Abstract
Oral lichen planus (OLP) is a common chronic inflammatory autoimmune disease with unclear etiology. The aim of the present study was to identify the expression profiles of circulating exosomal miRNAs, which have been shown to be potent stimulators of inflammatory and immune responses, in OLP patients. Plasma exosomes were isolated from the patients and healthy individuals, and RAE scoring system was used to evaluate the severity of OLP. Differentially deregulated exosomal miRNAs associated with inflammatory response and autoimmunity in OLP were identified by miScript® miRNA PCR Array, and the results were confirmed by RT-PCR. The relationship between exosomal miRNAs and RAE scores was then analyzed, and bioinformatics analysis was used to predict the target genes and pathways of the differentially expressed exosomal miRNAs. Expression profiling showed that circulating exosomal miR-34a-5p and miR-130b-3p were upregulated, while miR-301b-3p was downregulated in OLP patients. Exosomal miR-34a-5p was positively correlated with the severity of OLP. Bioinformatics analysis revealed that the target genes of miR-34a-5p were mainly involved in regulation of gene expression, cell communication, signaling, and metabolic process, and modulated OLP progression through the PI3K/Akt signaling pathway. In conclusion, circulating exosomal miR-34a-5p could be a potential biomarker for evaluating the severity of OLP.
Keywords: Oral lichen planus, exosomes, miRNAs, plasma, microarray
Introduction
Oral lichen planus (OLP) is a common T-cell-mediated chronic inflammatory autoimmune disease with unclear etiology [1]. It is reported to affect about 0.1 to 4% of the global population, and the WHO has recognized it as an oral potentially malignant disorder [2]. Histopathologically, OLP is characterized by dense subepithelial infiltration of T lymphocytes, increased numbers of intraepithelial T lymphocytes, and degeneration of basal keratinocytes [3].
Exosomes are extracellular vesicles 30-150 nm in diameter and derived from the fusion of multi-vesicular bodies (MVB) with the cell plasma membrane [4,5]. They are shed from most all cell types, like T cells, B cells, hematopoietic cells, reticulocytes, dendritic cells, and tumor cells [6,7]. The exosomes secreted from immune cells regulate immune responses, and are associated with the pathogenesis of autoimmune diseases [6,8]. They act as natural nanocarriers and intercellular messengers by transferring proteins, lipids, DNAs, and RNAs to neighboring or distant cells, and thus mediate the cell-to-cell communication [9].
MicroRNAs (miRNAs) are small non-coding RNA molecules approximately 18-25 nucleotides in length, and participate in the transcriptional and post-transcriptional regulation of gene expression by provoking degradation or functional inhibition of target mRNA [10-12]. Exosomal miRNAs influence the proliferation, apoptosis, differentiation, and migration of recipient cells, and therefore key players in the modulation, pathogenesis, diagnosis and therapeutics of many autoimmune diseases [8,13,14]. Cristina Sole et al. showed that exosomal miR-29c in patients with lupus nephritis was significantly decreased, which correlated with renal function and the degree of renal fibrosis, suggesting its roles in the early prediction of histological fibrosis [15].
Emerging evidence, including our own previous studies, show aberrant expression of miR-146a, miR-155, miR-21, miR-125b and miR-203 in OLP [16-19]. However, it is unclear whether circulating exosomal miRNAs participate in OLP pathogenesis. The aim of the present study was to determine the differentially expressed circulating exosomal miRNAs in OLP and their correlations with the clinical characteristics.
Materials and methods
Patients and characteristics
Nineteen OLP patients and 11 age-sex-matched healthy individuals (P > 0.05) were recruited from the Department of Oral Medicine, School and Hospital of Stomatology, Wuhan University. Informed consent was obtained from each subject. The inclusion criteria of patients with OLP have been described in our previous study [20]. Briefly, the patients who did not suffer from any other systematic disorders or received any treatment within 3 months were included. The clinical characteristics of the participants are listed in Table 1. The study was approved by the Ethical Committee Board of the School and Hospital of Stomatology, Wuhan University, according to the Declaration of Helsinki on human subject protection.
Table 1.
Clinical characteristics of the OLP patients and healthy individuals
| OLP (n = 19) | Control (n = 11) | |
|---|---|---|
| Gender | ||
| Female | 10 | 7 |
| Male | 9 | 4 |
| Ages (years) | ||
| Range | 32-60 | 35-59 |
| Mean ± SEM | 47.3 ± 8.0 | 47.6 ± 6.1 |
Clinical assessment
The severity of OLP was assessed with the RAE (reticular, atrophic, and erosive lesion) scoring system, as described in our previous study [21].
Isolation and characterization of exosomes
Plasma exosomes were isolated using the exoEasy Maxi Kit (Qiagen, GmbH, Hilden, Germany) according to the manufacturer’s instructions. Briefly, prefiltered plasma was mixed 1:1 with 2 × binding Buffer XBP, and loaded onto the exoEasy membrane affinity column. The columns were centrifuged once and the flow-through was discarded, followed by a wash with 10 ml Buffer XWP to remove any non-specific retained material. The spin column was then transferred to a fresh collection tube and exosomes were eluted with 400 µl Buffer XE after centrifugation.
