Abstract
Background: Glioma is the most common malignant tumor in the adult human brain and has one of the lowest patient survival rates. MicroRNAs (miRNAs) play important roles in the development of cancers, including glioma, and potentially have valuable therapeutic applications in glioma; however, their specific functions and mechanisms of action have yet to be fully defined. Here, we report that miR-129-5p directly targets DNA (cytosine-5)-methyltransferase 3A (DNMT3A) and functions as a tumor-suppressor in glioma. Method: We analyzed the expression profiles of miR-129-5p and DNMT3A in glioma-related databases. Quantitative reverse transcription-PCR was applied to detect the level of miR-129-5p in glioma specimens and cell lines. Western blotting was applied to detect the level of DNMT3A. We examined the effect of miR-129-5p on the cell cycle and proliferation of glioma cells using CCK-8 and EDU assays and flow cytometry. TargetScan software predicted DNMT3A to be a target of miR-129-5p, which we confirmed by means of luciferase reporter assays and rescue experiments. Result: miR-129-5p was expressed at low levels in glioma and negatively correlated with glioma grade. Over-expression of miR-129-5p in U87and LN229 cells inhibited proliferation and blocked the cell cycle in G1 Phase. DNMT3A is a direct target of miR-129-5p, and miR-129-5p affects glioma cell proliferation by targeting DNMT3A. Conclusion: Taken together, our results demonstrate that miR-129-5p plays a significant role in glioma suppression through inhibition of DNMT3A, which may provide a novel therapeutic strategy for treatment of glioma and other DNMT3A-driven cancers.
Keywords: miR-129-5p, DNMT3A, proliferation, glioma
Introduction
Glioma are neuro-epithelial tumors or neuro-ectodermal tumors because they occur in neuro-ectodermal regions. Glioma cases are divided into four categories based on histological classification: two low-grade astrocytomas (WHO grade I-II), anaplastic astrocytomas (WHO grade III), and glioblastoma (GBM, WHO grade IV [1]. The incidence rate for all glioma types ranges from 4.67 to 5.73 per 100,000 people [2]. Patients with GBM have a low survival rate, and their median survival is only about 15 months [3-6]. Despite large improvements in microsurgical procedures and the increased availability of advanced chemotherapy, the improvement of patient survival has been limited. This is attributed to the highly malignant and invasive nature of glioma and because the molecular mechanisms underlying the development of glioma are not clear. Although a number of molecular markers have been discovered, their prediction power is ambiguous [7]. Therefore, a full understanding of the molecular mechanisms underlying the occurrence, development and evolution of glioma is urgently needed to enable development of novel and effective therapeutic treatments.
MiRNAs are small non-coding RNAs 19-24 nucleotides in length that were discovered by Victor Ambros and colleagues. Imperfect complementary sequence pairing between the miRNA seed region and the 3’-untranslated region (UTR) of target mRNAs, orchestrates the negative regulation of target genes by either mRNA degradation or translational repression and, thus, directly or indirectly affects almost all cellular pathways [8,9]. As endogenous regulators of gene expression, miRNAs play vital roles in diverse physiological processes, including proliferation [10,11], invasion [12], apoptosis [13], cell identity [14] and stem cell maintenance [15]. There is increasing evidence to indicate that specific miRNAs can be downregulated or upregulated in different types of tumor [16-18]. Understanding miRNA biogenesis mechanisms in normal physiology and in pathological states is important to understand the role that miRNAs play in carcinogenesis, a situation that will result in the development and improvement of tools for diagnosis, risk evaluation and follow up of cancer patients.
MiR-129 is a miRNA family containing three members, miR-129-5p, miR-129-2-3p and miR-129-3p. Among them, miR-129-5p has been reported as a tumor suppressor in many types of carcinoma, including breast cancer [19], hepatocellular malignancy [20], and lung adenocarcinoma [21]. Some researchers have also explored the mechanisms by which miR-129 family members affect glioma cell processes. For example, Zeng et al reported that miR-129-5p targets Wnt5a to affect the behavior of glioma cells [22]. Despite this and subsequent studies, the specific mechanism of miR-129-5p regulation in glioma cells is still unclear.
