Abstract
Inflammation is indispensable for host defense, whereas excessive inflammation often develop inflammatory diseases. Autophagy is thought to be engaged in many extracellular stress responses, such as starvation and innate immunity. Thus, autophagy plays an important role in maintaining homeostasis. The purpose of this study was to elucidate the function of BRF1 in the regulation of inflammation and autophagy response in macrophages. We found that BRF1 inhibited the LPS-induced inflammatory factors expression and the autophagy flux in macrophage. Furthermore, inhibition autophagy with 3-MA can attenuate the suppressive effect of BRF1 on LPS-mediated inflammation. In addition, MAPK/ERK signaling pathway was involved in the BRF1 inhibition inflammation and autophagy in macrophages. These findings indicate that BRF1 attenuates LPS-induced inflammatory factors secretion through autophagy, at least in part, through MAPK/ERK signaling pathway.
Keywords: Autophagy, BRF1, Inflammation, Macrophage, MAPK/ERK
Introduction
Butyrate response factor1(BRF1) belongs to RNA-binding proteins of the ZFP36 family (TTP, BRF-1/BRF-2) which have a highly conserved CCCH tandem zinc-finger (Zn-finger) motif and are best known for mRNA decay through binding to AU-rich elements in the 3′ untranslated region.1, 2 BRF1, as a circadian gene,3 was also reported to bind ARE-mRNAs to processing bodies (PBs) or cytoplasmic assemblies of mRNAs which promote translational silencing.4 Meanwhile, the p38 MAPK/MAPK- activated protein kinase 2 (MK2) and PKB/AKT pathways was confirmed to inhibit its ARE-binding activity of BRF1 through phosphorylation and binding to 14-3-35, 6. Furthermore, it has been shown that BRF-1 can promote the carcinoma cell death and prevent an evolutionarily conserved posttranscriptional regulation from promoting transition into the S phase of the cell cycle. BRF1 also played an crucial role in PTH-dependent bone remodeling and deletion of BRF1 is lethal at embryonic day 117, 8. Most importantly, BRF1 was participated in the immune system regulation.9
Autophagy is concerned as a principal catabolic Pathway for cellular homeostasis through engulfing cytoplasmic components and subsequently fusing with lysosomes to degrade them,10 which also plays a part in degrading and recycling long-lived proteins and organelles. It is thought to be engaged in many extracellular stress responses, such as starvation and innate immunity, SQSTM1(p62) can binds inflammasome subunit ASC to suppress inflammation.11 Moreover, the autophagosome can engulfs IL-1β in a same way for degradation, whereas deficiency of autophagy lead to inflammasome hyperactive.12
Inflammasome, a cytosolic protein complex, plays an essential role in immune responses and inflammation, such as NLRP3(NOD-like receptor protein3) inflammasome which keeps IL-1β and IL-18 expressions and subsequent inflammation in check. It is considered to be the most clinically implicated inflammasome.13, 14 Inflammation is indispensable for immune defense, excessive inflammation develop inflammatory diseases.15 So, intensive monitoring of the innate immune system is necessary. It has been reported that LPS-induced SQSTM1 can recruitment of NLRP3 inflammasome to autophagosomes for degradation.16, 17, 18
Given the important role that ZFP36 proteins play in the regulation of inflammation.19 In this study, we examined whether BRF1, an important molecule in the ZFP36 family, influenced the inflammation is associated with autophagy; which signaling pathway is involved in this process. Importantly, the data presented here clarified that BRF1 attenuated LPS-induced inflammation through autophagy, and ERK signaling pathway.
Materials and methods
Cell culture
RAW264.7 cells were a gift with Prof. Chuanju Liu, New York University School of Medicine, New York, USA. Then cultured in DMEM with 10% FBS.20 Incubated in standard incubator for 12 h Before stimulation, resuspended cells in a 60 mm tissue culture dish or seed the cells on sterile glass coverslips, then incubated in 37 °C with 5% CO2, And the cells were also viewed by their typical morphology of cobblestone under a microscope before treatment.
Adenovirus and plasmid
The cDNA of BRF1 was amplified from 293 cells and subcloned into pAd-Track-CMV (presented by Professor Chuauju Liu of New York University), and then recombinated with the supercoiled backbone vector in BJ5183 bacterial cells, Finally, recombinant adenovirus was generated and amplified in 293 cells by transfecting the recombinant adenovirus DNA. Primer of BRF1 was designed as the following: forward:5′-GTCAGATCCGCTAGAGATCTCGACACACCAGATCCT CGC-3'and reverse: 5′-GATATCTTATCTA GAAGCTTCCCTCCCTACCCTGGC TTA-3′.
The cDNA of BRF1 was amplified from 293 cells with another primer as the following: Forward:5′- GTTTAAACGGGCCCTCTAGACGACACACCAGATCCTC GC-3′ and Reverse:5′-TGG AATTCTGCAGATATCCCCTCCCTACCCTGGCTTA- 3’. Then subcloned into pcDNA3.1 (−).
