Abstract
Autophagy is critical for the survival of cancer cells. It has been reported that long noncoding RNA (lncRNA) neuroblastoma associated transcript 1 (NBAT1) exerts as a tumor suppressor in some cancers. However, the role of NBAT1 in autophagy of non-small cell lung cancer (NSCLC) remains unknown. Here, it was reported that NBAT1 overexpression inhibited autophagy, while knockdown of NBAT1 induced autophagy in NSCLC cells. Further mechanistic study showed that NBAT1 interacted with PSMD10 and promoted its degradation, and then inhibited the occupancy of PSMD10 and HSF1 in the ATG7 promoter to suppress ATG7 transcription. A significantly negative correlation between NBAT1 and ATG7 levels was observed in NSCLC tissue. The prognoses of NSCLC patients with low expression of NBAT1 were much worse than those with high-level NBAT1. Moreover, NBAT1 negatively regulated cell viability, clonogenicity, and chemoresistance through inhibition of autophagy. Our findings suggest that the NBAT1-PSMD10-ATG7 axis may be an attractive strategy in NSCLC treatment by suppressing autophagy and chemoresistance.
Keywords: NBAT1, PSMD10, HSF1, protein stability
Introduction
Lung cancer is the leading cause of cancer-related death with 226,000 new cases in the United States in 2012 [1]. Non-small cell lung cancer (NSCLC), including large cell carcinoma, squamous cell carcinoma and adenocarcinoma, accounts for approximately 85% of lung cancer cases. Due to the distant metastasis, the 5-year survival rate remains below 15% [2]. Therefore, a thorough understanding of the molecular mechanism involved in the progression of NSCLC could provide more effective therapeutic targets for NSCLC treatment.
Autophagy is a highly conserved cellular self-degradative process which delivers long-lived proteins and damaged organelles for degrading and recycling these materials and promoting a cell survival response to environmental stress or nutrient starvation [3]. A lot of evidence suggests the important roles of autophagy in cancer initiation and development. However, autophagy exerts opposite functions in the different stages of cancer. Autophagy functions refer to a tumor suppressor at the early stage of tumorigenesis, which protects cells from oxidative stress and harmful genomic mutations [4]. In contrast, autophagy protects cancer cells against environmental stress, nutrient starvation and anticancer treatment at advanced stages of cancer development [5,6].
Recent transcriptomic researches have demonstrated that only approximately 2% of the human genome is protein coding genes, while the majority of the rest is noncoding genes [7]. Among these noncoding transcripts, long noncoding RNAs (lncRNAs) are a class of noncoding RNAs with larger than 200 nucleotides in length and without protein-coding potential [8]. Emerging evidence has proved that lncRNAs play crucial roles in development and progression of diseases, including NSCLC. For example, lncRNA linc00460 is significantly upregulated in NSCLC tissue and closely associated with poor prognoses of NSCLC patients. linc00460 promotes NSCLC cell migration and invasion and induces epithelial-mesenchymal transition through association with hnRNP K [9]. Increased expression of lncRNA MetaLnc9 also correlates with poor prognoses and promotes NSCLC metastasis. Mechanistically, MetaLnc9 physically associates with the glycolytic kinase PGK1, inhibits its ubiquitination, and then activates the AKT/mTOR signaling pathway [10]. Therefore, the identification of functional lncRNAs in NSCLC is important to elucidate the molecular pathogenesis of NSCLC in order to establish better NSCLC therapy.
Recently, it has been reported that lncRNAs are novel regulators of gene expression involved in the regulation of cell autophagy [11,12]. LncRNA neuroblastoma associated transcript 1 (NBAT1), transcribed from the intron of chromosome 6p22, is a newly identified tumor-suppressing lncRNA in neuroblastoma, ovarian cancer, osteosarcoma, and breast cancer. NBAT1 expression is frequently downregulated in these cancers. Loss of NBAT1 contributes to the aggressiveness of these cancers by enhancing cell proliferation, differentiation, migration and invasion through associating with EZH2 and miR-21, or targeting ERK1/2 and AKT [13-16]. However, its role in NSCLC and its relationship with autophagy have not been investigated. In the present study, it was showed that NBAT1 inhibited autophagy through suppression of ATG7 expression to promote NSCLC progression.
Materials and methods
Cell lines
A549 and 95D NSCLC cell lines were cultured in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37°C in a humidified air atmosphere containing 5% CO2.
Construction of stable cells
For lentivirus preparation, 293T cells were transfected with pLV-NBAT1 or pLKO.1-shNBAT1 plasmid, packaging plasmid pMD2G and psPAX2 (Addgene). Then the medium containing lentivirus was harvested and transfected into NSCLC cells. The stable cells were selected by puromycin. The target sequence of shNBAT1 was shown as follow: CCCACAGAGATGAAGTAAC.