Epigenetic modifications including DNA methylation play a significant role in regulating gene transcription [23,24]. DNA methylation participates in many physiological processes, including cell differentiation [25], oncogenic transformation [26], and long-term memory [27]. It is a covalent modification of DNA that, in mammals, is tightly regulated by a group of three DNA methyltransferases (DNMT), DNMT1, DNMT3A, and DNMT3B [28,29]. DNMT1 primarily maintains existing DNA methylation patterns, whereas DNMT3A and DNMT3B are de novo methyltransferases and are essential for introducing methyl groups onto CG sites [30-32]. Aberrant DNA methylation and overexpressed DNMTs are observed in many cancer types. For example, DNMT1 and DNMT3b play a specific role in tumorigenesis by silencing tumor suppressors [33-36]. Meanwhile, several inactivating mutations of DNMT3A in myeloid malignancies [37-39] and loss of DNMT3A activity at advanced tumor stages [40] were recently identified. However, the expression status and the role of DNMT3A in the development of glioma have not been fully elucidated.
In the present study, we investigated miR-129-5p expression in both clinical specimens and commonly used glioma databases. We show that miR-129-5p was expressed at low levels in glioma, and that its expression was negatively correlated with levels of the DNMT3A protein. Upregulation of miR-129-5p inhibited the proliferation of glioma cells and resulted in decreased expression of DNMT3A. Moreover, we demonstrated that DNMT3A is a direct target of miR-129-5p. Our findings provide new insights into the molecular mechanism of glioma development and will help in the development of unique miRNA-based therapies for GBM management.
Materials and methods
Ethics statement
The study has been conducted in accordance with the ethical standards and according to the Declaration of Helsinki and national and international guidelines. Informed consent was obtained from all patients involved in this study, and the study protocol was approved by the Clinical Research Ethics Committee of Haimen People’s Hospital.
Public datasets
Microarray miRNA expression data was downloaded from the Chinese Glioma Genome Atlas (CGGA) data portal (http://www.cgga.org.cn.portal.phpg). Whole genome mRNA expression microarray data and clinical information of 158 glioma samples were obtained from CGGA database (http://www.cgga.org.cn). The Cancer Genome Atlas database (TCGA) was download from http://tcga-data.nci.nih.gov/, Repository of Molecular Brain Neoplasia Data (REMBRANDT, was download from http://caintegrator.nci.nih.gov/rembrandt/ and GSE4290 data was download from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4290.
Cell culture and reagents
The human GBM cell line U118, LN229, H4, A172, U87 and U251 (American Type Culture Collection, VA, USA) were identified by the American Type Culture Collection using the short tandem repeat genotyping method and cultured according to the manufacturer’s recommendations.Human astrocyte cells NHAs were purchased from Lonza (Basel, Switzerland) and cultured in the astrocyte growth medium supplemented with rhEGF, insulin, ascorbic acid, GA-1000, L-glutamine and 5% fetal bovine serum. All the cells were incubated at 37°C in a humidified atmosphere with 5% CO2. Antibodies against DNMT3A (ab2850) and Actin (ab8226) were obtained from Abcam (Cambridge, UK). Antibodies against Cyclin A2 (#4656) and CDK2 (#2546) were purchased from Cell Signaling Technology (Massachusetts, USA).
Tissue samples
Nine normal brain tissues (NBT) consecutively recruited from patients undergoing internal decompression surgery following severe traumatic brain injury between December 2015 and December 2016 from the Department of Neurosurgery of Haimen People’s Hospital. Seventeen glioblastoma multiform tissues were obtained between December 2015 and December 2016 from the Department of Neurosurgery, The Affiliated Hospital of Nantong University. Glioma specimens were verified and classified according to the WHO standard classification of central nervous system tumors.
Western blot analysis
Western blotting analysis was performed according to our previous study [41]. Briefly, Equal amount of protein lysates were subjected to 12% SDS-PAGE gel and then transferred to PVDF membrane (Millipore, MA, USA) and probed with primary antibodies (DNMT3A, CDK2, Cyclin A2 and Actin) at 4°C overnight and secondary antibodies at room temperature for 2 h. Bound antibodies were detected by the ECL Plus western blotting substrate (Thermo Fisher, MA, USA) and the results were recorded by Biorad ChemiDoc MP Gel Imaging System, The band density of specific proteins was quantified after normalization with the density of Actin.