RNA extraction and real-time PCR
The RNeasy Mini Kit (Qiagen, Hilden, Germany) and the PrimeScript RT reagent Kit with gDNA Eraser (TARAKA) was used to isolate and reverse transcribe total mRNA of RAW264.7 macrophages.21 And RT-QPCR was performed. The specific primer sequences were designed as follows: TNFα: Forward 5- CCTGTAGCCCACGTCG TAG-3 and Reverse 5-GGGAGTAGACAAGGTACAACCC-3; IL6: Forward 5-CTGCAAGAGACTTCCATCCAG-3 and Reverse 5-AGTGGTATAGACAGGTC TGTTGG-3; GAPDH:Forward 5- AGG TCGGTGTGAACGGATTTG-3 and Reverse 5- GGGGTCGTTGATGGCAACA-3 COX2/Ptgs2: Forward 5-TGTGACTGTACC CGGACTGG-3 and Reverse 5-TGCACATTGTAAGTAGGTGGAC-3; Nos2: Forward 5-ACATCGACCCGTCCACAGTAT-3 and Reverse 5-CAGAGGGGTA GGCTTGTCTC-3; Zfp36l1: forward 5-GACCACCACCCTCGTGTCT-3 and reverse 5-GG TGCCCACTGCCTTTCT-3. The Real-Time PCR system was used and the data were normalized with GAPDH according to the 2–ΔΔCt relative quantitation method in the manufacturer's manual.21
Western blot analysis and antibodies
Lysis buffer containing PMSF was used to extract protein from the cells. Proteins was transformed to PVDF membrane by separation and transmembrane. 5% nonfat dry milk was used to reduce it nonspecific binding and primary antibodies was incubated overnight at 4 °C: Anti-SQSTM1 (p62), anti-caspase-12, anti-caspase-3, anti-ATG12, anti-BRF1, anti-ATG7, anti-β-actin, anti-ERK1/2, anti-p-ERK1/2 were bought from CST corresponding secondary antibody was incubated for 2 h at 25 °C22. Then proteins were visualized by film exposure or ECL.
Immunofluorescence
Cells were fixed and permeabilized for 30 min at room temperature separately with 4% PF and 0.1% Triton X-100. 5% BSA for 1 h to block membrane and primary antibody incubated overnight at 4 °C. The antibodies used in this research were: anti-LC3 (1:200, novus, Rabbit IgG) and anti-LAMP1 (1:1000, novus, mouse IgG). The cells were stained with corresponding secondary IgG antibody (1:200, abbkine) for 1 h at dark. Cells were then incubated with DAPI for 15 min. Finally, cells were analyzed with a microscope.20
Flow cytometry (FCM)
Following incubation with Ad-BRF1/GFP or LPS + Ad-BRF1/GFP, cells were then collected for FCM analysis. Cellular apoptotic rate was detected by flow cytometer. The experiments were performed in triplicate.
Statistical analysis
After 3 independent experiments, GraphPad Prism 5 software (GraphPad) was used to analysis the data. Student's t-test was used and P < 0.05 was considered statistically significant.
Results
LPS induced inflammation and autophagy in dose- and time-dependence in RAW264.7 cells
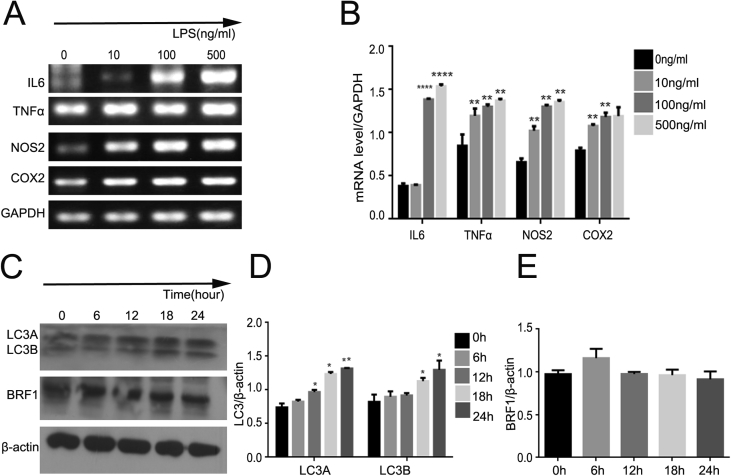
Lipopolysaccharide (LPS) has been recognized as a crucial inducer of inflammation, most of proinflammatory cytokines can be induced after treated with LPS in macrophages. To test the effect of LPS on inflammatory factors, we performed different concentrations (0, 10, 100, 500 ng/mL) of LPS in RAW264.7 for 24 h, mRNA level was measured by real-time PCR. As show in Fig. 1A and B, the expression of interleukin6 (IL6), TNFα, NOS2 and COX2 were significantly increased in a dose-dependent manner comparing with control. Meanwhile, numerous studies have confirmed that LPS can increased autophagy through TRIF-dependent MyD88-independent TLR4 signaling and PI3K pathway as well. Then we conducted LPS (500 ng/mL) to RAW264.7 in a time-dependent manner (0, 6, 12, 18, 24 h) to verify whether the ratio of LC3/β-actin was increased, western blot as show in Fig. 1C and D represented that the expression of LC3A/B was enhanced obviously at 24 h. Surprisingly, the expression of BRF1 was highest at 6 h (Fig. 1C and E).
