Abstract
Dysregulated metabolism in the form of aerobic glycolysis occurs in many cancers including breast carcinoma. Here, we report PDK4 (pyruvate dehydrogenase kinase 4) as key enzyme implicated in the control of glucose metabolism and mitochondrial respiration is relatively highly expressed in breast cancers, and its expression correlates with poor patient outcomes. Silencing of PDK4 and ectopic expression of miR-211 attenuates PDK4 expression in breast cancer cells. Interestingly, low miR-211 expression is significantly associated with shorter overall survival and reveals an inverse correlation between expression of miR-211 and PDK4. We have found that depletion of PDK4 by miR-211 shows an oxidative phosphorylation-dominant phenotype consisting of the reduction of glucose with increased expression of PDH and key enzymes of the TCA cycle. miR-211 expression causes alteration of mitochondrial membrane potential and induces mitochondrial apoptosis as observed via IPAD assay. Further, by inhibiting PDK4 expression, miR-211 promotes a phenotype shift towards a pro-glycolytic state evidenced by decreased extracellular acidification rate (ECAR); increased oxygen consumption rate (OCR); and increased spare respiratory capacity in breast cancer cell lines. Taken together this data establishes a molecular connection between PDK4 and miR-211 and suggests that targeting miR-211 to inhibit PDK4 could represent a novel therapeutic strategy in breast cancers.
Keywords: PDK4, miR-211, altered metabolism, apoptosis, Warburg effect
Introduction
Breast cancer is the most common solid tumor in women with an estimated 2.4 million cases in 2015 [1]. It is also the second leading cause of cancer death in women. Breast cancer is a heterogeneous disease classified by expression or absence of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) [2]. The standard treatment regimen for breast cancer includes chemotherapy, radiation, surgical resection and hormone therapy directed at the receptors outlined above. Existing therapies still fall short in adequately treating breast cancer, especially for triple-negative breast cancers (TNBC) that lack ER, PR, and HER2 expression. Thus, identifying novel therapeutic strategies for breast cancer is necessary to improve patient outcomes.
Tumor cells undergo metabolic reprogramming to utilize glycolysis even in the presence of abundant oxygen while shifting energy derivation away from mitochondrial oxidative phosphorylation (OXPHOS). This metabolic phenomenon is referred to as the “Warburg effect”, or aerobic glycolysis. This anomaly is now widely accepted as one of the hallmarks of aggressive cancers, and understanding the mechanistic steps of this process might be beneficial in identifying potential targets for novel breast cancer therapies [3,4]. Pyruvate dehydrogenase kinase isoform 4 (PDK4) is a member of PDK family located in the mitochondrial matrix of cells. PDK4 inhibits the entry of pyruvate into the TCA cycle by inhibiting pyruvate dehydrogenase activity, thereby switching energy derivation to cytoplasmic glycolysis in lieu of mitochondrial OXPHOS, consistent with the aforementioned Warburg effect. PDK isoforms are highly upregulated in various other cancers including glioblastoma, lung carcinoma, pancreatic cancer, amongst others [5-8].
Attention has recently been focused on the role of small, non-coding RNA molecules in cancer development known as microRNAs (miRNAs) [9-11] and in breast cancer [12]. miRNAs are a class of short (~22) nucleotide non-coding RNA molecules that bind to the 3’ untranslated region (3’UTR) of target messenger RNA to suppress gene expression [13,14] involved in the modulation of diverse biological processes [15-17]. As a tumor suppressor, miR-211 regulates several biological processes, and its downregulation is associated with increased tumorigenicity in many cancers [18,19]. However, the regulatory mechanisms underlying miR-211 in breast cancer and in glucose metabolism is not well reported.
In this study, we show that PDK4 is highly expressed in all molecular subtypes of breast cancer. MIR-211 expression is more highly expressed in normal breast tissue compared to breast cancer tissue as seen with in situ hybridization experiments. Overexpression of miR-211 supresses the Warburg effect in breast cancer cell lines, resulting in increased pyruvate entry to OXPHOS as shown in metabolic studies conducted using the Seahorse Bioanalyzer. Further, miR-211 overexpression regulates the proliferative and apoptotic behavior of MDA-MB-468 and BT-474 cells. Collectively, we demonstrate that PDK4 is a potential target of miR-211 and our results establish miR-211 as a robust inhibitor of the Warburg effect and promising potential therapeutic target for breast cancer treatment.
Materials and methods
Cell culture
Two breast cancer cell lines, BT-474 and MDA-MB-468, were obtained from the American Tissue Cell Culture (ATCC) and were cultured in RPMI and DMEM high-glucose media containing 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, in a 5% CO2 humidified incubator at 37°C.
Plasmids, chemicals and antibodies
A PLKO-mcherry-luc-puro-miR-211 construct and empty vector were purchased from Addgene (Cambridge, MA) and transfected as described previously [20]. The siRNA construct used to silence PDK4 was purchased from Santa Cruz (sc-39030). The miRCURY LNATM microRNA ISH Optimization Kit (FFPE) and miR-211 probe was obtained from Exiqon (Woburn, MA). A human breast cancer tissue microarray (TMAH-BRC-03) was obtained from Ray Biotech. The JC-1 kit was obtained from Cayman Chemicals (Ann Arbor, MI). In situ cell death detection kit and Fluorescein kits were purchased from Sigma (11684795910). Proteome Profiler Human Apoptosis Array kits (ARY009) and the human pluripotent stem cell antibody array (ARY010) were purchased from R&D Biosystems. Seahorse XFp cell energy phenotype test kits (103275-100), Mito Stress test kits (103010-100) and Glycolysis Stress kits (103346-100) were purchased from Agilent Technologies. Protein concentration was estimated using the bicinchonic acid (BCA) assay (Pierce, Rockford, IL). All antibodies used in this study were purchased from Santa Cruz Biotechnology, (Santa Cruz, CA). The primer sequences were synthesized by IDT and the primer list was provided as listed in Table S1.
TCGA cohort analysis
We collected mRNA expression profiles from The Cancer Genome Atlas (TCGA) invasive breast carcinoma cohort and miRNA cohort using the TCGA data portal (http://cancergenome.nih.gov/). To gather information on ER, PR, and HER2 status, we also downloaded clinical information from the TCGA data portal.
Quantitative real-time PCR
Total RNA was isolated from BT-474 and MDA-MB-468 cell lysates using the Trizol method (Life Technologies). Subsequent quantification was performed using a CFX-96 analyzer. SYBR green primers used in this study were provided as outlined in Table S1. GAPDH was used as the endogenous control. A total RNA concentration of 2 ng/μl was reverse-transcribed to cDNA using miR-211 primers from miRCURY LNATM miRNA polymerase chain reaction system (Exiqon, Vedbaek, Denmark).
Western blot analysis
Proteins extracted from BT-474 and MDA-MB-468 cells obtained from both control and respective treatments were assayed with multiple antibodies using the immunoblot approach as described previously [5]. GAPDH was used to confirm equal loading.