Oligonucleotides, plasmid construction, and transfection
Hsa-miR-129-5p mimic and hsa-miR-ctrl were chemically synthesized and authenticated by Ribobio (Guangzhou, China). Small interfering RNAs (siRNAs) targeting DNMT3A (sc-37757) was purchased from Santa Cruz Biotechnology (TX, USA). The DNMT3A-overexpression plasmids were generated by cloning the Corresponding cDNA into the expression vector pENTER at the AsisI and M1uI restriction sites. The plasmid was sequenced verified by Genepharma (Shanghai, Chinna). All oligonucleotides and plasmids were transfected into cells using Lipofectamine 3000 Transfection Reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions.
Lentiviral packaging and establishment of stably transduced cell lines
A lentiviral packaging kit was purchased from Genechem (Shanghai, China). A lentivirus carrying hsa-miR-129-5p or hsa-miR-negative control (miR-ctrl) was packaged in the human embryonic kidney cell line, 293T, and the virions were collected according to the manufacturer’s instructions. Stable cell lines were established by infecting U87-MG cells and LN229 cells with lentiviruses, followed by puromycin selection.
RNA isolation and quantitative real-time PCR (qPCR)
Total RNA from glioma cells or tissues was isolated using TRIzol reagent (Invitrogen, CA, USA) following the product manual. RNA (1 μg) was reverse-transcribed using a Bulge-loopTM miRNA qRT-PCR Starter Kit (RIBOBIO, Guangzhou, China) and converted to cDNA. PCR was performed using 2X SYBR Green Mix (RIBOBIO, Guangzhou, China) in a Bio-Rad instrument; U6 was used as controls to compare relative RNA expression between samples. Bulge-loop primers were purchased from Ribobio. Data were analyzed using the 2-ΔΔCt method.
Cell counting kit-8 assay
After transfection for one day, U87 or LN229 cells were placed into a 96 well culture plate with 1×103 cells/well and incubated for 1, 2, 3, or 4 days. Cell proliferation was measured using a cell counting kit-8 (CCK-8) (Dojindo, Japan) in accordance with the protocol. After adding appropriate amount of reagent into wells, the cell plate was incubated at 37°C for two hours. Absorbance with 450 nm wavelength was detected after incubation. Cell growth curve was plotted based on the measured experimental values.
EdU incorporation assay
After transfection for one day, U87 or LN229 cells were placed into a 24 well culture plate with 5×104 cells/well and incubated for 24 days. Cell proliferation was then measured by 5-ethynyl-20-deoxyuridine (EdU) assay using an EdU assay kit (Life Technologies, MA, USA) according to the manufacturer’s protocol. Briefly, cells were incubated with 10 μM EDU for 3 h at 37°C, and then the cells were fixed with 4% paraformaldehyde. After permeabilization with 0.5% Triton X-100 20 min, the cells were then stained with the Alexa-Fluor 594 reaction cocktail for for 30 min. Subsequently, the cells nuclei were stained with Hoechst33342 for 10 min. Finally, samples were visualized under a fluorescent microscope (Olympus).
Flow cytometric analysis of the cell cycle
After transfected for 48 h, transfected cells were washed by PBS twice and then fixed with 75% ethanol at -20°C overnight. After washing with PBS again, cells were incubated with PBS containing 50 μg/ml propidium iodide (Sigma-Aldrich, MO, USA) in the presence of 100 μg/ml RNaseA (Sigma-Aldrich, MO, USA) for 20 min at room temperature. The FACScan flow cytometer (BD Bioscience, NJ, USA) was adopted to analyze the cell cycle. Data was expressed by the cell percentage in each phase of cell cycle.