Figure 1.
LPS activated inflammation and autophagy in dose- and time-dependence in RAW264.7 cells. (A) RAW264.7 macrophages were treated with LPS with different concentrations (0, 10, 100, 500 ng/mL) for 24 h. And realtime PCR was used to detect the mRNA expression of various inflammatory factors such as TNFα, IL6, NOS2, COX2. GAPDH was performed as a controls. (B) Quantitative analysis of endogenous IL6,TNFα, NOS2 and COX2 expressions were shown in bar graphs. Data are showed as mean ± SEM and repeated for three times. (C) Cells were exposed to LPS with 500 ng/ml at different interval times (0,6,12,18,24 h), LC3 and BRF1 protein expression was examined by Western bloting. β-actin expression was regarded as control for similar loading of proteins in each lane. (D–E) Bar graphs exhibited the relative protein level of LC3A/β-actin, LC3B/β-actin (D) and BRF1/β-actin (E) by Western blot analysis.
BRF1 decreased autophagy in RAW264.7 cells
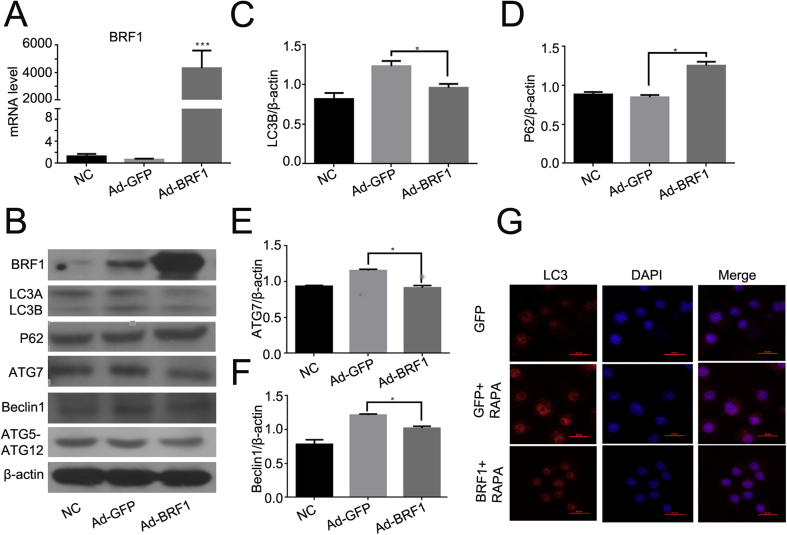
Lots of studies showed that PI3K/AKT pathway plays an important roles in ARE-binding activity of BRF1 which promoting mRNA decay by binding to ARE domains.5 Meanwhile, PI3K/AKT exerts great effects on activation of autophagy. To further study the relationship between BRF1 and autophagy. We detected the expression of BRF1 after infected with Ad-BRF1 in RAW264.7 for 24 h. The mRNA level of BRF1 was markedly increased compared with Ad-GFP by qPCR (Fig. 2A). We next examined whether BRF1 influenced the autophagy. As show in Fig. 2B, the ratio of LC3B/β-actin was decreased after Ad-BRF1 infection for 24 h, while the SQSTM1/P62 abundance was increased. Besides, the mRNA level of ATG7 and Beclin1 were declined significantly compared with the control (Fig. 2C–F). Furthermore, it was demonstrated that incubating Ad-BRF1 and Rapamycin (50 nmoL/ml) reduced the expression of LC3 comparing with the presence of Ad-GFP and Rapamycin by the technique of immunofluorescence (Fig. 2G).
Figure 2.
BRF1 reduced autophagy in RAW264.7 macrophage. (A) Relative mRNA level of BRF1 was measured by qPCR after infection with Ad-GFP or Ad-BRF1 for 24 h in RAW264.7 macrophages. (B) Determination of BRF1 and autophagy related protein LC3, SQSTM1/p62, ATG7, Beclin1, ATG5- ATG12 protein expression level after infection with Ad-GFP or Ad-BRF1 in RAW264.7 macrophage for 24 h by Western blotting analysis. β-actin expression monitored as controls for similar loading of proteins in each lane. (C–F) Quantitative analysis of endogenous LC3(C),SQSTM1/p62 (D), ATG7 (E), Beclin1 (F) expression were shown in bar graphs. Y axis presented the ratio of protein level compared to β-actin respectively. Experiments were repeated at least three times. (G) Cells were incubated with Ad-BRF1/Ad-GFP and LPS for 24 h in 24-wells dish, followed with 50 nmol/ml of autophagy activator Rapamycine for 3 h. Next, it was stained by LC3 (red) and DAPI (blue) and performed by confocal microscopy (400x).