Apoptosis protein arrays and stem cell antibody arrays
An apoptosis array (ARY009) detecting the expression level of 35 apoptotic proteins and a human pluripotent stem cell antibody array (ARY010) detecting the relative expression of 15 pluripotent stem cell markers were purchased from R&D Systems (Minneapolis, MN). Approximately 200 µg of total-cell lysates were taken for these arrays and were performed in accordance with the manufacturer’s instructions. Signal intensity of each spot was quantified with Image J (Bethesda, MD) and averaged for each specific protein.
IR treatment
For combination treatment with miR-211, an IR dose of 8 Gy was given 48 h after plasmid transfection in both BT-474 and MDA-MB-468 cells using the RS 2000 Biological Irradiator X-ray unit (Rad Source Technologies Inc., Boca Raton, FL, USA). Irradiated cells were further incubated for 24 h before harvesting for Western blot analysis.
Mitochondrial dysfunction and TUNEL assay
Mitochondrial membrane potential was detected using JC-1 fluorescent dye (Cayman Chemicals; Ann Arbor, MI) as described previously (21). Both the EV- and miR-211-treated BT474 and MDA-MB-468 cells were washed once with PBS and subjected to JC-1 staining for 10 min at 37°C. Cells were washed again with PBS and imaged for the JC-1 monomer and aggregate with fluorescence microscopy. The shift from red to green fluorescence indicates the collapse of mitochondrial membrane potential. To measure cell death, the TUNEL assay (Roche; Indiana polis, IN) was performed in both BT-474 and MDA-MB-468 cells treated with miR-211 following the manufacturer’s instructions.
Metabolic analysis
BT-474 and MDA-MB-468 cells were plated at a density of 20,000/well in an XFp 8-well plate. Two days before, cells were transfected with miR-211. On third day, cells were transferred to an 8-well moat chamber and allowed to grow overnight with media exchange occurring 1 hour before the assay with XFp media to determine the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). For the cell energy phenotype assay, FCCP and oligomycin were diluted into XFp media and loaded into an accompanying cartridge to achieve final concentrations of 10 μM; for the mitostress assay, oligomycin, FCCP and rotenone/antimycin A were diluted to achieve final concentrations of 1.0 μM and 0.5 μM, respectively. Injection of drugs into the medium occurred at the time points specified. These experiments were done to define a basal OCR, ATP-linked OCR, proton leak, maximal respiratory capacity, reserve respiratory capacity, and non-mitochondrial oxygen consumption. All reagents were purchased from Seahorse Biosciences.
Immuno-paired antibody detection (IPAD) analysis
The IPAD assay (ActivSignal, LLC, Natick, MA) was used to study the expression and phosphorylation status of 70 different human protein targets covering 20 major signaling pathways. In this assay, we used paired antibodies for each target protein and detection was contingent upon whether both antibodies in a pair bound to a specific target protein. Detection of the paired antibodies was facilitated by special DNA barcodes conjugated to antibodies, which were quantified using Next Generation Sequencing or Fluidigm digital PCR.
In situ hybridization of miR-211
Breast cancer tissue microarrays (TMA) were used in this experiment. The TMA was de-paraffinized and digested with proteinase-K for 10 min. Next, the TMA was hybridized with a DIG-labeled mercury LNA miR-211 probe (Exiqon, MA, USA) for one hour at 55°C. An alkaline phosphatase-conjugated secondary antibody and an anti-DIG antibody were used to detect the digoxigenins as per the manufacturer’s instructions. Raw images were captured using the Olympus BX61 fluorescent microscope.
Immunohistochemistry
For immunohistochemical analysis, we used breast cancer TMA (TMAH-BRC-03). The TMAs were de-paraffinized in xylene and rehydrated in graded ethanol solutions. Antigen retrieval was carried out with 10 mM of citrate buffer (pH 6.0) at boiling temperature for 60 min and permeabilization was carried out in in 0.1% Triton-X-100. DAB staining was done to evaluate the expression of PDK4.
Statistical analysis
The results shown are represented as mean ± SD. Graph pad 5.0 was used to perform student’s t-tests to evaluate the differences between the control and treated groups. For multiple comparisons within TCGA and GOBO datasets, ANOVA tests were used. All p-values were considered statistically significant with a value <0.01; 0.005.
Results
PDK4 expression is upregulated in breast cancer patients
To study the clinical relevance of PDK4 expression in breast cancer and to define its possible prognostic value, we performed datamining studies using TGCA data. It is well established that clinical outcomes in breast cancer are dependent in part on molecular subtype (i.e., ER, PR, and HER2 status) [22]. Therefore, we plotted Kaplan-Meier survival curves from 1216 cases using mRNA and clinical data to define the significance of PDK4 on survival within previously defined subtypes. In Figure 1A, we plotted survival curves for the specimens with ER positivity (n = 887; P<0.0001) and ER negativity (n = 255; P = 0.0063). Figure 1B shows data for PR positive (n = 763; P<0.0001) and PR negative (n = 368; P = 0.01) cases; Figure 1C shows the plot for HER2-positive (n = 184; P = 0.0033) and HER2-negative (n = 614; P = 0.0001) cases. Figure 1D shows the data plotted from all 1216 patients (P<0.0001). Overall, high expression of PDK4 was significantly associated with poor survival in each of the subtypes. A strong correlation was observed in the all-case curves shown in Figure 1D. Together, these results suggest that expression of PDK4 has potential as a prognostic marker and therapeutic target in breast cancer.
Figure 1.
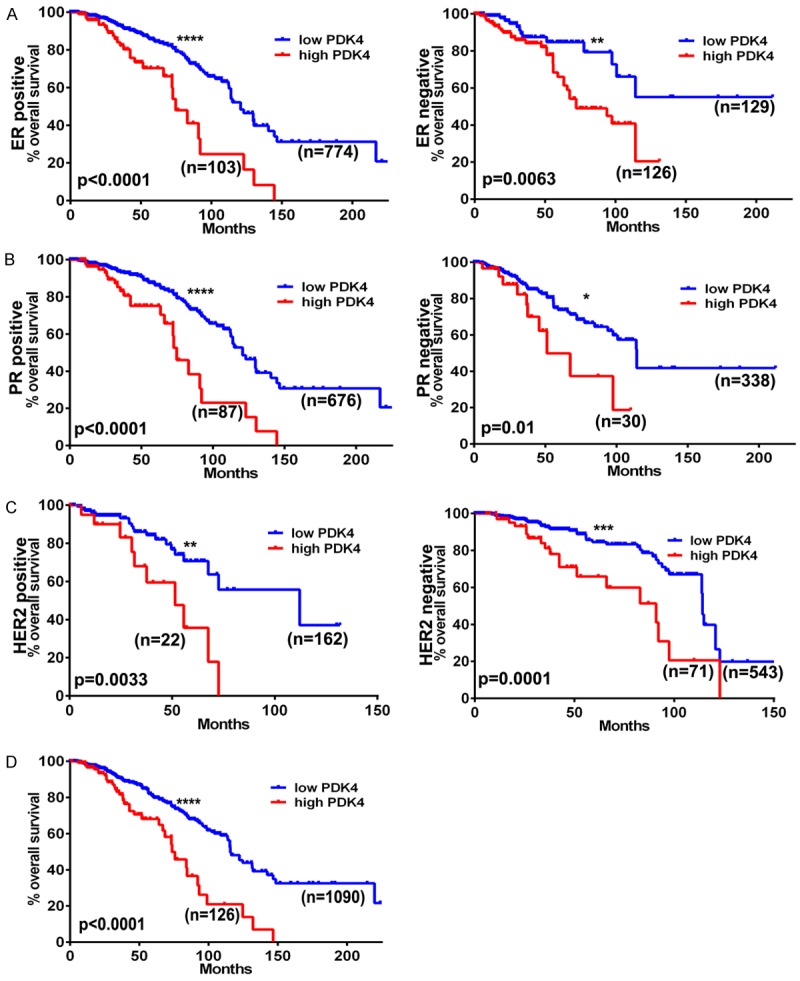
PDK4 status correlates with survivorship in various breast cancer genotypes. Kaplan-Meier curves generated from the cancer genome atlas database showing patient populations dichotomized by PDK4 expression (low = blue, high = red) graphing survival time given (A) estrogen receptor, (B) progesterone receptor, or (C) HER2 receptor status, and (D) overall survivorship.