Dual luciferase reporter assay
A dual-luciferase reporter vector was used to generate the luciferase constructs. The target genes of miR-129-5p were selected based on target scan algorithms (http://www.targetscan.org/). The putative binding sites of miR-129-5p and its homologous mutation sites in the 3’-UTR region of DNMT3A mRNA were amplified and cloned into the XbaI site pGL3 control luciferase reporter plasmid (Invitrogen, CA, USA). U87 and LN229 cells were seeded in a 96-well plate and con-transfected with WT or mutated 3’-UTR luciferase reporter and miR-129-5p mimics (RiboBio, Guangzhou, China). The pRL vector constitutively expressing Renilla luciferase was used to normalize for transfection efficiency. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, USA) after transfection at 48 h.
Statistical analysis
All experiments were performed at least three times, and all values were presented as the mean ± SD. Student t test and ANOVA test were used to detect the differences in the results between groups. KEGG pathway and GO analysis were performed via DAVID (http://david.abcc.ncifcrf.gov/). Spearman’s rank test was carried out to detect the correlation between the expression profile of miR-129-5p and DNMT3A gene. Heat map microarray analysis were implemented using Multiple Array Viewer 4.9 software (MEV). P<0.05 indicates a significant difference.
Results
MiR-129-5p is down-regulated in glioma tissues and cell lines
To explore the role of miRNAs in the development and progression of glioma, we analyzed the expression patterns of miRNAs in the CGGA database. We found that miR-129-5p levels in high-grade glioma (HGG) were significantly decreased compared with low-grade glioma (LGG) (Figure 1A). To investigate the expression level of miR-129-5p in clinical glioma tissues, we collected samples including nine non-tumor brain tissues (NBTs) and seventeen GBM tissues. RT-PCR showed that miR-129-5p was downregulated in glioma tissues (Figure 1B). To verify this result, we extracted RNA from the glioma cell lines, U118, LN229, H4, A172, U118 and U251 and from normal human astrocyte (NHAs). The expression levels of miR-129-5p were significantly lower in the glioma cell lines than that in NHAs, especially in U87 and LN229 cells (Figure 1C). Therefore, we concluded that: miR-129-5p was expressed at low levels in glioma and that the expression level of miR-129-5p was negatively correlated with the grade of glioma.
Figure 1.
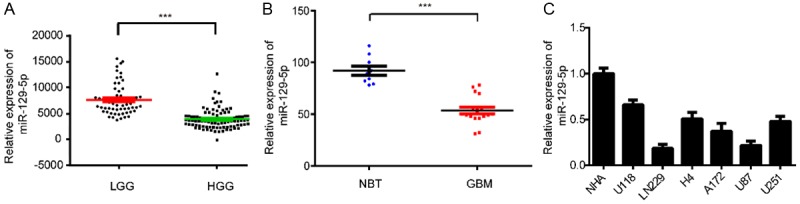
MiR-129-5p expression correlates negatively with malignant degrees of glioma. A. CGGA database showing reduced miR-129-5p expression in high-grade glioma tissues compared with that in low-grade glioma tissues. (***P<0.001). B. The expression of miR-129-5p in nine non-tumor brain tissues (NBTs) and seventeen glioblastoma (GBM) tissues was measured by real-time PCR, miR-129-5p levels in NBTs were indeed higher than in GBM specimens. (***P<0.001). C. The expression of miR-129-5p in normal human astrocytes (NHAs) and six glioma cell lines: U118, LN229, H4, A172, U87 and U251.