BRF1 depressed LPS-induced autophagy in RAW264.7 cells
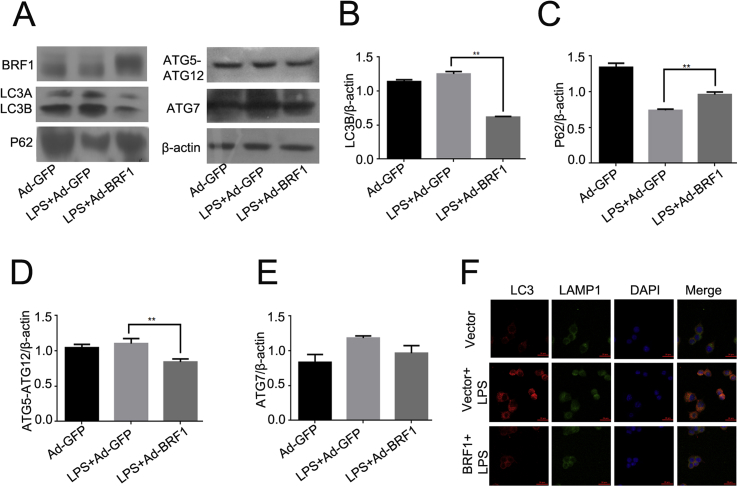
Previous studies have shown that LPS could activate autophagy in a time-dependent manner. Moreover, BRF1 was recognized as an anti-inflammatory factors.9 Next, we examined whether BRF1 can suppress autophagy induced by LPS in RAW264.7. Firstly, we detected the expression of autophagy-related protein, the ratio of LC3/β-actin was decreased, and the expression of ATG12-ATG5 and ATG7 were reduced after pretreated with Ad-BRF1+LPS (500 ng/ml) for 24 h (Fig. 3A,B,D,E). Conversely, P62(SQSTM1) which situated on the downstream of autophagy flux was increased compared with those infected with LPS and Ad-GFP in RAW264.7 (Fig. 3A and C). Furthermore, it was demonstrated that the incubation of pcDNA3.1 (−)-BRF1+LPS can significantly decrease the autophagosome formation comparing with the LPS and pcDNA3.1 (−) treatment with confocal microscopic images. Meanwhile, pcDNA3.1 (−)-BRF1+LPS can reduce the autophagy flux, according to the colocalization LC3 with lysosomal membrance protein LAMP1(Fig. 3F). Those results indicated that BRF1 could decrease LPS-induced autophagy in RAW264.7.
Figure 3.
BRF1 inhibited LPS-induced autophagy in RAW264.7 macrophages. (A) Determination of BRF1 and autophagy related protein LC3,SQSTM1/p62, ATG7 and ATG5-ATG12 protein expression level after infection with Ad-GFP or Ad-BRF1 and LPS (500 ng/ml) in RAW264.7 macrophage for 24 h by Western blotting analysis. β-actin was regarded as controls corresponding to protein above. (B–E) Quantitative analysis of the bar graphs represented endogenous LC3(B), SQSTM1/p62 (C), ATG5-ATG12 (D) and ATG7 (E) expression compared to β-actin respectively. *P < 0.05. Data are showed as mean ± SEM. Experiments were repeated at least three times. (F) Cells were pre-incubated with pcDNA3.1 (−)-BRF1/pcDNA3.1 (−) in the presence of LPS for 24 h, then stained LC3 (red), LAMP1(green) and DAPI(blue) by immunofluorescence. Image acquisited by microscopy (400x).
BRF1 inhibited LPS-induced inflammation and apoptosis
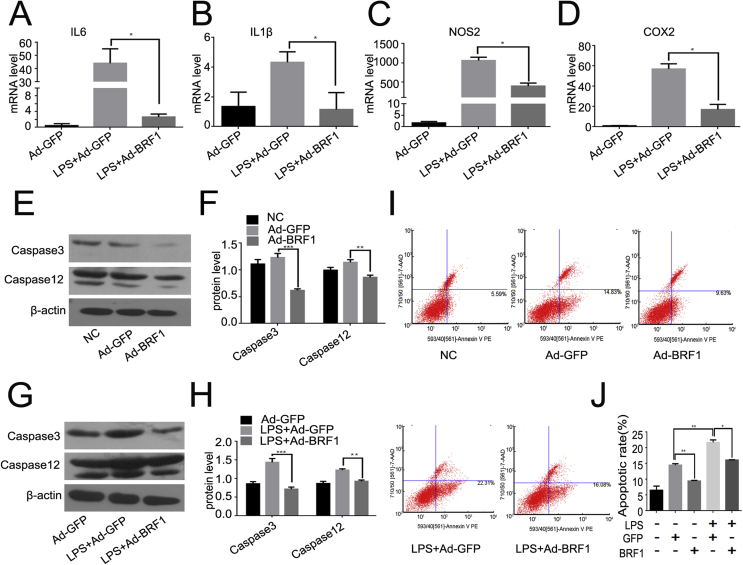
Then we examined the effect of BRF1 upon inflammatory activation. RAW264.7 macrophages were incubated with LPS alone or Ad-BRF1+ LPS respectively for 24 h, As shown in Fig. 4A–D, LPS enhanced the mRNA levels of interleukin6 (IL-6) interleukin1β(IL-1β), nitric oxide synthase2(NOS2) and cyclo-oxygenase2 (COX2). And BRF1 could significantly reduce the expression of IL-6, IL-1β, NOS2 and COX2, the LPS-induced inflammatory factors in RAW264.7. It is known that apoptosis coupled with autophagy was served as an essential character to maintain the cellular homeostasis. We also detected the apoptosis rate of macrophages after incubated with Ad-BRF1 or LPS + Ad-BRF1. It is showed that BRF1 inhibits not only the expression of caspase3 and caspase12 but also the expression of LPS-induced caspase3 and caspase12 comparing with their controls (Fig. 4E–H). FCM results also showed that BRF1 reduced the ratio of apoptosis with or without LPS in RAW264.7 (Fig. 4I and J). Taken together, BRF1 alleviates LPS-induced inflammation and cellular apoptosis rate.