Patient tissue samples
NSCLC tumors and their adjacent lung normal tissues were collected from the First Affiliated Hospital of Zhengzhou University, which was approved by the Ethics Committee of Zhengzhou University. Written informed consent was provided for all of the NSCLC patients.
Western blot
The total protein was extracted using RIPA lysis buffer (Beyotime Biotechnology) supplemented with protease inhibitors cocktail (Roche). The proteins were quantified using BCATM Protein Assay Kit (Pierce, Appleton, WI, USA). The western blot was performed according to the standard protocol.
RNA pull-down
NBAT1 was labeled with biotin by using Biotin RNA Labeling Mix, and then incubated with 1% formaldehyde-treated cell lysate overnight. Then, streptavidin beads (Sigma-Aldrich) were added and incubated for 4 hours. After wash, the cell lysate was added to the 5 × SDS-loading buffer, and then subjected to western blot.
Quantitative real-time PCR
Total RNAs were isolated by TRIzol reagent (Life Technologies) according to the manufacturer’s instruction. cDNA was synthesized by using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The qRT-PCR was carried out by using SYBR Green Master Mix in a StepOne Plus real-time PCR instrument (Applied Biosystems). GAPDH was taken as endogenous controls, the relative expression were calculated via the 2-ΔΔCt method.
RNA immunoprecipitation (RIP) and chromatin immunoprecipitation (ChIP)
RIP and ChIP assay was performed by using EZ-Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore) and EZ-Magna ChIPTM chromatin immunoprecipitation Kit (Millipore) according to the manufacturer’s instruction.
CCK-8 and colony formation assay
To detect cellular viability, 3 × 103 cells per well were seeded in a 96-well plate. Then, at different time point, 10 µL of CCK-8 (Dojindo) was added to each well and incubated at 37°C for 1.5 h. Absorbance values at 450 nm were tested by the microplate reader. For colony formation assay, 1 × 103 cells per cell well were seeded in 6-well plates. After two weeks of culture, colonies were fixed with methanol and then stained with 0.1% crystal violet.
Statistics
All statistical analyses were performed using SPSS software (version 19.0). Data were presented as the mean ± standard error of the mean (SEM) from three independent experiments. The comparison among different groups two groups or more than two groups was performed by Student t test or ANOVA followed by Dunnett’s multiple comparisons test. Overall survival rate were performed by the Kaplan-Meier method and log-rank test. P value less than 0.05 was considered statistically significant.
Results
NBAT1 inhibits autophagy in NSCLC cells
To investigate whether NBAT1 has an effect on autophagy in NSCLC cells, we constructed stable A549 cells that express NBAT1 shRNA and 95D cells that overexpress NBAT1, respectively (Figure 1A, 1B). Then it was found that knockdown of NBAT1 markedly increased the amount of LC3-II levels and decreased P62 expression in A549 cells, whereas knockdown of NBAT1 in 95D cells reversed it (Figure 1C, 1D). To further confirm the results, the immunofluorescence assay was performed to observe the changes of LC3 dots. As shown in Figure 1E, NBAT1 knockdown increased the accumulation of LC3 dots in A549 cells. In contrast, the amount of LC3 dots per cell was significantly decreased in NBAT1 overexpressing 95D cells as compared with the control group (Figure 1F). Taken together, our results demonstrated a suppressive role of NBAT1 in autophagy of NSCLC cells.
Figure 1.
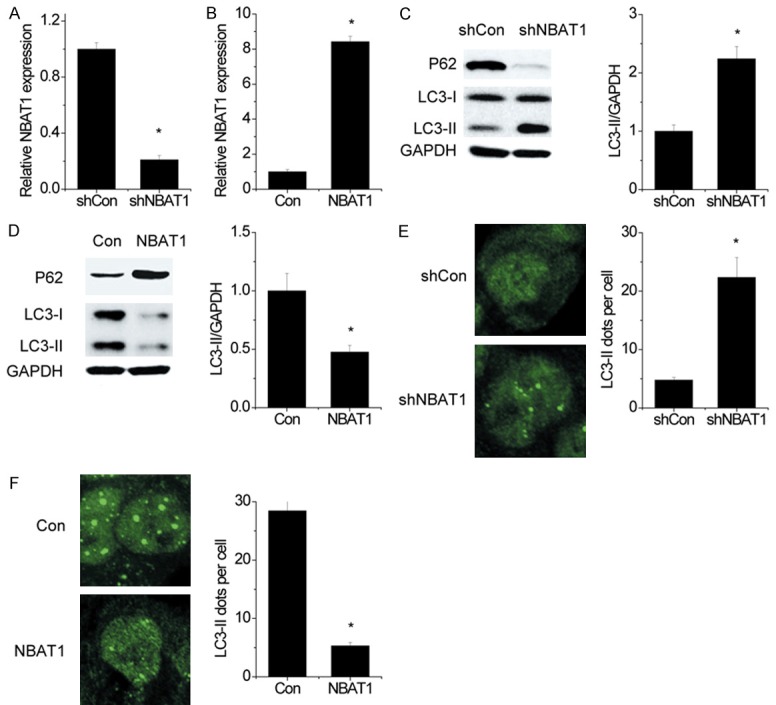
NBAT1 inhibits autophagy in NSCLC cells. (A) The NBAT1 expression was silenced in A549 cells, and the NBAT1 levels were detected by qRT-PCR. (B) The NBAT1 was overexpressed in 95D cells, and the NBAT1 levels were examined by qRT-PCR. (C and D) LC3, P62 and GAPDH were analyzed by western blot. Representative western blot and densitometric analysis normalized to GAPDH demonstrating the effect of NBAT1 silencing (C) and NBAT1 overexpression (D) on LC3-II levels. (E and F) Representative immunofluorescent images showing redistribution of autophagic marker LC3 in NBAT1 knockdown (E) and NBAT1 overexpressing (F) cells were taken on a confocal microscope. The average number of LC3 dots per cell was counted in more than 5 fields with at least 100 cells for each group. *P < 0.05.