The purified exosomes (10 µl) were loaded onto 200-mesh Formvar/carbon grids (Head Biotechnology Co. Ltd, Beijing, China) and then transferred to a 10 µl drop of 20 g/l uranyl acetate (Provided by School of Basic Medical Sciences, Wuhan University). The final exosomes were observed under an HT7700 transmission electron microscope (Hitachi, Japan) at 80 kV and images were captured by a digital camera.
Particle size distribution was analyzed by Nanoparticle Tracking Analysis (NTA; LM10; Nanosight) following the manufacturer’s instructions, and repeated thrice.
The exosomal biomarkers CD9 (1:1,000; BD Biosciences Pharmingen, USA) and CD63 (1:1,000; Millipore, Germany) were detected by flow cytometric analysis. Exosomes were coated onto 4-μm-diameter aldehyde/sulfate latex beads (Invitrogen), as previously described [22].
Exosomal RNA isolation and detection
Total RNA was extracted using miRNeasy Micro kit (Qiagen GmbH, Hilden, Germany) as per the manufacturer’s instructions. Briefly, exosomes were added to 700 µl Qiazol Lysis Reagent (Qiagen) and homogenized by pipetting. After 5 min of incubation at room temperature, 3.5 µl 1.6 × 108 copies/µl of exogenous miRNeasy Serum/Plasma Spike-In Control cel-miR-39 mimic (Qiagen) was added to each sample as a reference molecule for unbiased normalization [23-25]. After adding 140 µl chloroform and shaking vigorously for 15 s, the mixture was centrifuged for 15 min at 12,000 g at 4°C, and the upper aqueous phase was transferred into a new collection tube along with 1.5 volumes of 100% ethanol. The samples were then pipetted into an RNeasy Micro spin column and centrifuged. The flow-through was discarded and Buffer RWT, Buffer RPE and 80% ethanol were added sequentially to the spin column. Purified total RNA was extracted in 14 µl elution buffer and quantified by NanoDrop 2000 (Thermo Fisher Scientific, USA). Integrity of the total RNA was verified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Exosomal miRNA microarray analysis
Exosomal miRNA expression profiles were analyzed using a miScript miRNA PCR Array (Qiagen, MIHS-105Z) that includes 84 microRNAs associated with human inflammatory response and autoimmunity. cDNA was synthesized from the total RNA (25 ng) using the miScript II Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. The cDNA was preamplified with the miScript PreAMP PCR Kit, and then mixed with QuantiTect SYBR Green PCR Master Mix, miScript Universal Primer, and RNase-free water. The PCR master mix was then distributed in 25 µl aliquots across the microarray. The plate was sealed, centrifuged at 1000 g for 1 min to remove bubbles, and run in a CFX96TM real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA). The PCR parameters were as follows: initial activation step at 95°C for 15 min, followed by 15 s at 94°C for denaturation, 30 s at 55°C for annealing, and 30 s at 70°C for extension. The cycle number was set as 30 cycles. The relative expression of the miRNAs was analyzed by the 2-ΔΔCt method, where ΔCt = Ct miRNA - AVG Ct cel-miR-39 and ΔΔCt = ΔCt (OLP group) - ΔCt (Control group). Fold change > 2 or < 0.5 and t-test P value < 0.05 were used as the threshold for selecting differentially expressed miRNAs.
Quantitative real-time RT-PCR confirmation
The differentially expressed exosomal miRNAs in OLP were validated by quantitative real-time RT-PCR. The reaction parameters were as follows: HotStar Taq DNA Polymerase activation at 95°C for 15 min, 15 s at 94°C for denaturation, 30 s at 55°C for annealing, and 30 s at 70°C for extension. The cycle number was set as 35 cycles. The level of miRNA expression was normalized to cel-miR-39 using the 2-ΔΔCt method as described. The forward primer sequence (Qiagen) of the miRNAs were synthesized as shown in Table 2, and the miScript universal primer was provided in the PCR kit.
Table 2.
miRNA primer sequence for RT-PCR
| miRNA | Accession number | Primer sequence |
|---|---|---|
| hsa-miR-34a-5p | MIMAT0000255 | TGGCAGTGTCTTAGCTGGTTG |
| hsa-miR-130b-3p | MIMAT0000691 | ACAGTGCAATGATGAAAGGGCAT |
| hsa-miR-29c-3p | MIMAT0000681 | GCGTAGCACCATTTGAAATCGGTTA |
| hsa-miR-144-3p | MIMAT0000436 | CGCGCGTACAGTATAGATGATGTACT |
| hsa-miR-301b-3p | MIMAT0004958 | CGCAGTGCAATGATATTGTCAAAGC |
Target gene prediction by bioinformatics analysis
Target genes of the differentially expressed miRNAs that were correlated with OLP severity were predicted through the miRecord database (http://c1.accurascience.com/miRecords/prediction_query.php). Eleven established miRNA target prediction programs are integrated into the miRecord database, including diana, microinspector, miranda, mitarget2, mitarget, nbmirtar, pictar, pita, rna22, rnahybrid and targetscan. To further predict the functions of target genes, both gene ontology (GO) analysis and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis were performed by the online database for annotation, visualization and integrated discovery (DAVID) v6.8 (https://david.ncifcrf.gov/).
Statistical analysis
Data were compared by independent-samples t-test and one-way ANOVA analysis of variance using SPSS statistical software (SPSS 17.0; SPSS Inc., Chicago, IL, USA). Data were presented as means ± SEM, and statistical significance was defined as P < 0.05.