Figure 4.
BRF1 decreased LPS-induced inflammation and apoptosis in RAW264.7 cells. (A–D) RAW264.7 was pretreated with Ad-GFP/Ad-BRF1 and LPS (500 ng/ml) for 24 h. Relative mRNA level of inflammatory factors IL6 (A), IL1β (B), NOS2 (C), COX2 (D) was quantified by qPCR. Experiments were repeated at least three times. (E) Apotosis of cells was evaluated by western blot through detecting caspase3 and caspase12 after infected by Ad-BRF1/Ad-GFP for 24 h. (F) Quantitative analysis performed in bar graphs was the ratio of caspase3 and caspase12 compared to β-actin respectively. Experiments were repeated at least three times. (G) Western blot was used to detect caspase3 and caspase12 following by overexpression of BRF1 and LPS for 24 h in RAW264.7 macrophage. (H) Quantitative analysis of endogenous caspase3 and caspase12 expression were shown in bar graphs. Experiments were repeated at least three times. β-actin served as an internal control. *P < 0.05. Data are showed as mean ± SEM, and repeated at least three experiments. (I) The ratio of apoptosis were detected by Flow Cytometry while incubated Ad-GFP/Ad-BRF1 with macrophage for 24 h with or without LPS. (J) Quantitative analysis on cell apoptosis results. Data are mean ± SD for relative apoptosis normalized to control cells for three independent experiments. Columns mean of five separate experiments; *P < 0.05; **P < 0.01.
BRF1 inhibits inflammation through autophagy in RAW264.7 cells
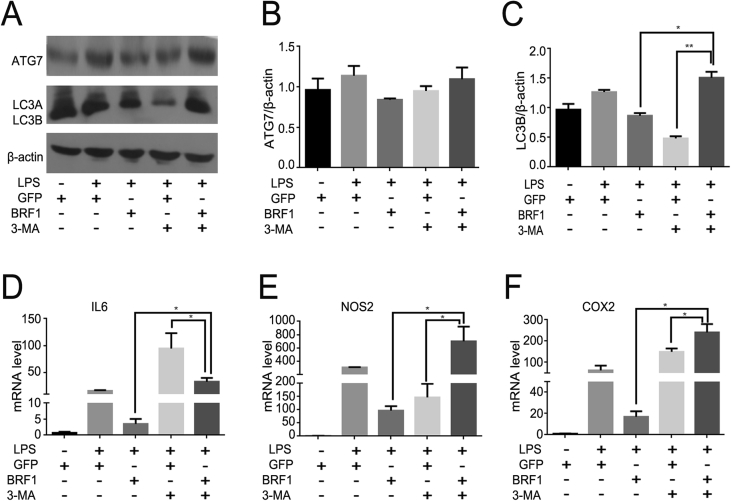
3-MA is an inhibitor of phosphoinositide 3-kinase (PI3K) signaling pathway that blocks formation of autophagolysosomes and degradation of proteins. To further demonstrate whether BRF1 inhibition inflammation is associated with autophagy, we performed macrophages with 3-MA (5 mM) for 3 h in the presence of Ad-BRF1 or LPS (500 ng/mL). As shown in Fig. 5A–C, the proteins level of LC3 and ATG7 were reduced in Ad-BRF1 infected RAW264.7 cells, while increased clearly after incubated with Ad-BRF1+3-MA, suggesting that the effect of BRF1 inhibition autophagy can be abolished after treatment with 3-MA. Then we detected whether LPS-induced inflammatory factors can be changed by BRF1+3-MA, Q-PCR showed that the endogenous expression of IL6, NOS2 and COX2 were reversed after incubated with Ad-BRF1+3-MA comparing with Ad-BRF1 alone (Fig. 5D–F). It is demonstrated that BRF1 inhibits inflammation through autophagy.