ATG7 is required for autophagy in response to NBAT1
Several autophagy-related genes (ATG) are critical for autophagic flux. ATG5 and ATG7 are involved in the initiation of autophagy, and Beclin1 is required for the formation of autophagosome. To determine whether ATG5, ATG7 and Beclin1 were related to the suppression of autophagy by NBAT1, their expression was detected by qRT-PCR and the western blot. Overexpression of NBAT1 significantly decreased both mRNA and protein levels of ATG7, but not ATG5 and Beclin1 (Figure 2A, 2B), while NBAT1 knockdown upregulated ATG7 expression (Figure 2C, 2D).
Figure 2.
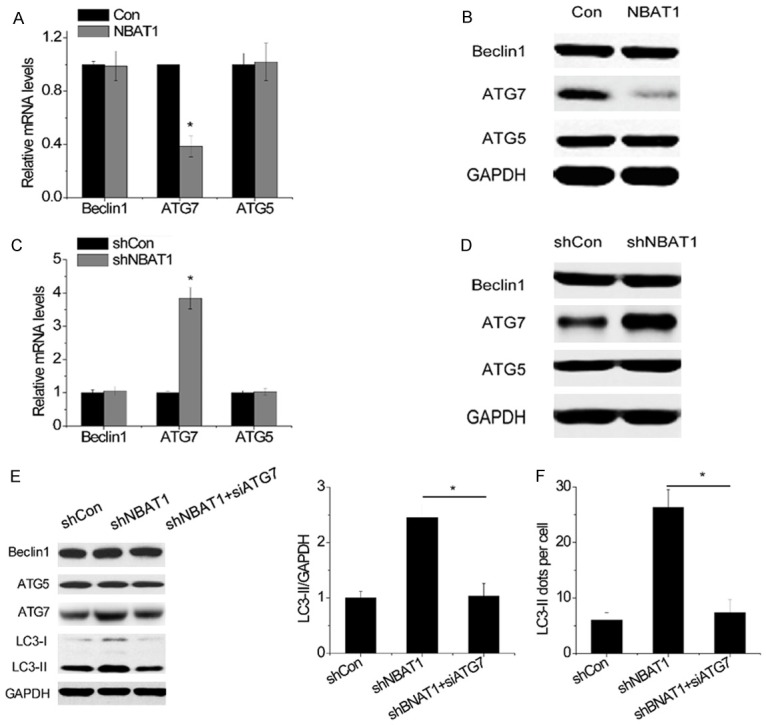
ATG7 is required for autophagy in response to NBAT1. (A and B) The mRNA (A) and protein (B) levels of Beclin1, ATG5 and ATG7 were detected in control and NBAT1-overexpressing 95D cells. (C and D) The mRNA (C) and protein (D) levels of Beclin1, ATG5 and ATG7 were detected in control and NBAT1 knockdown A549 cells. (E) LC3 and GAPDH were analyzed by western blot. Representative western blot and densitometric analysis normalized to GAPDH demonstrating the effect of ATG7 siRNA on LC3-II levels increased by NBAT1 knockdown. (F) The effects of ATG7 siRNA on LC3 dots accumulation induced by NBAT1 silencing. *P < 0.05.
To validate that ATG7 was essential for the effect of NBAT1 on autophagy, rescue experiments were performed. We transfected ATG7 siRNA into A549-shNBAT1 cells. Silencing ATG7 almost abolished the accumulation of LC3-II in A549-shNBAT1 cells (Figure 2E). Furthermore, a significant decrease of the number of LC3 dots per cell was observed after ATG7 downregulation in NBAT1 knockdown A549 cells (Figure 2F). To sum up, these results strongly suggested that NBAT1 suppressed autophagy in an ATG7-dependent manner.