PDK4 is a direct target of miR-211
To further our understanding on the use of miRNAs and to identify the potential targets in aerobic glycolysis, we reviewed available literature and public databases including TargetScan and miRanda and concluded that PDK4 is a potential target of miR-211 [23]. To study levels of endogenous miR-211 and levels after overexpression, we transfected BT-474 (ER+, PR+ and HER2+) and MDA-MB-468 (ER-, PR- and HER2-) [24] cells with a miR-211 overexpression plasmid and an empty vector (EV). RT-PCR studies showed a significant increase in miR-211 levels when compared to the empty vector. No miR-211 expression was seen in EV-transfected cells (Figure 2A). Next, we confirmed the presence of a seed sequence of the predicted miR-211 target binding site within the PDK4 mRNA sequence located at the 302-308 position in the 3’UTR region, which is highly conserved across four species [42]. This data was used to demonstrate PDK4 as a putative target of miR-211 (Figure 2B). To determine whether miR-211 influences PDK4 expression, we transfected BT-474 and MDA-MB-468 cells with the miR-211 plasmid. We observed significant silencing of PDK4 mRNA levels upon miR-211 transfected cells when compared to the empty vector transfected cells (Figure 2C). Because increased PDK4 levels have been associated with increased metabolic rate within tumors [25], we checked the existence of a similar correlation using a human breast cancer tissue microarray (TMAH-BRC-03, Ray Biotech) comprised of 39 metastatic and 38 benign breast tumor tissues using immunohistochemistry. DAB-stained tissues showed intensive staining of PDK4 in breast carcinoma tissues (C-2) when compared to normal breast tissues (I-4). To test the expression of miR-211 in clinical breast cancer specimens, we performed in situ hybridization assay using the human breast cancer tissue microarray (TMA). Tumor specimens showed increased positivity for PDK4 expression when compared to the normal breast tissue. Likewise, spots from normal breast tissue demonstrated increased expression of miR-211 when compared to the tumor counter parts (Figures 2D and S1A). To corroborate our TMA and in situ findings we plotted the Kaplan-Meier survival curve for miR-211 using previously described TCGA dataset in Figure 1. High miR-211 expression promotes increased survival when compared to its low expression (Figures S1B and S1C). These results show that both PDK4 and miR-211 levels are inversely correlated in breast cancer cells, humanderived tissues and in the TCGA datasets.
Figure 2.
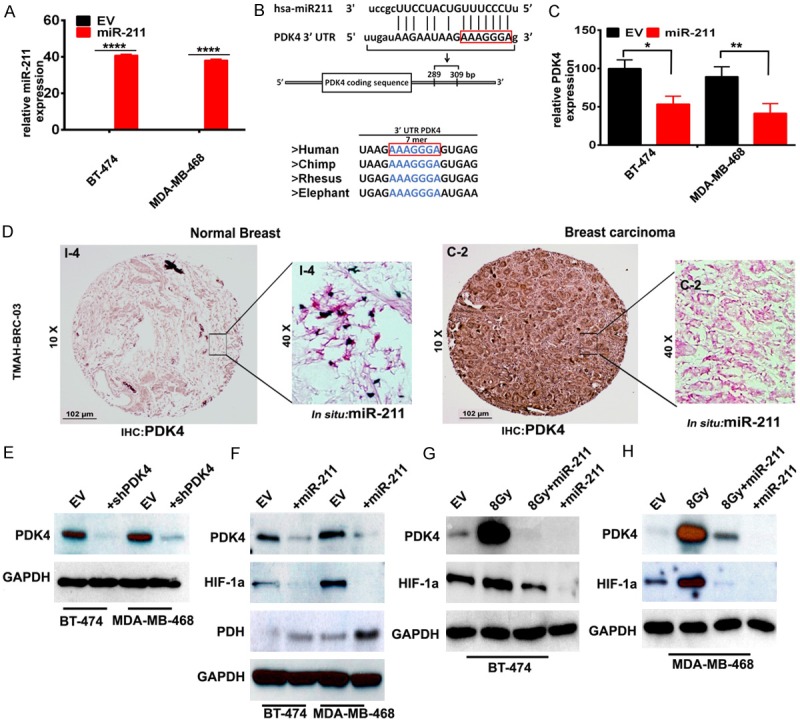
miR-211 is down regulated in breast cancer. (A) RT-PCR analysis to detect miR-211 endogenous levels in BT-474 and MDA-MB-468 cell lines. (B) Schematic representation of miR-211 sequence alignment with 3’UTR of human PDK4. The nucleotides in red box are putative positions of interaction. The region of miR-211 binding to PDK4 is conserved across the species. (C) RT-PCR analysis to detect mRNA levels of PDK4 in empty vector and miR-211 over expressing plasmid transfected BT-474 and MDA-MB-468 cells. (D) Negative IHC of normal breast tissue for PDK4 protein alongside inset in situ hybridization showing expression of miR-211 RNA (left panel); IHC of breast carcinoma demonstrating PDK4 positivity and inset image showing relative absence of miR-211 RNA via in situ hybridization (right panel). (E) Western blot analysis of PDK4 in PDK4 silenced (shPDK4) cells (F) miR-211 transfected cells probed for PDK4, HIF1α and PDH proteins using specific antibodies (EV = Empty vector). Total protein lysates obtained after 8 Gy radiation, miR-211 transfection, and 8 Gy radiation + miR-211 transfection from (G) BT-474 and (H) MDA-MB-468 cells were collected after 72 h. Immuno blot analysis of PDK4 and HIF1α levels were shown in Panel (G and H) respectively. GAPDH was used as a loading control.
We next conducted a transient transfection of shPDK4 plasmid and observed decreased levels of PDK4 as demonstrated by Western blot (Figure 2E). We conducted this experiment to compare the levels of PDK4 in both shPDK4 and miR-211 treatments. Since we observed negligible amounts of endogenous miR-211 (Figure 2A), we overexpressed miR-211 using transient transfection. Transfected miR-211 cells showed significantly decreased expression levels of both PDK4, HIF1α and the results obtained are comparable shPDK4 treatments. Interestingly, the levels of PDH are observed to be highly expressed in the miR-211 transfected cells (Figure 2F).