Results
Characterization of circulating exosomes
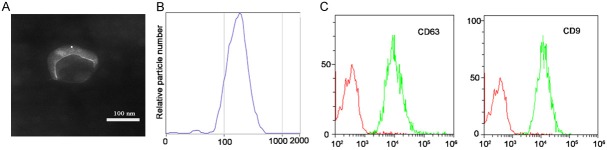
Exosomes were isolated and identified on the basis of morphology, size distribution, and membrane composition. TEM images showed the canonical cup-shaped morphology (Figure 1A). NTA showed a homogenous distribution of the exosomes with average size 178.7 ± 68.1 nm and a diameter peak at 175 nm, thus confirming their expected size profiles (Figure 1B). The established exosome-associated protein markers CD9 and CD63 were detected by flow cytometric analysis (Figure 1C).
Figure 1.
Isolation and validation of plasma-derived exosomes. A. The canonical cup-shaped morphology of exosomes was displayed by TEM. B. Size distribution profile by nanoparticle tracking analysis (NTA) demonstrated a homogeneous distribution of exosomes with peak diameter of 175 nm and an average size of 178.7 ± 68.1 nm. C. Identification of exosomal specific biomarkers. Both CD9 and CD63, the commonly acknowledged exosomal markers, were identified by flow cytometry.
Differential expression of circulating exosomal miRNAs in OLP
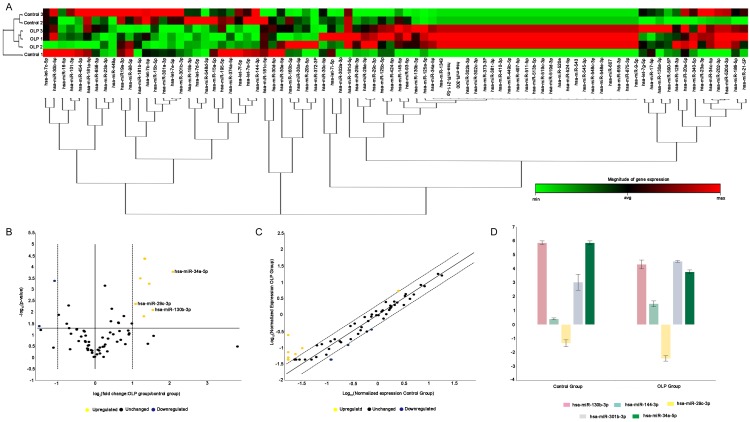
Total RNA extracted from the exosomes isolated from both OLP patients and healthy individuals were enriched in 18-25 nucleotide, indicating the presence of miRNAs (Figure 2). Microarray analysis revealed 83 differentially expressed miRNAs, 28 miRNAs down-regulated and 55 miRNAs upregulated, in the OLP exosomes relative to the healthy individuals (Figure 3A). Based on the criteria of fold change > 2 or < 0.5 and significance P < 0.05, 2 miRNAs were significantly downregulated and 3 miRNAs were significantly upregulated (Figure 3B-D). Specifically, significant increase was seen in the levels of miR-34a-5p (fold change = 4.24, P = 0.000165), miR-130b-3p (fold change = 2.92, P = 0.007933), and miR-29c-3p (fold change = 2.12, P = 0.0426), but miR-301b-3p (fold change = 0.35, P = 0.040683) and miR-144-3p (fold change = 0.47, P = 0.00414) were significantly decreased in OLP exosomes.
Figure 2.
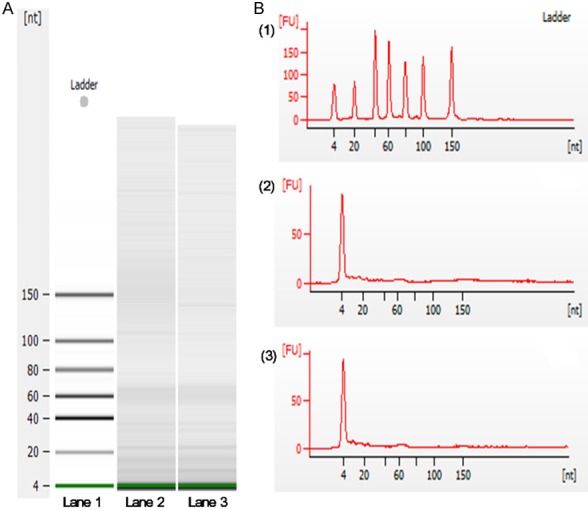
Plasma derived exosomal RNA analysis. A. Digital gel electropherograms of RNA from plasma exosome. Lane 1, RNA ladder shows the sizes of the nucleotides. Lane 2 and lane 3 display the size of the exosomal RNA from OLP patient and normal individual respectively. Results demonstrated that small RNAs were dominant in the exosomal RNAs. B. (1) Profile of RNA standard; (2) Total RNA from an OLP patient; (3) Total RNA from a normal individual. The data showed that the samples from OLP patients and normal individuals were enriched in nucleotide < 25 nt, indicating the presence of miRNAs.
Figure 3.