PSMD10 is a target of NBAT1
To investigate the mechanism, by which NBAT1 suppressed ATG7 transcription, we performed the RNA pull-down assay with biotin-labeled NBAT1 and mass spectral analysis to search the endogenous proteins associated with NBAT1. Among these pull-down proteins, PSMD10 arouses our interests due to its pro-autophagy function via activation of ATG7 expression [17]. To verify this result, a RIP assay was conducted to validate the association between NBAT1 and PSMD10. Intriguingly, it was observed that NBAT1 could be pulled down by the PSMD10 antibody to a great extent, instead of negative control antibody IgG (Figure 3A). For further confirmation, the RNA pull-down assay was performed, and then an interaction between NBAT1 and PSMD10 was found. From the deletion mapping assay, it was also identified that the 5’-end fragment of NBAT1 (0 to 710) was key to the PSMD10 binding (Figure 3B).
Figure 3.
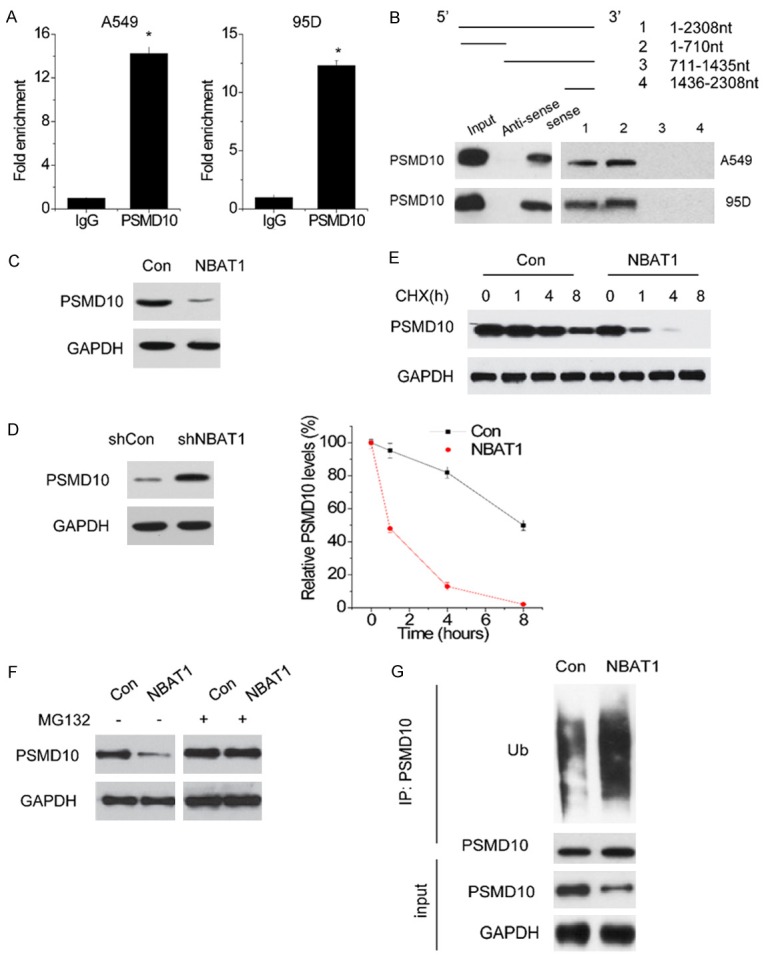
PSMD10 is a target of NBAT1. (A) Results from RIP and subsequent qPCR assays. Relative quantification of NBAT1 in RNA-protein complexes immunoprecipitated with negative control IgG or PSMD10 antibody from nuclear extracts of A549 and 95D cells. (B) Deletion mapping of PSMD10-binding domain in NBAT1. (Top) The schematic diagram of full-length and deleted fragments of NBAT1; (Bottom) western blot of PSMD10 in protein samples pulled down by different NBAT1 fragments. (C amd D) The effect of NBAT1 overexpression (C) and knockdown (D) on PSMD10 protein levels was determined by western blot. (E) 95D cells with NBAT1 overexpression was treated with CHX (100 mg/ml) for the indicated times. The cell extracts were analyzed by western blot (Up). A plot of normalized amount of PSMD10 protein is shown (Down). (F) PSMD10 expression in control and NBAT1-overexpressing 95D cells treated with vehicle control or MG132. (G) Western blot of PSMD10 ubiquitination level in control and NBAT1-overexpressing 95D cells treated with MG132. *P < 0.05.