Recent reports suggesting the decline of tumor recurrence with radiation is encouraging but is not consistent from patient to patient [26]. Our laboratory previously showed that miR-211 overexpression treatment caused DNA fragmentation in glioma stem cells even at sub-lethal (i.e., 5 GY) doses of ionizing radiation (IR) [20]. Thus, as a proof of concept, we examined the effects of miR-211 transfection on BT-474 and MDA-MB-468 cells exposed to 8 Gy IR. After 72 h, we harvested the samples and checked expression levels of PDK4 and HIF1α. IR treatment increased PDK4 and HIF1α levels, whereas miR-211 treatment reduced levels of these proteins. Interestingly, the combination treatment (8 Gy+miR-211 treatment) showed comparable results of miR-211 overexpression. These results confirm PDK4 as a target of miR-211 in in vitro breast cancer models (Figure 2G and 2H).
miR-211 overexpression promotes mitochondrial apoptosis in BT-474 cells
Our previous study conducted on the role of miR-211 in glioblastoma demonstrated that miR-211 regulated the tumor growth by inducing apoptosis [20]. In this study, we aimed to delineate the effects of miR-211 on mitochondrial-mediated apoptosis in the BT-474 cell line. For this experiment, BT-474 cells were transfected with either an EV or a miR-211 overexpression plasmid. To verify if miR-211 transfection induced apoptosis, we loaded the human apoptosis array with total protein lysates of EV and miR-211 treated BT-474 cells. A comparison of pictographs of the spots between EV-treated and miR-211-treated arrays showed increased expression of mitochondrial apoptosis-related proteins including Bad, Bax, FADD, SMAC, p21, p27 and p53s46, and p53s392. In total, 16 out of 35 proteins were upregulated in miR-211-treated cells when compared to the EV treatment (Figures 3A, 3B and S2A). To further study the effects of miR-211 treatments on its downstream signaling, we performed an IPAD assay on BT-474 and MDA-MB-486 cells. The IPAD assay is a relatively new platform that can simultaneously study the activation of approximately 20 major active signaling pathways in cancer. We observed increased phosphorylation of retinoblastoma protein (Rb), p38MAPK, pGSKβ, and p53 in both the cell lines tested, implicating the role of miR-211 as a potential tumor suppressor in cancer pathways (Figure 3C). As we observed the induction of mitochondrial apoptosis in miR-211-treated cells (Figure 3A, 3B), we next measured the mitochondrial membrane potential using JC-1 dye. EV-treated BT-474 and MDA-MB-468 cells showed J-aggregates as shown in red fluorescence. The miR-211-treated cells showed green fluorescence, indicating an onset of apoptosis. These results confirm that miR-211 may induce apoptosis through mitochondrial dysfunction (Figures 3D, S2B and S2C). To further asses the percent apoptosis, we conducted the Tunel assay and showed increased cell death in miR-211-treated cells compared to EV treatments in both BT-474 and MDA-MB-468 cells (Figures 3E, S2D). These results suggest that miR-211 may control breast cancer progression by inducing mitochondrial apoptosis and by influencing the expression of p53, pRb, and p38.
Figure 3.
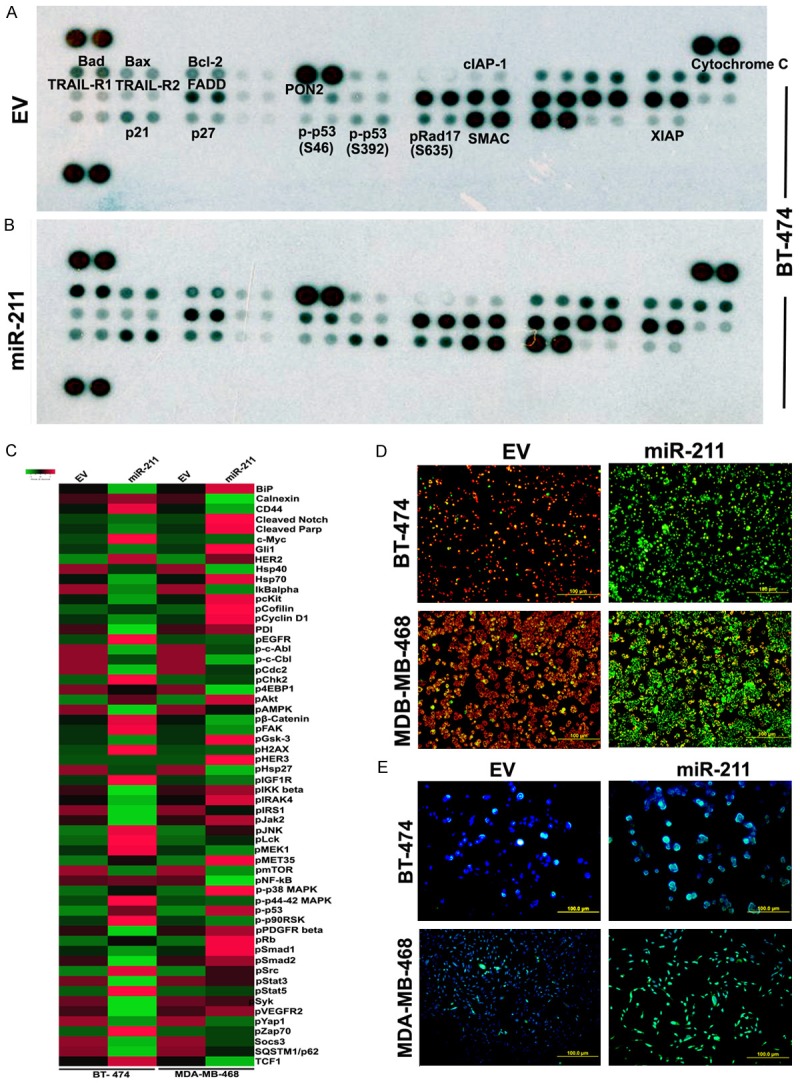
miR-211 promote mitochondrial apoptosis of breast cancer cells in vitro. Representative expression levels of various apoptosis -related proteins in BT-474 cells transfected with EV (A and B) miR-211. Illustrated images showed increased levels of several apoptotic proteins after mir-211 silencing. (C) BT-474 and (D) MDA-MB-468 cells were transfected with EV and miR-211 were subjected to IPAD assay, which measure simultaneous expression or activation of 70 proteins involving various activated signaling pathways in tumors. The differential expression is presented as heatmaps using a free software available online (www.shinyheatmap.com). (D) Both BT-474 and MBA-MD-468 cells transfected with miR-211 and EV were treated with JC-1 dye to record the alteration in mitochondrial membrane potential (Red = live cells; Green = dead cells). (E) Cells were stained for apoptosis using TUNEL assay.