Differentially expressed exosomal miRNAs in OLP patients based on miRNA microarray data analysis. A. The columns and rows of hierarchical cluster indicated samples and specific miRNAs. miRNA cluster tree is shown at the bottom of the figure. Red to green color indicated the magnitude of gene expression change. B and C. Volcano plot and scatter plot depicting the miRNAs expression level. Fold change > 2 and P < 0.05 are shown in yellow; fold change < 0.5 and P < 0.05 are shown in blue. D. Multigroup plot displaying the top five significantly differentially expressed miRNAs: miR-34a-5p (fold change = 4.24, P = 0.000165), miR-130b-3p (fold change = 2.92, P = 0.007933), and miR-29c-3p (fold change = 2.12, P = 0.0426) were significantly increased, while miR-301b-3p (fold change = 0.35, P = 0.040683) and miR-144-3p (fold change = 0.47, P = 0.00414) were significantly decreased.
Validation of microarray results by RT-PCR
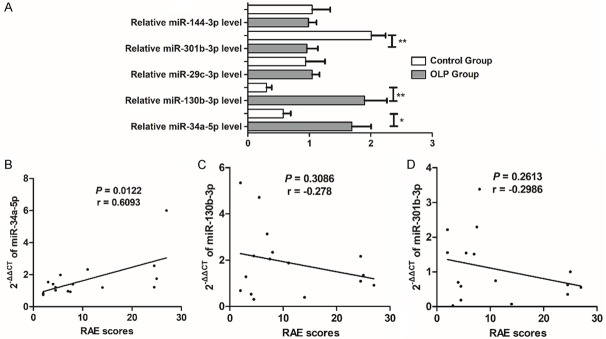
As shown in Figure 4A, exosomal miR-34a-5p and miR-130b-3p were significantly upregulated, whereas miR-301b-5p was significantly downregulated in OLP patients. However, no significant differences were seen in the expression of exosomal miR-29c-3p or miR-144-3p in the OLP group compared to the control group.
Figure 4.
Correlations between differentially expressed exosomal miRNAs and clinical characteristics of OLP. (A) Differential expression of exosomal miRNAs in OLP patients and normal controls. All the results were analyzed by the 2-ΔΔCt method, and spike-in control cel-miR-39 was used as internal reference. Exosomal miR-34a-5p and miR-130b-3p were upregulated, whereas miR-301b-5p was significantly downregulated in OLP. However, no significant difference was found in the expression of exosomal miR-29c-3p or miR-144-3p. (B-D) The correlation between the expression level of circulating exosomal miRNAs and RAE scores: miR-34a-5p (B), miR-130b-3p (C), and miR-301b-3p (D). Significantly positive correlation was found between the expression of exosomal miR-34a-5p and RAE scores, indicating that exosomal miR-34a-5p was correlated to the severity of OLP. *P < 0.05; **P < 0.01.
Differentially expressed exosomal miRNA-34a-5p is correlated to OLP severity
The RAE scoring system was used to evaluate the severity of OLP patients and significant positive correlation was seen between the expression of exosomal miR-34a-5p and RAE scores (Figure 4B), indicating that exosomal miR-34a-5p was associated with the severity of OLP patients. However, miR-130b-3p, and miR-301b-3p displayed no significant correlation with RAE scores (Figure 4C and 4D).
Target gene prediction of exosomal miRNA-34a-5p
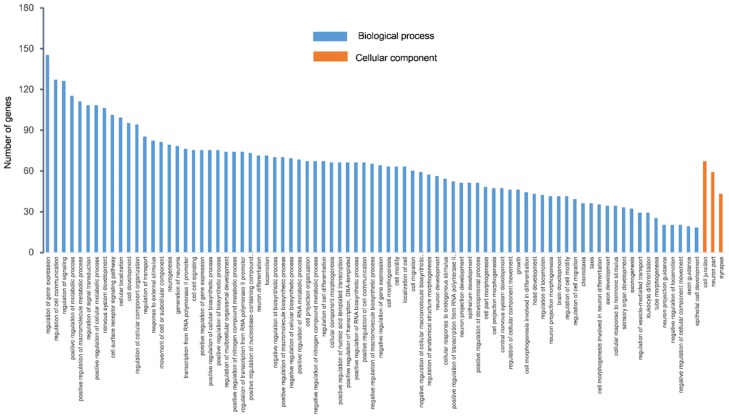
Eleven bioinformatics programs targeted 33063 genes in the miRecord database. In order to decrease false positive rate, 547 genes which were supported by more than 4 programs were selected for GO and KEGG analyses. GO analysis describes gene function with respect to three aspects including biological processes, cellular components, and molecular functions [26]. GO analysis of the miRNA targets indicated that functions of miR-34a-5p were mainly enriched in biological processes (Figure 5). The top 10 functions are shown in Table 3, and mainly involved a regulatory role in gene expression, cell communication, signaling, metabolic processes, macromolecule metabolic processes, signal transduction, cellular metabolic processes, nervous system development, cell surface receptor signaling pathway, and cellular location. KEGG analysis showed 27 enriched signaling pathways (Figure 6), and among the top 5 predicted pathways, PI3K/Akt signaling pathway is likely to participate in OLP progression.
Figure 5.
GO analysis of the target genes predicted by miR-34a-5p. GO category is displayed on the x-axis, and the number of genes is shown on the y-axis. The target genes were mainly enriched in the biological processes and included a variety of processes associated with gene expression, intercellular communication, and metabolic process. Cellular components were enriched in cell junction, neuron part, and synapse.
Table 3.