Figure 5.
3-MA reversed the inhibition effect of BRF1 on autophagy and inflammation. (A) Cells were pretreated with or without LPS (100 ng/mL), followed by infection with Ad-GFP or Ad-BRF1 for 24 h, in the presence of autophagy inhibitor 3 MA (5 mM) for 3 h. Then the protein levels of LC3, ATG7 were examined by western blot. (B–C) Bar graphs analyzed the relative protein levels of ATG7(B), LC3 (C) compared to β-actin above in RAW264.7.*P < 0.05. Data are showed as mean ± SEM and repeated at least three experiments. (D–F) RNA was extracted from RAW264.7 with the same treatment as before. And quantitative analysis of endogenous IL6(D), NOS2 (E), COX2 (F) expression performed by qPCR were shown in bar graphs. Data are expressed as mean ± SEM and repeated at least three times.
BRF1 influences inflammation and autophagy through ERK signal pathway
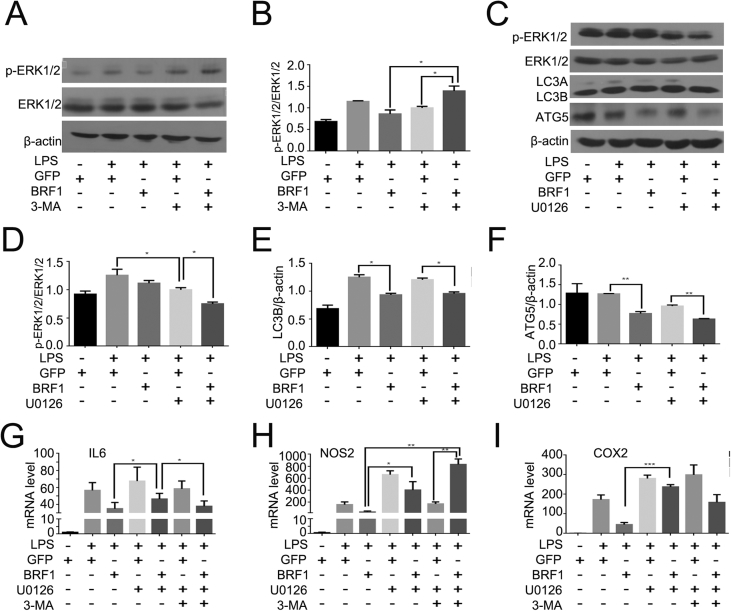
Later, we searched for signaling pathway that participated in this process. As LPS stimulated TLR4 can activate MAPK/ERK pathway and FGF/ERK MAP kinase signaling can regulate the expression of BRF16, 23. To further demonstrate whether BRF1 inhibition autophagy is associated with ERK signal pathway, we firstly performed macrophages with 3-MA (5 mM) for 3 h in the presence of Ad-BRF1 or LPS (500 ng/mL). As displayed in Fig. 6A–B, BRF1 inhibited ERK signal pathway, while the ratio of p-ERK1/2/ERK1/2 was increased after incubated with Ad-BRF1+3-MA comparing with Ad-BRF1. These results indicated that MAPK/ERK signaling pathway may be involved in autophagy in RAW264.7 cells. Then we used U0126 as an specific inhibitor of ERK to test whether BRF1 reduction inflammation was through ERK signaling pathway. Macrophages were incubated with the U0126 (5uM) for 2 h or Ad-BRF1 for 24 h in the presence of LPS. As revealed in Fig. 6C–F, it was showed that U0126 obviously reduced the phosphorylation of ERK and BRF1 promotes this phosphorylation process furtherly, however, U0126 can not obviously influence the BRF1 inhibition effect on autophagy. We next determined that the endogenous expression of IL6, NOS2 and COX2 were increased significantly after co-incubating Ad-BRF1 and U0126 compared with Ad-BRF1 alone in the presence of LPS. BRF1 inhibited the expression of IL6, NOS2 and COX2, however, U0126, 3-MA and U0126+3-MA can reverse this inhibition. Endogenous expression of IL6, NOS2 and COX2 were reversed after incubated with Ad-BRF1+ U0126, Ad-BRF1+3-MA or Ad-BRF1+U0126+3-MA (Fig. 6G–I). Taken together, BRF1 inhibits autophagy and inflammation through ERK signaling pathway.
Figure 6.