Overexpression of miR-129-5p inhibits the proliferation of glioma cells
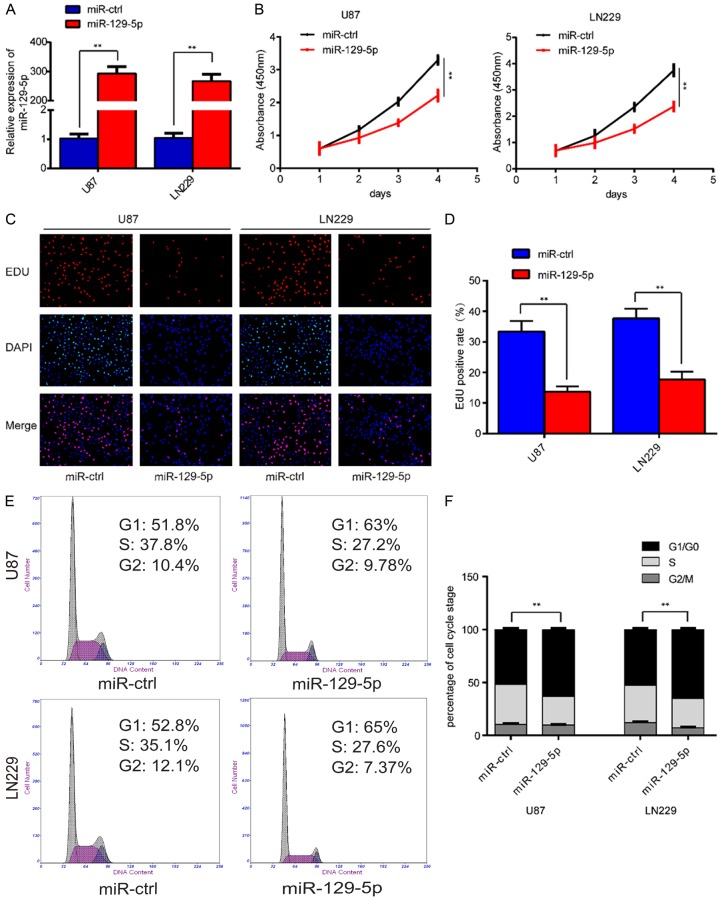
Having determined that miR-129-5p was down-regulated in glioma cells and negatively correlated with the pathological grade of glioma. We then investigated the role of miR-129-5p in glioma development. We selected the two glioma cell lines with the lowest levels of miR-129-5p expression, U87 and LN229. In these two cell lines we overexpressed miR-129-5p. We verified the overexpression by RT-PCR (Figure 2A). We then planted these overexpressing cells into 96-well plates at 1000 cells per well. At the time point of 0, 24, 48, 72, and 96 h we detected cell proliferation by CCK-8 assay and found that miR-129-5p overexpression inhibited the proliferation of U87 and LN229 cells (Figure 2B). To verify this phenomenon, we plated u87 and LN229 cells transfected with miR-129-5p lentivirus into 24-well plates. After 24 hours of culture, we detected cell proliferation using a EDU kit and the results were consistent with those of the CCK-8 assay (Figure 2C, 2D). It is well-known that the cell cycle is closely related to cell proliferation. What effect does miR-129-5p have on the cell cycle? Through flow cytometry we found that overexpression of miR-129-5p arrested the cell cycle in the G1 phase (Figure 2E, 2F), which may be one of the reasons that miR-129-5p causes a decrease in cell proliferation.
Figure 2.
Overexpression of miR-129-5p inhibits glioma cell proliferation in vitro. A. The relative expression level of miR-129-5p in U87-MG and LN229 cells was analyzed by qRT-PCR after transfection with lentivirus carrying has-miR-129-5p. (**P<0.01). B. Overexpression of miR-129-5p inhibited cell proliferation detected by CCK8 assays. Data are presented as the means of triplicate experiments. (**P<0.01). C, D. EdU assays show that miR-129-5p up-regulation inhibited cell proliferation in both U87 and LN229 cells. Representative images were shown (original magnification, 200×). (**P<0.01). E, F. The cell cycle phase of U87-MG and LN229 cells transfected with miR-129-5p or negative control (miR-ctrl) lentivirus analyzed by flow cytometry. (**P<0.01).