The top 10 biological processes of 547 target genes
| ID | Term | Count | P-Value | FDR |
|---|---|---|---|---|
| GO:0010468 | Regulation of gene expression | 145 | 2.33E-05 | 0.044690617 |
| GO:0010646 | Regulation of cell communication | 127 | 2.27E-10 | 4.35E-07 |
| GO:0023051 | Regulation of signaling | 126 | 1.35E-09 | 2.59E-06 |
| GO:0009893 | Positive regulation of metabolic process | 115 | 1.09E-06 | 0.0020927 |
| GO:0010604 | Positive regulation of macromolecule metabolic process | 111 | 4.02E-07 | 7.70E-04 |
| GO:0009966 | Regulation of signal transduction | 108 | 2.48E-07 | 4.75E-04 |
| GO:0031325 | Positive regulation of cellular metabolic process | 108 | 1.83E-06 | 0.003509574 |
| GO:0007399 | Nervous system development | 106 | 2.49E-12 | 4.77E-09 |
| GO:0007166 | Cell surface receptor signaling pathway | 101 | 5.73E-06 | 0.010980524 |
| GO:0051641 | Cellular location | 99 | 2.07E-06 | 0.00396585 |
Figure 6.
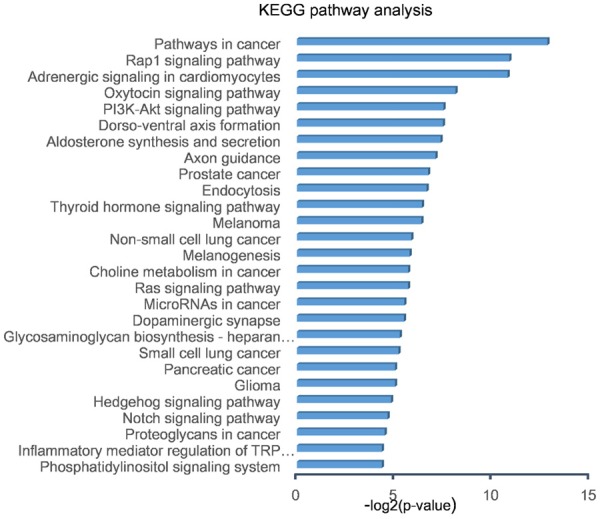
KEGG pathway analysis of target genes predicted by exosomal miR-34a-5p. Twenty-seven signaling pathways were selected as significantly enriched (P < 0.05). The -log2 (p-value) is displayed on the x-axis, and specific signalling pathways are shown on the y-axis. Among the top 5 predicted pathways, PI3K/Akt signaling pathway is likely to participate in OLP progression.
Discussion
Circulating exosomes are taken up by different cell types, then influence their biological functions and contribute to intercellular signaling transmission [27,28]. Emerging evidence have shown that exosomal miRNAs are potent stimulators of inflammatory and immune response, and thus potential biomarkers and therapeutic targets for autoimmune diseases [8,29,30]. Our study is the first to profile the differentially expressed circulating exosomal miRNAs in OLP patients, and correlate them with the clinical severity of OLP.
Aberrant expression of miR-34a-5p has been observed in multiple cell types, including T cells, dendritic cells, B cells and tumor cells, and regulates the apoptosis and survival of these cells [31-34]. In the present study, circulating exosomal miR-34a-5p was significantly increased in OLP patients. This might be partly due to the transmission of exosomal miR-34a-5p into T cells, which enhanced their activation and proliferation following T cell receptor (TCR) cross-linking in a diacylglycerol kinase ζ (DGKζ) suppression pathway which triggered a T cell response [31]. DGKζ negatively regulates the TCR by selectively interfering with the Ras-ERK pathway [35], and CD8+ miR-34a-deficient T cells have a lower proliferation rate compared to the wild-type T cells due to downregulation of DGKζ mRNA [35]. In addition, the increased expression of circulating exosomal miR-34a-5p was positively correlated with RAE scores, suggesting a potential diagnostic and prognostic use of this circulating exosomal miRNA in evaluating the severity of OLP.
Circulating exosomal miR-130b-3p was also significantly upregulated in OLP, but was not correlated to the RAE scores. Wanpeng Wang et al. demonstrated that serum miR-130b-3p was also up-regulated in patients with systemic lupus erythematous but did not correlate with the disease severity [36]. The peroxisome proliferator-activated receptor γ (PPARγ) and phosphatase and tensin homolog (PTEN) are the direct target genes of miR-130b-3p [37-40]. Overexpressed miR-130b-3p binds to the 3’-UTR sequence of PPARγ and PTEN, and represses their expression [37,39]. PPARγ is a typical anti-inflammatory factor which plays an important role in adaptive immunity and autoimmune disease [41]. One study showed that PPARγ agonist pioglitazone downregulated Th1 associated proinflammatory cytokine IFN-γ and upregulated Th2 associated anti-inflammatory cytokine IL-4 [42]. Seung Hoon Lee et al. found that increased PTEN ameliorated experimental autoimmune arthritis by decreasing the activation of T cells and modulating reciprocal differentiation of Th17 and Treg cells [43]. Therefore, we hypothesize that after the upregulated circulating exosomal miR-130b-3p is transferred into T cells, it modulates the expression of PPARγ and PTEN and eventually interferes with T cell differentiation.