miR-211 reverses the Warburg effect in BT-474 and MDA-MB-468 cells
The data presented in Figure 2 shows that miR-211 treatment reduces PDK4 and increased the expression of PDH expression in miR-211 transfected cells. It is widely accepted that silencing of PDK4 and overexpression of PDH reverses metabolic reprogramming consistent with the Warburg Effect. To study if miR-211 plays such a role in breast cancer cells, we conducted cell energy phenotype assays using the XFp Seahorse Bioanalyzer system. This assay delineated the metabolic phenotypes of BT-474 and MDA-MB-468 under both baseline and miR-211-overexpressed conditions. BT-474 and MDA-MB-468 cells demonstrated a glycolytic phenotype, while miR-211 treatment induced an energetic phenotype in both lines. These results indicate that miR-211 overexpression reduces the ECAR (extracellular acidification rate) of breast cancer cells and shifts their energy derivation toward OXPHOS (Figure 4A, 4B). Next, we queried alterations of the major respiratory chain complexes in BT-474 and MDA-MB-468. In this experiment, both BT-474 and MDA-MB-468 cells treated with EV and miR-211 overexpression were exposed to the mitochondrial inhibitors rotenone and antimycin A. MiR-211-treated cells increased spare respiratory capacity compared to test controls (Figure 4C and 4D). Further, we extrapolated glycolytic flux by measuring the extracellular acidification rate (ECAR) (i.e., the secretion of protons into the extracellular medium) in BT-474 and MDA-MB-468 cells. Under normal conditions, ECAR profiles were reciprocal of OCR profiles (Figure 4E, 4F), in both cell lines. These results indicate that miR-211 could metabolically control breast cancer by favoring oxidative phosphorylation over glycolysis. We next conducted RT-PCR analysis on enzymes of TCA cycle and observed their increased expression confirming the aforementioned effects of miR-211 on breast cancer cells (Figure 4G, 4H).
Figure 4.
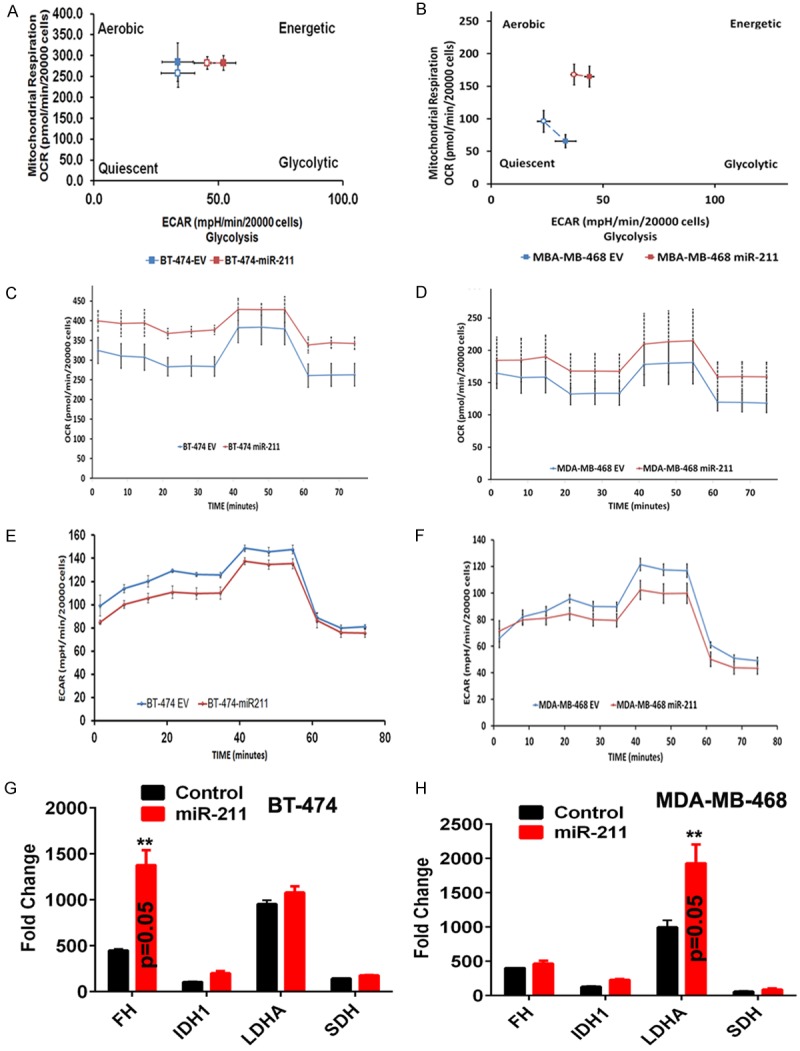
miR-211 suppresses the glycolytic pathway; modulate cellular bioenergetics. Both the EV- and miR-211 transfected cells were plated in Seahorse XFp 8 well plates. For all the experiments that involve Seahorse XFp, cell medium is replaced with bicarbonate free medium, one hour before use. Cell energy phenotype (A and B) assay was performed to measure the metabolic phenotype in EV and miR-211 transfected cells. Mitostress test (C and D) and Glycolytic stress test (E and F) were performed to assess the spare respiratory capacities, oxygen consumption rates (OXPHOS) and extracellular acidification rates (glycolysis) in EV and miR-211 transfected BT-474 and MDA-MB-468 cells. All the experiments were performed in duplicates. (G and H) RT-PCR analysis.
miR-211 reduces stemness in breast cancer cells
Earlier reports suggest that overexpression of certain miRNAs in breast cancer cell lines induces stemness [27]. To analyze the possible role of miR-211 in inducing a stem-like phenotype, we overexpressed miR-211 in BT-474 and MDA-MB-468 cells and analyzed various stem cells markers using a stem cell array (Figure 5A). Treatment of BT-474 and MDA-MB-468 cells with miR-211 reduced the expression of several transcription factors known to regulate stemness. Among these were Oct3/4, Otx2, and Nanog, which form a transcriptional complex in embryonic stem cells with Sox2. To further investigate the effect of miR-211 on properties of stemness, we checked the real-time PCR analysis of stem-cell markers Sox2, Twist and SNAIL. RT-PCR analysis showed that cells overexpressing miR-211 have reduced expression of these markers compared to EV-treated cells (Figure 5B, 5C). Collectively, the results obtained in this present study emphasize the role of miR-211 overexpression in the breast cancer by induction of mitochondrial apoptosis, stemness; promoting radio sensitivity and reversal of Warburg Effect (Figure 6).
Figure 5.
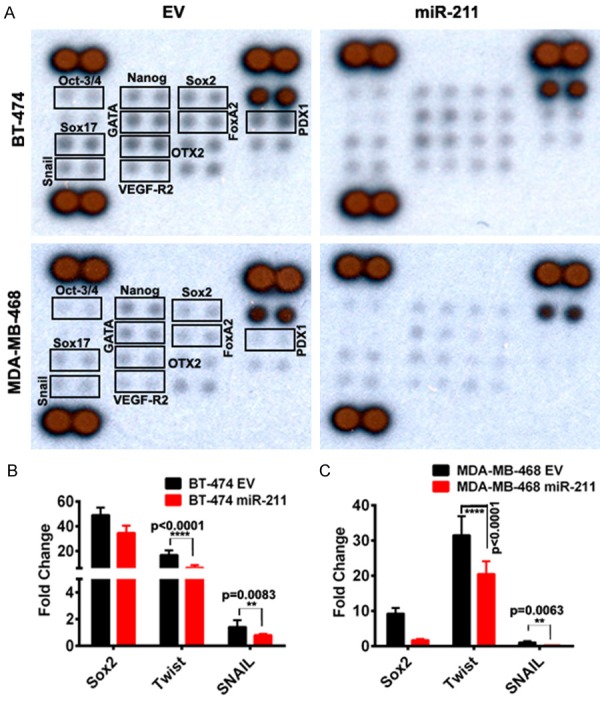
miR-211 overexpression reduced breast cancer stemness in vitro. (A) Around ~200 mg of total protein was extracted from EV and miR-211 transfected breast cancer cell. The experiment was conducted in accordance with the manufacturer’s instructions. Human pluripotent stem cell antibody array (#ARY010) was used to measure the change in the expression profiles of stemness related markers. Representative pictographs showed decreased levels of stem cell markers in miR-211 treatments. (B and C) Validation of differentially expressed stem cells markers in (A) using RT-PCR analysis.