U0126 blocked the effect of BRF1 on inflammation in the presence of LPS. (A) Cells were pretreated with or without LPS (100 ng/mL), followed by infection with Ad-GFP or Ad-BRF1 for 24 h, in the presence of autophagy inhibitor 3-MA (5 mM) for 3 h The protein levels of phospho-ERK1/2 (p-ERK1/2), total ERK1/2 were determined with the use of western blot. (B) Bar graph showed the relative protein levels of p-ERK1/2/ERK1/2 above in RAW264.7.*P < 0.05. Data are showed as mean ± SEM and repeated at least three experiments. (C) Cells were pretreated with or without LPS(100 ng/mL), followed by infection with Ad-GFP orAd-BRF1 for 24 h, in the presence of ERK inhibitor U0126 (5uM) for 2 h. Then the protein levels of ERK1/2, p-ERK1/2, LC3, ATG5 were examined by western blot analysis. (D–F) Bar graphs showed the relative protein levels of p-ERK1/2/ERK1/2 (D), LC3 (E), ATG5 (F) compared to β-actin with the same treatments above in RAW264.7.*P < 0.05. Data are showed as mean ± SEM and repeated at least three experiments. (G–I) Cells were pretreated with or without LPS(100 ng/mL), followed by infection with Ad-GFP or Ad-BRF1 for 24 h, in the presence of U0126 for 2 h and 3-MA for 3 h. Relative mRNA level of inflammatory factors IL6 (G), NOS2 (H), COX2 (I) was quantified by qPCR. Quantitative analysis of endogenous IL6, NOS2, COX2 expression were shown in bar graphs. Data are expressed as mean ± SEM and repeated at least three times.
Discussion
The ZFP36 family have been considered as key role in regulation of ARE-mediated decay. One of the member of TIS11 family TTP has already been recognized as an anti-inflammatory factors because of its destabilizing pro-inflammatory cytokines, such as TNFα and GM-CSF.8 At present studies, BRF1 has been reported to bind to FXR which promoted lipid absorption.24 Moreover, BRF1 was also proved to destabilize LDLR mRNA which could inhibited by ERK/RSK1 signaling through phosphorylation of BRF1.25 However, whether BRF1 has the same effects on inflammatory as TTP remains unknown. In our present study, we found that BRF1 could suppress the secretion of IL6, NOS2 and COX2 induced by RAW264.7 cells. Meanwhile, BRF1 decreased autophagy and apoptosis at the same time. Here, we further investigated the potential mechanism of BRF1 in the control of inflammation and autophagy.
The innate immune system is served as a protective system to host after infection by Pathogens, which can form an inflammasome and recognize pathogen-associated molecular patterns. But once beyond of control, it will cause serious tissue damage and inflammatory diseases. Thus, activation of inflammasomes is often followed by autophagy which act as intracellular degradation systems. SQSTM1 was reported to target ASC and result in inactivation.12 Our study showed that LPS induced autophagy and inflammation in a time gradient in RAW264.7 macrophages, which was confirmed by increasing the expression of LC3 and inflammatory factors (Fig. 1).
Autophagy can destroy and recycle long-living proteins and damage organelles to protect cell from dead. The process of autophagy activation could be divided into four stages: formation, elongation, maturation and degradation. Once ULK1 (mammalian ATG1) was activated which phosphorylated by mechanistic target of rapamycin complex 1 (mTORC1), beclin-1 (mammalian ATG6) will be phosphorylated and released from an inhibitory interaction with BCL-2 to activate the class III PI3K (PI3KC3) complex1, then activate the formation of autophagosome.26 Rapamycin, as an important inhibitor of mTOR, suppresses the SASP by specifically down-regulating MAPKAPK2 translation which can phosphorylates and inhibits BRF1, and BRF1 was proved to prevent SASP induction which could activate the immune system.27 Thus there has large possibility that mTOR inhibition prevented SASP from activation correlated to the dephosphorylation of BRF1. As downstream of mTOR pathway factor, overexpression of BRF1 may activate mTOR pathway so that decrease the formation of autophagosome. Herein, we testified BRF1 inhibits autophagy and LPS-induced autophagy, as revealed by decreasing expressions of LC3B, ATG7, ATG5-ATG12 and autophagy flux reduction (Figure 2, Figure 3). Meanwhile, PKB as a downstream target of PI3-K could efficiently stabilized ARE-containing reporter transcripts. Recent studies showed that BRF1 was the potential target which was phosphorylated by PKB at serine 92 (S92). S92 phosphorylation of BRF1 was induced complex formation with the scaffold protein 14-3-3 rather than impair ARE binding.10 In our study, we found that BRF1 inhibits LPS-induced inflammation and cell apoptosis (Fig. 4). The effect of inflammation reduced by BRF1 was enhanced in the presence of 3-MA, an inhibitor of autophagy. We further proved that the effect of BRF1 on inflammation is required for autophagy (Fig. 5). Bourcier et al pointed that TTP could decreased autophagy and it was phosphorylated by ERK. In this study, we found that MAPK/ERK signaling pathway may be involved in autophagy in RAW264.7 cells. BRF1 inhibits autophagy and inflammation through ERK Signaling. Interestingly, co-suppression of ERK and autophagy can reverse the effect of BRF1 on inflammation (Fig. 6).