Our previous study on OLP showed a positive correlation between high NF-κB and TNF-α expression in epithelial keratinocytes and disease severity, indicating a positive regulatory loop between NF-κB and TNF-α which may contribute to the inflammation seen in OLP [44]. In addition, upregulation of NF-κB may also protect the OLP keratinocytes from TNF-induced apoptosis [44]. TP63, a member of the P53 tumor suppressor gene family, regulates NF-κB transcription [45], and is a direct target gene of miR-301b-3p [46]. Consistent with these observations, NF-κB could be activated via the suppression of TP63 by miR-301b, indicating a positive regulation loop between miR-301b and NF-κB [46]. In the present study, circulating exosomal miR-301b-3p was significantly decreased in OLP, and we hypothesize that this prohibits NF-κB activity in OLP.
According to the results of GO analysis, target genes of miR-34a-5p participated in regulation of gene expression, cell communication, signaling, and metabolic process among others. KEGG pathway analysis indicated that the target genes of exosomal miR-34a-5p modulates OLP progression through the PI3K-Akt signaling pathway. Interestingly, Georgios Prodromidis et al. showed that a subset of OLP patients expressed cytoplasmic p-Akt, p-mTOR, and p-pS6 proteins, pointing to the activation of the Akt/mTOR/pS6 pathway in OLP [47]. We previously showed that the activated Akt/mTOR-autophagy signaling pathway likely plays a role in local T cell-mediated immune-regulation in OLP on the basis of the increased p-Akt, p-mTOR, ULK1 and LC3B [48]. Furthermore, Toll-like receptor 4-mediated upregulation of B7-H1 in keratinocytes is correlated with the PI3K/mTOR pathway [49]. Therefore, we hypothesize that the aberrantly expressed circulating exosomal miR-34a-5p might regulate OLP progression through the PI3K/Akt signaling pathway.
In conclusion, circulating exosomal miR-34a-5p was significantly upregulated in OLP and positively correlated with clinical RAE scores, indicating that miR-34a-5p might be a useful biomarker to evaluate the severity of OLP. Our future studies will mainly focus on the underlying mechanisms of exosomal miRNAs mediated OLP progression.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 81771080, No. 81371147) to Professor Zhou Gang.
Disclosure of conflict of interest
None.
References
- 1.Kurago ZB. Etiology and pathogenesis of oral lichen planus: an overview. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;122:72–80. doi: 10.1016/j.oooo.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Peng Q, Zhang J, Ye XJ, Zhou G. Tumor-like microenvironment in oral lichen planus: evidence of malignant transformation? Expert Rev Clin Immunol. 2017;13:635–643. doi: 10.1080/1744666X.2017.1295852. [DOI] [PubMed] [Google Scholar]
- 3.Lu R, Zhang J, Sun W, Du G, Zhou G. Inflammation-related cytokines in oral lichen planus: an overview. J Oral Pathol Med. 2015;44:1–14. doi: 10.1111/jop.12142. [DOI] [PubMed] [Google Scholar]
- 4.Xu AT, Lu JT, Ran ZH, Zheng Q. Exosome in intestinal mucosal immunity. J Gastroenterol Hepatol. 2016;31:1694–1699. doi: 10.1111/jgh.13413. [DOI] [PubMed] [Google Scholar]
- 5.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. 2015;40:72–81. doi: 10.1016/j.semcdb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Huang A, Dong J, Li S, Wang C, Ding H, Li H, Su X, Ge X, Sun L, Bai C, Shen X, Fang T, Li J, Shao N. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int J Biol Sci. 2015;11:961–969. doi: 10.7150/ijbs.11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan L, Wu H, Liu Y, Zhao M, Li D, Lu Q. Recent advances of exosomes in immune modulation and autoimmune diseases. Autoimmunity. 2016;49:357–365. doi: 10.1080/08916934.2016.1191477. [DOI] [PubMed] [Google Scholar]
- 9.Ibrahim A, Marban E. Exosomes: fundamental biology and roles in cardiovascular physiology. Annu Rev Physiol. 2016;78:67–83. doi: 10.1146/annurev-physiol-021115-104929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang WY, Liu W, Zhou YM, Shen XM, Wang YF, Tang GY. Altered microRNA expression profile with miR-27b down-regulation correlated with disease activity of oral lichen planus. Oral Dis. 2012;18:265–270. doi: 10.1111/j.1601-0825.2011.01869.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Wang X, Liu Y, Wei M, Chen L. A combinative analysis of gene expression profiles and microRNA expression profiles identifies critical genes and microRNAs in oral lichen planus. Arch Oral Biol. 2016;68:61–65. doi: 10.1016/j.archoralbio.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Das S, Halushka MK. Extracellular vesicle microRNA transfer in cardiovascular disease. Cardiovasc Pathol. 2015;24:199–206. doi: 10.1016/j.carpath.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siles-Lucas M, Morchon R, Simon F, Manzano-Roman R. Exosome-transported microRNAs of helminth origin: new tools for allergic and autoimmune diseases therapy? Parasite Immunol. 2015;37:208–214. doi: 10.1111/pim.12182. [DOI] [PubMed] [Google Scholar]