Figure 6.
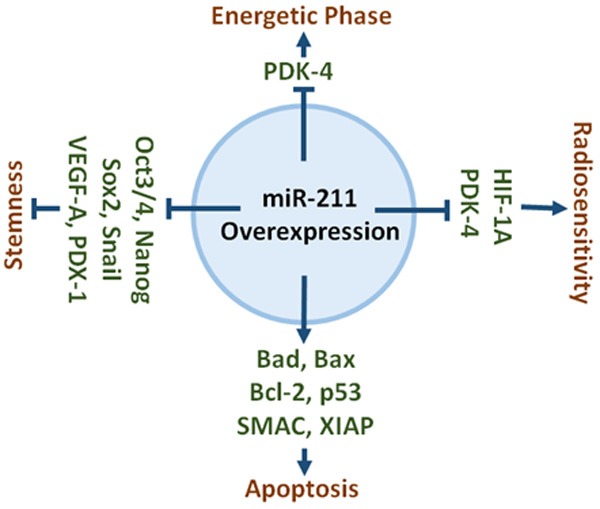
Schematic representation of role of miR-211 in different functions. Overexpression of miR-211 increase the expression of apoptotic genes, reduce the expression of stemness markers, and decrease the expression of metabolic markers.
Discussion
Micro RNAs that post-transcriptionally modify the expression of millions of genes are an interesting therapeutic target that have the potential to alter cellular processes characteristic of malignancy [28-31]. In this study, we have provided evidence that the glycolytic enzyme PDK4 is overexpressed in breast cancer tissue and drives a functional metabolic shift toward glycolysis. We have shown that PDK4’s overexpression in breast cancer patients is correlated with poor prognoses for patients irrespective of their ER, PR, and HER2 status and irrespective of their histological subtype. Furthermore, we have demonstrated that miR-211 can be utilized to alter the expression of HIF1α and PDK4 in BT-474 & MDA-MB-468 breast cancer cell lines in vitro by targeting the 3’UTR of PDK4. The application of miR-211’s ability to reduce HIF1α and PDK4 expression seems to have particular utility in the context of concurrent ionizing radiation, which we demonstrated causes an increase in PDK4 and HIF1α levels. Even when cells were treated with IR and miR-211 simultaneously, PDK4 and HIF1α levels were reduced.
The Warburg Effect is a shift observed in cancer cells from oxidative phosphorylation to glycolysis that allows for cancers to disproportionately utilize available nutrient stores [32]. A 2016 study by Han et al. showed another miRNA’s ability to block the Warburg Effect through post-transcriptional modification of PDK4 in hepatocellular carcinoma [33]. Our evidence provided by cell energy phenotype assays run using the Seahorse Bioanalyzer provides quantitative evidence suggests that a similar end result can be accomplished with the use of miR-211 in breast cancer. Initially, both cell lines demonstrated a glycolytic phenotype consistent with the Warburg effect that would be expected in cancer. However, with miR-211 overexpression, a more energetic phenotype as demonstrated by reduced ECAR and increased OCR. This demonstration of miR-211 as a controller of metabolism in breast cancer cell lines can be connected back to its effects on expression levels of PDK4 and HIF1α, as PDK4 and HIF1α are both implicated in mediating glycolysis, while their inhibition shifts metabolism toward an energetic phenotype.
There is additionally evidence that HIF1α and PDK upregulation may contribute to the evasion of apoptosis following radiation therapy in hypoxic malignancies [34,35]. A 2012 study by Duru et al. demonstrated a HER2-mediated survival in which radioresistance is developed by HER2-positive breast cancer stem cells [36]. Our treatment of the BT-474 & MDA-MB-468 cell lines with 8 Gy radiotherapy seems to suggest that increases in PDK4 & HIF1α protein expression occur regardless of HER2 status, as BT-474 is HER2+ while MDA-MB-468 is HER2-. More importantly, our studies indicate that addition of miR-211 may attenuate the expression of these two proteins in both cell lines, thereby inhibiting the typical changes that occur following radiotherapy and allow for a pro-apoptotic state. The maintenance of radio-sensitivity during ionizing radiation would make this treatment more efficacious, suggesting that miR-211 could be a good adjunct to the treatment protocol for breast cancer patients if these changes are in fact conserved in vitro.
Our TUNEL assay results demonstrated that miR-211 treatment was associated with a much higher degree of apoptosis in both cell lines. When discussing potential mechanisms by which apoptosis could occur because of miR-211 treatment, several of the factors that miR-211 increased on apoptosis assay have been well established as pro-apoptotic targets in other cancer treatment. TRAIL-R2, also known as death receptor 5, is involved with activating apoptotic pathways, and, when given exogenously, is indicated for increasing immune defense and surveillance against cancer cells. Its monoclonal antibody, tigatuzumab, has been approved for the treatment of ovarian cancer, and has been clinically studied as an adjunct for paclitaxel in triple negative breast cancer [37] as well as in colorectal [38] and non-small cell lung [39] cancer. P21 is a cyclin-dependent kinase inhibitor and is associated with p53 to respond to membrane or DNA damage and promote cell cycle arrest [40]. p27 is another cyclin-dependent kinase inhibitor that acts to decrease proliferation and differentiation of cells by limiting cells’ ability to enter S phase. Dephosphorylation of BAD, which is also known as Bcl-2 associated death promoter, is proapoptotic. Targeting this protein pharmacologically is currently being studied in preclinical trials [41]. Through its induction of apoptosis through these proposed mechanisms, miR-211 treatment created a pro-apoptotic state in both the BT-474 and MDA-MB-468 cell lines. The IPAD assay highlighted how miR-211 affects the phosphorylation status of RB and MAPK proteins, which are known tumor suppressors. These results show miR-211’s expression attenuates tumor progression in a multifactorial manner.
In conclusion, we have demonstrated PDK4 is highly expressed in breast cancers irrespective of their molecular or histologic subtype. Furthermore, PDK4’s overexpression has a negative impact on prognosis for breast cancer patients. On the other hand, miR-211 is upregulated in normal breast tissue and downregulated in breast cancer, and that miR-211 is a suppressor of Warburgian aerobic glycolysis. MiR-211 treatment of breast cancer cell lines caused an attenuation in PDK4 expression including in cells treated with ionizing radiation, and was found to increase apoptosis, purportedly by the increased expression of pro-apoptotic factors and alteration of various tumor pathways. These studies elucidate PDK4 as an important target of miR-211 and reveal miR-211 as a robust inhibitor of the Warburg effect and promising therapeutic target for breast cancer treatment.