The lncRNA could interact with RNA binding proteins to posttranslationally regulate their expression. Then, the PSMD10 expression in control and NBAT1-overexpressing 95D cells were detected. The results showed that NBAT1 overexpression significantly reduced PSMD10 expression in 95D cells (Figure 3C). On the contrary, the NBAT1 knockdown A549 cell expressed higher PSMD10 when compared with control group cells (Figure 3D). After that, the stability of PSMD10 in response to NBAT1 expression overexpression was analyzed by treating cells with the inhibitor of protein synthesis cycloheximide (CHX). NBAT1 overexpression significantly shortened the half-life of PSMD10 (Figure 3E). Consistent with this result, when proteasome degradation inhibitor MG132 was added into the culture medium, the PSMD10 protein expression in 95D-NBAT1 cells was significantly increased, and reached a level that was comparable to that in control cells (Figure 3F). In addition to that, a higher PSMD10 ubiquitination level was also observed in 95DNBAT1 cells with MG132 treatment (Figure 3G). In this case, these results suggest that NBAT1 is important for the stability of PSMD10 protein.
NBAT1 attenuates the binding of PSMD10 and HSF1 to ATG7 promoter
Given that PSMD10 bounded cooperatively with HSF1 onto the ATG7 promoter to activate ATG7 expression [17], we examined whether NBAT1 affected PSMD10 and HSF1 occupancy of the ATG7 promoter regions. The effect of NBAT1 on the binding levels of PSMD10 and HSF1 in the ATG7 promoters was evaluated using the chromatin immunoprecipitation (ChIP) assay followed by qRT-PCR. The results showed that overexpression of NBAT1 markedly decreased the binding levels of both PSMD10 and HSF1 to ATG7 promoter regions (Figure 4A), and NBAT1 knockdown presented the opposite effect (Figure 4B). In addition, the interaction between PSMD10 and HSF1 was significantly decreased in response to NBAT1 overexpression (Figure 4C), while knockdown of NBAT1 enhanced the PSMD10-HSF1 interaction (Figure 4D). Then, our observation indicates that NBAT1 inhibits autophagy through suppressing the occupancy of PSMD10 and HSF1 of the ATG7 promoter.
Figure 4.
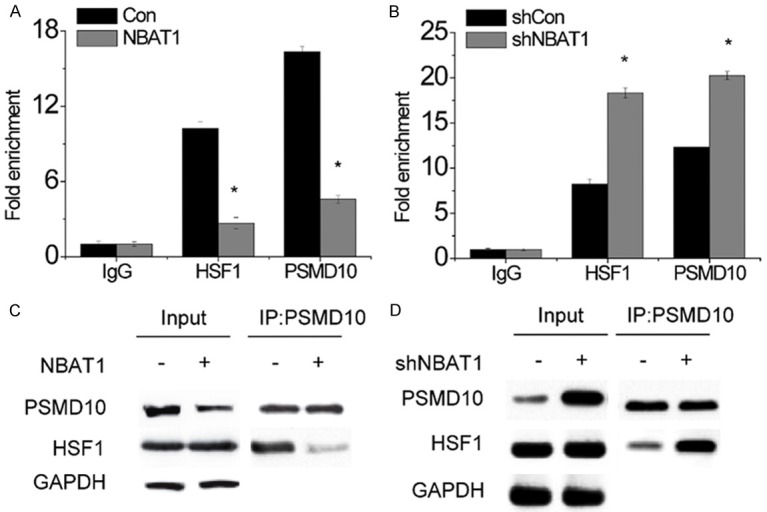
NBAT1 attenuates the binding of PSMD10 and HSF1 to ATG7 promoter. (A and B) The occupancy changes of PSMD10 and HSF1 in ATG7 promoter after NBAT1 overexpression (A) and NBAT1 knockdown (B) were determined by ChIP-qPCR. (C and D) The interaction changes between HSF1 and PSMD10 after NBAT1 overexpression (C) and NBAT1 knockdown (D) were detected by co-IP assay. *P < 0.05.
NBAT1 negatively correlates with ATG7 expression in NSCLC
Sixty NSCLC patient specimens were collected, and a possible correlation of NBAT1 with ATG7 was evaluated by qRT-PCR. As shown in Figure 5A, NBAT1 expression negatively correlated with ATG7 levels in NSCLC tissue (R2 = 0.3131, P < 0.0001). Furthermore, a significant downregulation of NBAT1 expression in NSCLC tissue was observed compared with matched non-tumor tissue (Figure 5B). The overall survival was markedly reduced in NSCLC patients with low-level NBAT1 (Figure 5C). These results further support a critical role of NBAT1 in modulating ATG7, and resulting in autophagy.
Figure 5.