- 15.Sole C, Cortes-Hernandez J, Felip ML, Vidal M, Ordi-Ros J. miR-29c in urinary exosomes as predictor of early renal fibrosis in lupus nephritis. Nephrol Dial Transplant. 2015;30:1488–1496. doi: 10.1093/ndt/gfv128. [DOI] [PubMed] [Google Scholar]
- 16.Arao TC, Guimaraes AL, de Paula AM, Gomes CC, Gomez RS. Increased miRNA-146a and miRNA-155 expressions in oral lichen planus. Arch Dermatol Res. 2012;304:371–375. doi: 10.1007/s00403-011-1197-x. [DOI] [PubMed] [Google Scholar]
- 17.Yang JG, Sun YR, Chen GY, Liang XY, Zhang J, Zhou G. Different expression of microRNA-146a in peripheral blood CD4 T cells and lesions of oral lichen planus. Inflammation. 2016;39:860–866. doi: 10.1007/s10753-016-0316-4. [DOI] [PubMed] [Google Scholar]
- 18.Hu JY, Zhang J, Ma JZ, Liang XY, Chen GY, Lu R, Du GF, Zhou G. MicroRNA-155-IFN-gamma feedback loop in CD4(+)T cells of erosive type oral lichen planus. Sci Rep. 2015;5:16935–16944. doi: 10.1038/srep16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danielsson K, Wahlin YB, Gu X, Boldrup L, Nylander K. Altered expression of miR-21, miR-125b, and miR-203 indicates a role for these microRNAs in oral lichen planus. J Oral Pathol Med. 2012;41:90–95. doi: 10.1111/j.1600-0714.2011.01084.x. [DOI] [PubMed] [Google Scholar]
- 20.Tan YQ, Zhang J, Du GF, Lu R, Chen GY, Zhou G. Altered autophagy-associated genes expression in T cells of oral lichen planus correlated with clinical features. Mediators Inflamm. 2016;2016:4867368–4867377. doi: 10.1155/2016/4867368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Wei MH, Lu R, Du GF, Zhou G. Declined hTERT expression of peripheral blood CD4(+) T cells in oral lichen planus correlated with clinical parameter. J Oral Pathol Med. 2016;45:516–522. doi: 10.1111/jop.12399. [DOI] [PubMed] [Google Scholar]
- 22.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zangari J, Ilie M, Rouaud F, Signetti L, Ohanna M, Didier R, Romeo B, Goldoni D, Nottet N, Staedel C, Gal J, Mari B, Mograbi B, Hofman P, Brest P. Rapid decay of engulfed extracellular miRNA by XRN1 exonuclease promotes transient epithelial-mesenchymal transition. Nucleic Acids Res. 2017;45:4131–4141. doi: 10.1093/nar/gkw1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beltrami C, Besnier M, Shantikumar S, Shearn AI, Rajakaruna C, Laftah A, Sessa F, Spinetti G, Petretto E, Angelini GD, Emanueli C. Human pericardial fluid contains exosomes enriched with cardiovascular-expressed microRNAs and promotes therapeutic angiogenesis. Mol Ther. 2017;25:679–693. doi: 10.1016/j.ymthe.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akamatsu M, Makino N, Ikeda Y, Matsuda A, Ito M, Kakizaki Y, Saito Y, Ishizawa T, Kobayashi T, Furukawa T, Ueno Y. Specific MAPK-associated microRNAs in serum differentiate pancreatic cancer from autoimmune pancreatitis. PLoS One. 2016;11:e0158669–e0158679. doi: 10.1371/journal.pone.0158669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vicencio JM, Yellon DM, Sivaraman V, Das D, Boi-Doku C, Arjun S, Zheng Y, Riquelme JA, Kearney J, Sharma V, Multhoff G, Hall AR, Davidson SM. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J Am Coll Cardiol. 2015;65:1525–1536. doi: 10.1016/j.jacc.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Ludwig S, Floros T, Theodoraki MN, Hong CS, Jackson EK, Lang S, Whiteside TL. Suppression of lymphocyte functions by plasma exosomes correlates with disease activity in patients with head and neck cancer. Clin Cancer Res. 2017;23:4843–4854. doi: 10.1158/1078-0432.CCR-16-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyriakidis NC, Kapsogeorgou EK, Tzioufas AG. A comprehensive review of autoantibodies in primary Sjogren’s syndrome: clinical phenotypes and regulatory mechanisms. J Autoimmun. 2014;51:67–74. doi: 10.1016/j.jaut.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Lv LL, Cao Y, Liu D, Xu M, Liu H, Tang RN, Ma KL, Liu BC. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci. 2013;9:1021–1031. doi: 10.7150/ijbs.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin J, Xie D, Zhong XP. MicroRNA-34a enhances T cell activation by targeting diacylglycerol kinase zeta. PLoS One. 2013;8:e77983–e77988. doi: 10.1371/journal.pone.0077983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurowska-Stolarska M, Alivernini S, Melchor EG, Elmesmari A, Tolusso B, Tange C, Petricca L, Gilchrist DS, Di Sante G, Keijzer C, Stewart L, Di Mario C, Morrison V, Brewer JM, Porter D, Milling S, Baxter RD, McCarey D, Gremese E, Lemke G, Ferraccioli G, McSharry C, McInnes IB. MicroRNA-34a dependent regulation of AXL controls the activation of dendritic cells in inflammatory arthritis. Nat Commun. 2017;8:15877–15889. doi: 10.1038/ncomms15877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen F, Hu SJ. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: a review. J Biochem Mol Toxicol. 2012;26:79–86. doi: 10.1002/jbt.20412. [DOI] [PubMed] [Google Scholar]