Acknowledgements
This research was supported by OSF Foundation and UICOMP. The authors also thank Dr. Marcelo Bento Soares, Chairman of Cancer Biology and Pharmacology, UIC, Peoria Campus for providing bridge funding to complete this project. We are thankful to Dr. Sudipta Seal and Dr. Soumen Das, University of Central Florida for their help in data preparation. The authors declare that no conflict of interest exists with this manuscript and thank Mary Allyson Finger for the manuscript preparation.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, Fleming T, Forouzanfar MH, Hancock J, Hay RJ, Hunter-Merrill R, Huynh C, Hosgood HD, Johnson CO, Jonas JB, Khubchandani J, Kumar GA, Kutz M, Lan Q, Larson HJ, Liang X, Lim SS, Lopez AD, MacIntyre MF, Marczak L, Marquez N, Mokdad AH, Pinho C, Pourmalek F, Salomon JA, Sanabria JR, Sandar L, Sartorius B, Schwartz SM, Shackelford KA, Shibuya K, Stanaway J, Steiner C, Sun J, Takahashi K, Vollset SE, Vos T, Wagner JA, Wang H, Westerman R, Zeeb H, Zoeckler L, Abd-Allah F, Ahmed MB, Alabed S, Alam NK, Aldhahri SF, Alem G, Alemayohu MA, Ali R, Al-Raddadi R, Amare A, Amoako Y, Artaman A, Asayesh H, Atnafu N, Awasthi A, Saleem HB, Barac A, Bedi N, Bensenor I, Berhane A, Bernabé E, Betsu B, Binagwaho A, Boneya D, Campos-Nonato I, Castañeda-Orjuela C, Catalá-López F, Chiang P, Chibueze C, Chitheer A, Choi JY, Cowie B, Damtew S, das Neves J, Dey S, Dharmaratne S, Dhillon P, Ding E, Driscoll T, Ekwueme D, Endries AY, Farvid M, Farzadfar F, Fernandes J, Fischer F, G/Hiwot TT, Gebru A, Gopalani S, Hailu A, Horino M, Horita N, Husseini A, Huybrechts I, Inoue M, Islami F, Jakovljevic M, James S, Javanbakht M, Jee SH, Kasaeian A, Kedir MS, Khader YS, Khang YH, Kim D, Leigh J, Linn S, Lunevicius R, El Razek HMA, Malekzadeh R, Malta DC, Marcenes W, Markos D, Melaku YA, Meles KG, Mendoza W, Mengiste DT, Meretoja TJ, Miller TR, Mohammad KA, Mohammadi A, Mohammed S, Moradi-Lakeh M, Nagel G, Nand D, Le Nguyen Q, Nolte S, Ogbo FA, Oladimeji KE, Oren E, Pa M, Park EK, Pereira DM, Plass D, Qorbani M, Radfar A, Rafay A, Rahman M, Rana SM, Søreide K, Satpathy M, Sawhney M, Sepanlou SG, Shaikh MA, She J, Shiue I, Shore HR, Shrime MG, So S, Soneji S, Stathopoulou V, Stroumpoulis K, Sufiyan MB, Sykes BL, Tabarés-Seisdedos R, Tadese F, Tedla BA, Tessema GA, Thakur JS, Tran BX, Ukwaja KN, Uzochukwu BSC, Vlassov VV, Weiderpass E, Wubshet Terefe M, Yebyo HG, Yimam HH, Yonemoto N, Younis MZ, Yu C, Zaidi Z, Zaki MES, Zenebe ZM, Murray CJL, Naghavi M. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–48. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gajria D, Chandarlapaty S. HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev Anticancer Ther. 2011;11:263–75. doi: 10.1586/era.10.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L, Kang L, Zhao W, Feng Y, Liu W, Wang T, Mai H, Huang J, Chen S, Liang Y, Han J, Xu X, Ye Q. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated warburg effect. Cancer Lett. 2017;400:89–98. doi: 10.1016/j.canlet.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 4.Miao Y, Zhang LF, Guo R, Liang S, Zhang M, Shi S, Shang-Guan CF, Liu MF, Li B. (18)F-FDG PET/CT for monitoring the response of breast cancer to miR-143-based therapeutics by targeting tumor glycolysis. Mol Ther Nucleic Acids. 2016;5:e357. doi: 10.1038/mtna.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velpula KK, Bhasin A, Asuthkar S, Tsung AJ. Combined targeting of PDK1 and EGFR triggers regression of glioblastoma by reversing the warburg effect. Cancer Res. 2013;73:7277–89. doi: 10.1158/0008-5472.CAN-13-1868. [DOI] [PubMed] [Google Scholar]
- 6.Takeda S, Okazaki H, Kudo T, Kakizoe K, Himeno T, Matsumoto K, Shindo M, Aramaki H. Bongkrekic acid as a warburg effect modulator in long-term estradiol-deprived MCF-7 breast cancer cells. Anticancer Res. 2016;36:5171–82. doi: 10.21873/anticanres.11087. [DOI] [PubMed] [Google Scholar]
- 7.Michelakis ED, Gurtu V, Webster L, Barnes G, Watson G, Howard L, Cupitt J, Paterson I, Thompson RB, Chow K, O’Regan DP, Zhao L, Wharton J, Kiely DG, Kinnaird A, Boukouris AE, White C, Nagendran J, Freed DH, Wort SJ, Gibbs JSR, Wilkins MR. Inhibition of pyruvate dehydrogenase kinase improves pulmonary arterial hypertension in genetically susceptible patients. Sci Transl Med. 2017;9:10. doi: 10.1126/scitranslmed.aao4583. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, He J, Hunter T, Wang L, Lu Z. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell. 2016;61:705–19. doi: 10.1016/j.molcel.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 10.Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 11.Schmittgen TD. Regulation of microRNA processing in development, differentiation and cancer. J Cell Mol Med. 2008;12:1811–9. doi: 10.1111/j.1582-4934.2008.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asghari F, Haghnavaz N, Baradaran B, Hemmatzadeh M, Kazemi T. Tumor suppressor microRNAs: targeted molecules and signaling pathways in breast cancer. Biomed Pharmacother. 2016;81:305–17. doi: 10.1016/j.biopha.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Izaurralde E. GENE REGULATION. breakers and blockers-miRNAs at work. Science. 2015;349:380–2. doi: 10.1126/science.1260969. [DOI] [PubMed] [Google Scholar]
- 15.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 16.Verghese ET, Drury R, Green CA, Holliday DL, Lu X, Nash C, Speirs V, Thorne JL, Thygesen HH, Zougman A, Hull MA, Hanby AM, Hughes TA. MiR-26b is down-regulated in carcinomaassociated fibroblasts from ER-positive breast cancers leading to enhanced cell migration and invasion. J Pathol. 2013;231:388–99. doi: 10.1002/path.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim BG, Kang S, Han HH, Lee JH, Kim JE, Lee SH, Cho NH. Transcriptome-wide analysis of compression-induced microRNA expression alteration in breast cancer for mining therapeutic targets. Oncotarget. 2016;7:27468–78. doi: 10.18632/oncotarget.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G, Huang P, Xie J, Li R. microRNA211 suppresses the growth and metastasis of cervical cancer by directly targeting ZEB1. Mol Med Rep. 2018;17:1275–82. doi: 10.3892/mmr.2017.8006. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Gan C, Zhang J, Chen D. LPSinduced downregulation of microRNA204/211 upregulates and stabilizes Angiopoietin1 mRNA in EA.hy926 endothelial cells. Mol Med Rep. 2017;16:6081–7. doi: 10.3892/mmr.2017.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asuthkar S, Velpula KK, Chetty C, Gorantla B, Rao JS. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 2012;3:1439–54. doi: 10.18632/oncotarget.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velpula KK, Guda MR, Sahu K, Tuszynski J, Asuthkar S, Bach SE, Lathia JD, Tsung AJ. Metabolic targeting of EGFRvIII/PDK1 axis in temozolomide resistant glioblastoma. Oncotarget. 2017;8:35639–55. doi: 10.18632/oncotarget.16767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011;378:1812–23. doi: 10.1016/S0140-6736(11)61539-0. [DOI] [PubMed] [Google Scholar]