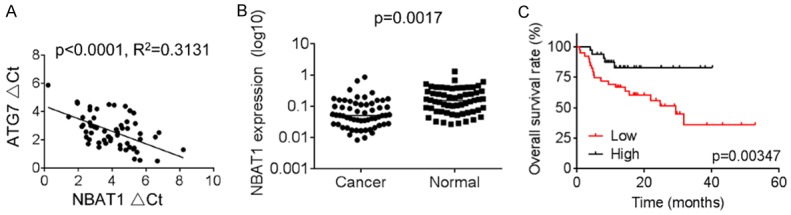
NBAT1 negatively correlates with ATG7 expression in NSCLC. A. The correlation between NBAT1 and ATG7 expression in NSCLC tissues. B. The NBAT1 expression in NSCLC and paired adjacent normal lung tissues was examine by qRT-PCR. C. Kaplan-Meier analyses of the overall survival in NSCLC patients with different levels of NBAT1. The median expression level of NBAT1 in NSCLC was used as the cutoff.
NBAT1-mediated suppression of autophagy inhibits NSCLC survival and chemoresistance
Given the fact that autophagy promotes cell survival, the role of autophagy inhibited by NBAT1 in NSCLC cells was investigated. Then, it was found that knockdown of NBAT1 facilitated cell viability and colony formation (Figure 6A, 6B), while overexpression of NBAT1 reversed these effects (Figure 6C, 6D). Disruption of autophagy by knockdown of ATG7 reduced cell viability in A549-shNBAT1 cells (Figure 6E). Similar to this result, the blockage of autophagic flux 3-Methyladenine (3-MA) attenuated the increase of cell viability increased by NBAT1 shRNA (Figure 6F). These results indicate that the cell-survival function of autophagy contributed to the NBAT1-mediated suppression of cell viability in NSCLC cells.
Figure 6.
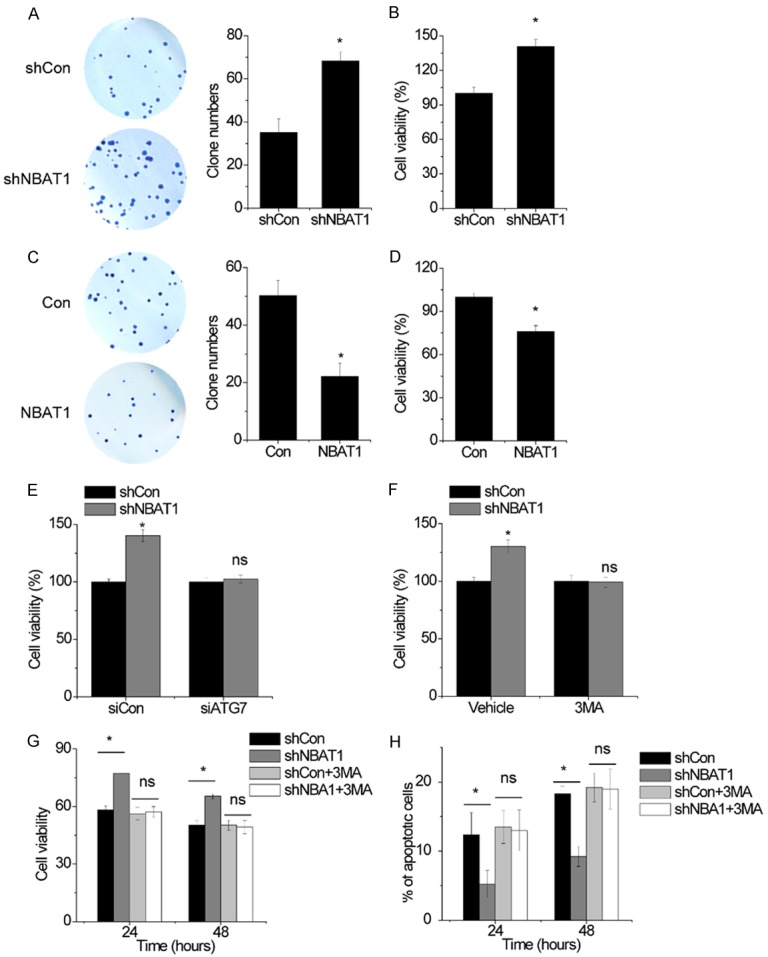
NBAT1-mediated suppression of autophgay inhibits NSCLC survival and chemoresistance. (A) The colony formation in control and NBAT1 knockdown A549 cells. (B) The cell viability in control and NBAT1 knockdown A549 cells was detected by CCK-8 assay. (C) The colony formation in control and NBAT1-overxpressing 95D cells. (D) The cell viability in control and NBAT1-overxpressing 95D cells was detected by CCK-8 assay. (E) The effects of ATG7 siRNA on the increase of cell viability induced by NBAT1 overexpression. (F) The effects of 3-MA on the increase of cell viability induced by NBAT1 overexpression. (G) Control and NBAT1 knockdown A549 cells were subjected to cisplatin for 24 or 48 h in the presence or absence of either 3-MA (10 mM) before performing CCK-8 assays. (H) Cells from (G) were analyzed for apoptosis. *P < 0.05; ns, no significance.
