenbeck SG, Huang J, Rimawi M, Schiff R, Arteaga C, Osborne CK, Chang JC. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J. Clin. Oncol. 2011;29:166–173. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakrabarty A, Rexer BN, Wang SE, Cook RS, Engelman JA, Arteaga CL. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29:5193–5203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanker AB, Pfefferle AD, Balko JM, Kuba MG, Young CD, Sanchez V, Sutton CR, Cheng H, Perou CM, Zhao JJ, Cook RS, Arteaga CL. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci U S A. 2013;110:14372–14377. doi: 10.1073/pnas.1303204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.