- 23.Romano G, Kwong LN. miRNAs, melanoma and microenvironment: an intricate network. Int J Mol Sci. 2017:18. doi: 10.3390/ijms18112354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang G, Zhang S, Yazdanparast A, Li M, Pawar AV, Liu Y, Inavolu SM, Cheng L. Comprehensive comparison of molecular portraits between cell lines and tumors in breast cancer. BMC Genomics. 2016;17(Suppl 7):525. doi: 10.1186/s12864-016-2911-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Daemen A, Hatzivassiliou G, Arnott D, Wilson C, Zhuang G, Gao M, Liu P, Boudreau A, Johnson L, Settleman J. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelialmesenchymal transition and drug resistance in tumor cells. Cancer Metab. 2014;2:20. doi: 10.1186/2049-3002-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solin LJ. Breast conservation treatment with radiation: an ongoing success story. J. Clin. Oncol. 2010;28:709–11. doi: 10.1200/JCO.2009.26.1164. [DOI] [PubMed] [Google Scholar]
- 27.Roscigno G, Quintavalle C, Donnarumma E, Puoti I, Diaz-Lagares A, Iaboni M, Fiore D, Russo V, Todaro M, Romano G, Thomas R, Cortino G, Gaggianesi M, Esteller M, Croce CM, Condorelli G. MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget. 2016;7:580–92. doi: 10.18632/oncotarget.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Wainscott C, Xi Y. MicroRNA provides insight into understanding esophageal cancer. Thorac Cancer. 2011;2:134–42. doi: 10.1111/j.1759-7714.2011.00059.x. [DOI] [PubMed] [Google Scholar]
- 29.Lubov J, Maschietto M, Ibrahim I, Mlynarek A, Hier M, Kowalski LP, Alaoui-Jamali MA, da Silva SD. Meta-analysis of microRNAs expression in head and neck cancer: uncovering association with outcome and mechanisms. Oncotarget. 2017;8:55511–24. doi: 10.18632/oncotarget.19224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Guan DH, Bi RX, Xie J, Yang CH, Jiang YH. Prognostic value of microRNAs in gastric cancer: a meta-analysis. Oncotarget. 2017;8:55489–510. doi: 10.18632/oncotarget.18590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda T, Hayashi N, Kuroda Y, Ito S, Eguchi H, Mimori K. MicroRNAs as biomarkers in colorectal cancer. Cancers (Basel) 2017:9. doi: 10.3390/cancers9090124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courtnay R, Ngo DC, Malik N, Ververis K, Tortorella SM, Karagiannis TC. Cancer metabolism and the warburg effect: the role of HIF-1 and PI3K. Mol Biol Rep. 2015;42:841–51. doi: 10.1007/s11033-015-3858-x. [DOI] [PubMed] [Google Scholar]
- 33.Han H, Li W, Shen H, Zhang J, Zhu Y, Li Y. microRNA-129-5p, a c-myc negative target, affects hepatocellular carcinoma progression by blocking the warburg effect. J Mol Cell Biol. 2016 doi: 10.1093/jmcb/mjw010. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Fu Z, Chen D, Cheng H, Wang F. Hypoxiainducible factor-1alpha protects cervical carcinoma cells from apoptosis induced by radiation via modulation of vascular endothelial growth factor and p53 under hypoxia. Med Sci Monit. 2015;21:318–25. doi: 10.12659/MSM.893265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen H, Hau E, Joshi S, Dilda PJ, McDonald KL. Sensitization of glioblastoma cells to irradiation by modulating the glucose metabolism. Mol Cancer Ther. 2015;14:1794–804. doi: 10.1158/1535-7163.MCT-15-0247. [DOI] [PubMed] [Google Scholar]
- 36.Duru N, Fan M, Candas D, Menaa C, Liu HC, Nantajit D, Wen Y, Xiao K, Eldridge A, Chromy BA, Li S, Spitz DR, Lam KS, Wicha MS, Li JJ. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin Cancer Res. 2012;18:6634–47. doi: 10.1158/1078-0432.CCR-12-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forero-Torres A, Varley KE, Abramson VG, Li Y, Vaklavas C, Lin NU, Liu MC, Rugo HS, Nanda R, Storniolo AM, Traina TA, Patil S, Van Poznak CH, Nangia JR, Irvin WJ Jr, Krontiras H, De Los Santos JF, Haluska P, Grizzle W, Myers RM, Wolff AC Translational Breast Cancer Research Consortium (TBCRC) TBCRC 019: a phase II trial of nanoparticle albumin-bound paclitaxel with or without the anti-death receptor 5 monoclonal antibody tigatuzumab in patients with triple-negative breast cancer. Clin Cancer Res. 2015;21:2722–9. doi: 10.1158/1078-0432.CCR-14-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciprotti M, Tebbutt NC, Lee FT, Lee ST, Gan HK, McKee DC, O’Keefe GJ, Gong SJ, Chong G, Hopkins W, Chappell B, Scott FE, Brechbiel MW, Tse AN, Jansen M, Matsumura M, Kotsuma M, Watanabe R, Venhaus R, Beckman RA, Greenberg J, Scott AM. Phase I imaging and pharmacodynamic trial of CS-1008 in patients with metastatic colorectal cancer. J. Clin. Oncol. 2015;33:2609–16. doi: 10.1200/JCO.2014.60.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brigant JL, Labarbe T, Israel JM, Vincent JD. GH clonal cells as a model for the relation between peptide control of hypophysial secretion and intracellular modulation of membrane electrical activity. Regul Pept Suppl. 1985;4:74–7. [PubMed] [Google Scholar]
- 40.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–90. [PubMed] [Google Scholar]
- 41.Tavallai M, Booth L, Roberts JL, McGuire WP, Poklepovic A, Dent P. Ruxolitinib synergizes with DMF to kill via BIM+BAD-induced mitochondrial dysfunction and via reduced SOD2/TRX expression and ROS. Oncotarget. 2016;7:17290–300. doi: 10.18632/oncotarget.8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazar J, Qi F, Lee B, Marchica J, Govindarajan S, Shelley J, Li JL, Ray A, Perera RJ. MicroRNA 211 functions as a metabolic switch in human melanoma cells. Mol Cell Biol. 2016;36:1090–108. doi: 10.1128/MCB.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