Finally, the role of NBAT1 in autophagy-induced chemoresistance was investigated. The A549 cells with NBAT1 knockdown were adopted to evaluate the effect of NBAT1 on cisplatin treatment. Interestingly, NBAT1 silence promoted survival and inhibited apoptosis in the cisplatin treatment. However, NBAT1-silencing-mediated resistance to cisplatin was completely abolished by 3-MA (Figure 6G, 6H). Hence, these findings reveal that autophagy induced by NBAT1 knockdown contributes to NSCLC resistance to cisplatin.
Discussion
The increasing number of evidence has demonstrated that lncRNAs play important roles in NSCLC tumorigenesis, apoptosis, proliferation and metastasis through multiple signaling pathways. Recently, some lncRNAs have been reported to be involved in autophagy. For instance, lncRNA HOATIR activated autophagy to promote drug resistance by inhibiting phosphorylation of ULK1 [18]. lncRNA MALAT1 positively regulated autophagy to promote proliferation and induce apoptosis [19]. Moreover, lncRNA-XIST functioned as a competing endogenous RNA of ATG7 against miR-17 to facilitate autophagy and chemoresistense [20]. However, the effect of other lncRNAs on cell autophagy and corresponding molecular mechanisms in NSCLC remain unclear. Our current study showed that the tumor suppressor lncRNA NBAT1 repressed autophagy and inhibited NSCLC cell survival with or without therapeutic agents. Mechanistically, NBAT1 interacted with PSMD10, downregulated the PSMD10 protein level, and then inhibited ATG7 transcription, which suppressed autophagic flux.
Previous studies reported that lncRNA could posttranscriptionally regulate the expression of their interacting proteins. For example, lncRNA FAL1 regulated the protein stability of BMI1 to epigenetically silence growth-related genes in breast cancer cells [21]. lncRNA LUCAT1 promoted tumorigenesis through stabilizing DNA methyltransferse 1 (DNMT1) in esophageal squamous cell carcinoma [22]. In the present study, RIP and RNA pull-down demonstrated a direct interaction between NBAT1 and PSMD10. In addition, overexpression of NBAT1 significantly increased the PSMD10 ubiquitination level and then induced PSMD10 degradation. However, the mechanism, by which NBAT1 promotes the degradation of PSMD10, needs further investigation. It was suspected that NBAT1 may recruit the ubiquitin E3 ligase or ubiquinase for increasing the PSDM10 ubiquitination level and enhancing its degradation.
Autophagy-related genes are required for various types of autophagy. ATG7 associates with ATG10 to mediate conjugation of ATG12 to ATG5, when concerning the formation of lipid phosphatidylethanolmine [23]. Besides, it was found that ATG7 was upregulated in cancers, and poor prognosis of cancer patients was predicted. Inhibition of ATG7 also suppresses tumor growth through inactivation of oncogenic KRAS [24,25]. A previous study showed that PSMD10 regulated ATG7 to affect autophagy. PSMD10 entered into the nucleus and bound to the ATG7 promoter in coordination with HSF1, which activated ATG7 transcription [17]. Intriguingly, in this paper, it was demonstrated that NBAT1 suppressed PSMD10 expression, and then decreased the occupancy of PSMD10 and HSF1 concerning the ATG7 promoter, which induced transcriptional inhibition of ATG7. Remarkably, a strong negative correlation between NBAT1 and ATG7 was observed in NSCLC tissue samples. NBAT1 downregulation also significantly correlated with poor prognosis of NSCLC patients. To our knowledge, it is the first to reveal a novel molecular mechanism for the regulation of ATG7 by NBAT1-mediated PSMD10 degradation.
Chemotherapy-induced autophagy protects cancer cells against cell damage, thereby leading to chemoresistance. Emerging evidence has demonstrated that lncRNAs contribute to the chemoresistance of some cancers [26,27]. In the present study, we bridged the gap between lncRNA NBAT1 and autophagy-related chemoresistance in NSCLC. The combination of autophagy inhibitor 3-MA with cisplatin significantly blocked cell survival increased by NBAT1 knockdown and induced apoptosis of NSCLC cells, suggesting that NBAT1-mediated autophagy inhibition is one vital element in NSCLC response to the efficacy of cisplatin.
Disclosure of conflict of interest
None.