- 34.He X, Yang A, McDonald DG, Riemer EC, Vanek KN, Schulte BA, GY W. MiR-34a modulates ionizing radiation-induced senescence in lung cancer cells. Oncotarget. 2017;8:69797–69807. doi: 10.18632/oncotarget.19267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun YX, Li H, Feng Q, Li X, Yu YY, Zhou LW, Gao Y, Li GS, Ren J, Ma CH, Gao CJ, Peng J. Dysregulated miR34a/diacylglycerol kinase ζ interaction enhances T-cell activation in acquired aplastic anemia. Oncotarget. 2017;8:6142–6154. doi: 10.18632/oncotarget.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Mou S, Wang L, Zhang M, Shao X, Fang W, Lu R, Qi C, Fan Z, Cao Q, Wang Q, Fang Y, Ni Z. Up-regulation of serum miR-130b-3p level is associated with renal damage in early lupus nephritis. Sci Rep. 2015;5:12644–12656. doi: 10.1038/srep12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu JJ, Zhang JH, Chen HJ, Wang SS. MicroRNA-130b promotes cell proliferation and invasion by inhibiting peroxisome proliferatoractivated receptor-gamma in human glioma cells. Int J Mol Med. 2016;37:1587–1593. doi: 10.3892/ijmm.2016.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertero T, Cottrill K, Krauszman A, Lu Y, Annis S, Hale A, Bhat B, Waxman AB, Chau BN, Kuebler WM, Chan SY. The microRNA-130/301 family controls vasoconstriction in pulmonary hypertension. J Biol Chem. 2015;290:2069–2085. doi: 10.1074/jbc.M114.617845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang RM, Xu JF, Fang F, Yang H, Yang LY. MicroRNA-130b promotes proliferation and EMT-induced metastasis via PTEN/p-AKT/HIF-1alpha signaling. Tumour Biol. 2016;37:10609–10619. doi: 10.1007/s13277-016-4919-z. [DOI] [PubMed] [Google Scholar]
- 40.Yu T, Cao R, Li S, Fu M, Ren L, Chen W, Zhu H, Zhan Q, Shi R. MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer. 2015;15:29–37. doi: 10.1186/s12885-015-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park HJ, Park HS, Lee JU, Bothwell AL, Choi JM. Sex-based selectivity of PPARgamma regulation in Th1, Th2, and Th17 differentiation. Int J Mol Sci. 2016;17:1347–1357. doi: 10.3390/ijms17081347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasegawa H, Takano H, Zou Y, Qin Y, Hizukuri K, Odaka K, Toyozaki T, Komuro I. Pioglitazone, a peroxisome proliferator-activated receptor gamma activator, ameliorates experimental autoimmune myocarditis by modulating Th1/Th2 balance. J Mol Cell Cardiol. 2005;38:257–265. doi: 10.1016/j.yjmcc.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Lee SH, Park JS, Byun JK, Jhun J, Jung K, Seo HB, Moon YM, Kim HY, Park SH, Cho ML. PTEN ameliorates autoimmune arthritis through down-regulating STAT3 activation with reciprocal balance of Th17 and Tregs. Sci Rep. 2016;6:34617–34627. doi: 10.1038/srep34617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou G, Xia K, Du GF, Chen XM, Xu XY, Lu R, Zhou HM. Activation of nuclear factor-kappa B correlates with tumor necrosis factor-alpha in oral lichen planus: a clinicopathologic study in atrophic-erosive and reticular form. J Oral Pathol Med. 2009;38:559–564. doi: 10.1111/j.1600-0714.2009.00779.x. [DOI] [PubMed] [Google Scholar]
- 45.Sen T, Sen N, Huang Y, Sinha D, Luo ZG, Ratovitski EA, Sidransky D. Tumor protein p63/nuclear factor kappaB feedback loop in regulation of cell death. J Biol Chem. 2011;286:43204–43213. doi: 10.1074/jbc.M111.257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Funamizu N, Lacy CR, Parpart ST, Takai A, Hiyoshi Y, Yanaga K. MicroRNA-301b promotes cell invasiveness through targeting TP63 in pancreatic carcinoma cells. Int J Oncol. 2014;44:725–734. doi: 10.3892/ijo.2014.2243. [DOI] [PubMed] [Google Scholar]
- 47.Prodromidis G, Nikitakis NG, Sklavounou A. Immunohistochemical analysis of the activation status of the Akt/mTOR/pS6 signaling pathway in oral lichen planus. Int J Dent. 2013;2013:743456. doi: 10.1155/2013/743456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang N, Zhang J, Tan YQ, Du GF, Lu R, Zhou G. Activated Akt/mTOR-autophagy in local T cells of oral lichen planus. Int Immunopharmacol. 2017;48:84–90. doi: 10.1016/j.intimp.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Tan YQ, Wei MH, Ye XJ, Chen GY, Lu R, Du GF, Zhou G. TLR4-induced B7-H1 on keratinocytes negatively regulates CD4+ T cells and CD8+ T cells responses in oral lichen planus. Exp Dermatol. 2017;26:409–415. doi: 10.1111/exd.13244. [DOI] [PubMed] [Google Scholar]