In a word, this study offers novel insight into the function of BRF1 in regulating inflammation and autophagy. Our study supports the notion that overexpression of BRF1, a key RNA-binding protein of the ZFP36 family, inhibits LPS-induced inflammation and autophagy. The inhibition effect of BRF1 on inflammation depends on autophagy and crosstalking with ERK Signaling. Our study implies that BRF1 is a reverse regulator of inflammation and autophagy, and the interaction mechanism between inflammation and autophagy remains further study. Our subsequent work aims to understand the complexities of this interaction. In addition, this study also provides potential molecular targets for a variety of inflammatory conditions. New insight into the mechanistic basis of inflammation and autophagy will provide new perspective of molecular targeted therapies.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81672209).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Blackshear P.J., Perera L. Phylogenetic distribution and evolution of the linked RNA-binding and NOT1-binding domains in the tristetraprolin family of tandem CCCH zinc finger proteins. J Interferon Cytokine Res. 2014;34(4):297–306. doi: 10.1089/jir.2013.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanduja S., Blanco F.F., Dixon D.A. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2010;2(1):42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storch K.F., Lipan O., Leykin I. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 4.Franks T.M., Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidlin M., Lu M., Leuenberger S.A. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004;23:4760–4769. doi: 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan F.E., Elowitz M.B. Brf1 posttranscriptionally regulates pluripotency and differentiation responses downstream of Erk MAP kinase. PNAS. 2014:E1740–E1748. doi: 10.1073/pnas.1320873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reppe S., Olstad O.K., Rian E., Gautvik V.T., Gautvik K.M., Jemtland R. Butyrate response factor 1 is regulated by parathyroid hormone and bone morphogenetic protein-2 in osteoblastic cells. Biochem Biophys Res Commun. 2004;324:218–223. doi: 10.1016/j.bbrc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Carballo E., Lai W.S., Blackshear P.J. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 9.Perga S., Montarolo F., Martire S., Berchialla P., Malucchi S., Bertolotto A. Anti-inflammatory genes associated with multiple sclerosis: a gene expression study. J Neuroimmunol. 2015;279:75–78. doi: 10.1016/j.jneuroim.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Levine B., Klionsky D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 11.Dupont N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1b. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi C.S., Shenderov K., Huang N.N. Activation of autophagy by inflammatory signals limits IL-1b production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauernfeind F.G., Horvath G., Stutz A. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y., Hara H., Nunez G. Mechanism and regulation of nlrp3 inflammasome activation. Trends Biochem Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Yuk J.M., Jo E.K. Crosstalk between autophagy and inflammasomes. Mol Cell. 2013;36:393–399. doi: 10.1007/s10059-013-0298-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong Z., Sanchez-Lopez E., Karin M. Autophagy, nlrp3 inflammasome and auto-Inflammatory/Immune diseases. Clin Exp Rheumatol. 2016;34:12–16. [PubMed] [Google Scholar]
- 18.Schrijvers D.M., De Meyer G.R., Martinet W. Autophagy in atherosclerosis: a potential drug target for plaque stabilization. Arterioscler Thromb Vasc Biol. 2011;31:2787–2791. doi: 10.1161/ATVBAHA.111.224899. [DOI] [PubMed] [Google Scholar]
- 19.Patial S., Curtis A.D., Lai W.S. Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies. PNAS. 2016;113:1865–1870. doi: 10.1073/pnas.1519906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fei H., Qingqing X., Shi P. Atorvastain ameliorates LPS-induced inflammatory response by autophagy via AKT/Mor signaling pathway. J Cell Biochem. 2017:1–12. doi: 10.1002/jcb.26320. [DOI] [PubMed] [Google Scholar]
- 21.Diana R., Julia B., Henrike S., Jürgen E. Reduced DPP4 activity improves insulin signaling in primary human adipocytes. Biochem Biophys Res Commun. 2016;471:348–354. doi: 10.1016/j.bbrc.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 22.Weiwei B., Yaru L., Dan W., Jian L., Xi W., Wei M. Sodium salicylate modulates inflammatory responses through AMP-activated protein kinase activation in LPS-stimulated THP-1 cells. J Cell Biochem. 2018;119:850–860. doi: 10.1002/jcb.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie S., Liu B., Fu S. GLP-2 suppresses LPS-induced inflammation in macrophages by inhibiting ERK phosphorylation and NF-κb activation. Cell Physiol Biochem. 2014;34:590–602. doi: 10.1159/000363025. [DOI] [PubMed] [Google Scholar]
- 24.Tarling E.J., Clifford B.L., Cheng J. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J Clin Invest. 2017;127(10):3741–3754. doi: 10.1172/JCI94029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adachi S., Homoto M., Tanaka R. ZFP36L1 and ZFP36L2 control LDLR mRNA stability via the ERK–RSK pathway. Nucleic Acids Res. 2014;42(15):10037–10049. doi: 10.1093/nar/gku652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cadwell K. Crosstalk between autophagy and inflammatory signaling pathways: balancing host defence and homeostasis. Nat Rev Immunol. 2016;16(11):661–675. doi: 10.1038/nri.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herranz N., Gallage S., Mellone M. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol. 2015;17(9):1205–1217. doi: 10.1038/ncb3225. [DOI] [PMC free article] [PubMed] [Google Scholar]