References
- 1.Saika K, Machii R. Cancer mortality attributable to tobacco in Asia based on the WHO Global Report. Jpn J Clin Oncol. 2012;42:985. doi: 10.1093/jjco/hys154. [DOI] [PubMed] [Google Scholar]
- 2.Chheang S, Brown K. Lung cancer staging: clinical and radiologic perspectives. Semin Intervent Radiol. 2013;30:99–113. doi: 10.1055/s-0033-1342950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 4.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–16. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lian Y, Cai Z, Gong H, Xue S, Wu D, Wang K. HOTTIP: a critical oncogenic long non-coding RNA in human cancers. Mol Biosyst. 2016;12:3247–53. doi: 10.1039/c6mb00475j. [DOI] [PubMed] [Google Scholar]
- 9.Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y, Zhou JK, Guo C, Xia Z, Liu L, Li Q, Dai L, Peng Y. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi: 10.1016/j.canlet.2018.01.060. [DOI] [PubMed] [Google Scholar]
- 10.Yu T, Zhao Y, Hu Z, Li J, Chu D, Zhang J, Li Z, Chen B, Zhang X, Pan H, Li S, Lin H, Liu L, Yan M, He X, Yao M. MetaLnc9 facilitates lung cancer metastasis via a PGK1-Activated AKT/mTOR pathway. Cancer Res. 2017;77:5782–94. doi: 10.1158/0008-5472.CAN-17-0671. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Wu DD, Sang XB, Wang LL, Zong ZH, Sun KX, Liu BL, Zhao Y. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 2017;8:e3118. doi: 10.1038/cddis.2017.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Xia Y, Zhang H, Guo H, Feng K, Zhang C. Overexpression of long non-coding RNA H19 promotes invasion and autophagy via the PI3K/AKT/mTOR pathways in trophoblast cells. Biomed Pharmacother. 2018;101:691–7. doi: 10.1016/j.biopha.2018.02.134. [DOI] [PubMed] [Google Scholar]
- 13.Hu P, Chu J, Wu Y, Sun L, Lv X, Zhu Y, Li J, Guo Q, Gong C, Liu B, Su S. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget. 2015;6:32410–25. doi: 10.18632/oncotarget.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, Wang G, Yang J, Wang L. Long noncoding RNA NBAT1 negatively modulates growth and metastasis of osteosarcoma cells through suppression of miR-21. Am J Cancer Res. 2017;7:2009–19. [PMC free article] [PubMed] [Google Scholar]
- 15.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, Ostensson M, Akyurek LM, Abrahamsson J, Pfeifer S, Larsson E, Shi L, Peng Z, Fischer M, Martinsson T, Hedborg F, Kogner P, Kanduri C. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–37. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Yan C, Jiang Y, Wan Y, Zhang L, Liu J, Zhou S, Cheng W. Long noncoding RNA NBAT-1 suppresses tumorigenesis and predicts favorable prognosis in ovarian cancer. Onco Targets Ther. 2017;10:1993–2002. doi: 10.2147/OTT.S124645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo T, Fu J, Xu A, Su B, Ren Y, Li N, Zhu J, Zhao X, Dai R, Cao J, Wang B, Qin W, Jiang J, Li J, Wu M, Feng G, Chen Y, Wang H. PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy. 2016;12:1355–71. doi: 10.1080/15548627.2015.1034405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Jiang C, Yang Y, Guo L, Huang J, Liu X, Wu C, Zou J. Silencing of LncRNA-HOTAIR decreases drug resistance of Non-Small Cell Lung Cancer cells by inactivating autophagy via suppressing the phosphorylation of ULK1. Biochem Biophys Res Commun. 2018;497:1003–10. doi: 10.1016/j.bbrc.2018.02.141. [DOI] [PubMed] [Google Scholar]
- 19.Ma J, Wu K, Liu K, Miao R. Effects of MALAT1 on proliferation and apo- ptosis of human nonsmall cell lung cancer A549 cells in vitro and tumor xenograft growth in vivo by modulating autophagy. Cancer Biomark. 2018;22:63–72. doi: 10.3233/CBM-170917. [DOI] [PubMed] [Google Scholar]
- 20.Sun W, Zu Y, Fu X, Deng Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep. 2017;38:3347–54. doi: 10.3892/or.2017.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, Jean S, Li C, Huang Q, Katsaros D, Montone KT, Tanyi JL, Lu Y, Boyd J, Nathanson KL, Li H, Mills GB, Zhang L. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–57. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon JH, You BH, Park CH, Kim YJ, Nam JW, Lee SK. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett. 2018;417:47–57. doi: 10.1016/j.canlet.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 23.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 24.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desai S, Liu Z, Yao J, Patel N, Chen J, Wu Y, Ahn EE, Fodstad O, Tan M. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7) J Biol Chem. 2013;288:9165–76. doi: 10.1074/jbc.M112.422071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bischof J, Westhoff MA, Wagner JE, Halatsch ME, Trentmann S, Knippschild U, Wirtz CR, Burster T. Cancer stem cells: the potential role of autophagy, proteolysis, and cathepsins in glioblastoma stem cells. Tumour Biol. 2017;39:1010428317692227. doi: 10.1177/1010428317692227. [DOI] [PubMed] [Google Scholar]
- 27.Huang Z, Zhou L, Chen Z, Nice EC, Huang C. Stress management by autophagy: implications for chemoresistance. Int J Cancer. 2016;139:23–32. doi: 10.1002/ijc.29990. [DOI] [PubMed] [Google Scholar]