DNMT3A is a direct target of miR-5129-5p
MiRNAs repress transcription or induce mRNA degradation by binding to complementary sequences in the 3’-UTRs of their target mRNAs. To investigate the molecular mechanism underlying the miR-129-5p-induced inhibitory effect on glioma cell proliferation and the cell cycle, we used the bioinformatics analytical tool TargetScan to identify potential targets of miR-129-5p. Among the putative targets of miR-129-5p, DNMT3A captured our interest (Figure 3A). To determine the correlation between levels of miR-129-5p and DNMT3A le, we measured DNMT3A protein levels in glioblastoma specimens and non-cancerous brain tissues. The average levels of DNMT3A were significantly higher in tumor tissues than in the non-cancerous brain tissues (Figure 3B). We then determined the correlation between DNMT3A and miR-129-5p levels in the same glioma tissues. As shown in Figure 3C, DNMT3A levels in glioma samples were inversely correlated with miR-129-5p expression levels (Spearman’s correlation r=-0.71). We then performed luciferase reporter assays to determine whether miR-129-5p could directly bind to the 3’-UTR of DNMT3A. The possible binding site and related sequence of miR-129-5p predicted by Targetscan in the 3’-UTR of DNMT3A has been shown in Figure 3D. The wild-type and mutant reporter plasmids were designed and synthesized at Shanghai Genechem, which were then co-transfected with miR-129-5p mimic into U87 and LN229 cell lines. Luciferase activity in U87 cells was markedly decreased after transfection with wild-type vector and miR-129-5p mimics (Figure 3E). Luciferase activity was also significantly decreased when the poorly conserved binding site 1 (7mer) was mutated, whereas mutation of conserved binding site 2 (7mer) nearly rescued the decrease. Similar results were obtained in LN229 cells (Figure 3E). These data suggest that miR-129-5p directly regulates DNMT3A expression through its binding to site 2 (nt6393-6399) in the 3’-UTR of DNMT3A. These data suggested that miR-129-5p directly regulates DNMT3A expression through its binding to the 3’-UTR of DNMT3A.
Figure 3.
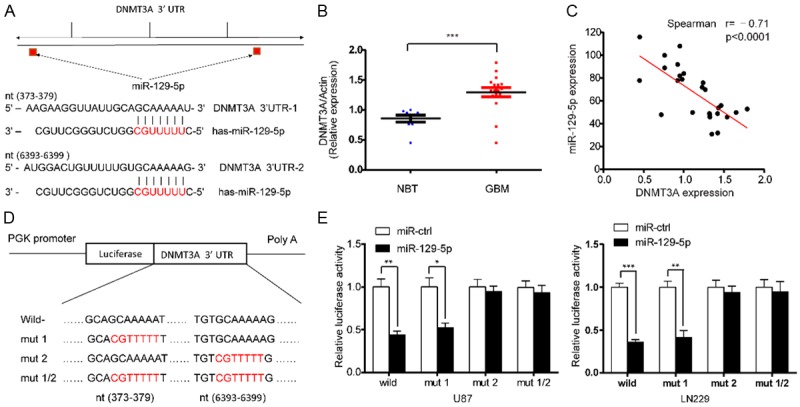
MiR-129-5p directly targets DNMT3A and negatively regulates cell cycle-related proteins. A. Predicted miR-129-5p binding sites in the 3’-UTR of the DNMT3A gene. B. The expression levels of DNMT3A in non-tumor brain tissues (NBTs) and glioma specimens were determined by western blotting; the fold changes were normalized to β-Actin. The NBTs (n=9) were collected from brain trauma surgery. The glioblastoma (GBM) tissues (n=17) were collected from Surgical specimens of patients with primary glioblastoma. Data represent the means ± SD from three independent experiments. (***P<0.001). C. Pearson’s correlation analysis of the relationship between the relative expression levels of miR-129-5p and the relative protein levels of DNMT3A in Clinical tissue samples. D. Wild-type and mutant DNMT3A 3’-UTR reporter constructs. E. Luciferase reporter assays were performed in U87 and LN229 cells co-transfected with the indicated wild-type or mutant 3’-UTR constructs and the miR-129-5p mimic. The data shown are representative of three independent experiments. Data shown are mean ± SD of three independent experiments. (*P<0.05, **P<0.01, ***P<0.001).
DNMT3A is highly expressed in gliomas and is associated with cell proliferation
We have demonstrated that miR-129-5p can target DNMT3A; therefore, what is the expression level and function of DNMT3A in glioma? First we analyzed the expression profile of DNMT3A in the glioma database: CGGA, TCGA, GSE4290 and Rembrandt (Figure 4A). We found a significant increase in the expression of DNMT3A in high-grade gliomas. We then performed western blot analysis on extracted protein from the clinical samples, and we found that levels of DNMT3A were significantly increased in GBM compared with NBT (Figure 4B). In summary, we have found that DNMT3A is highly expressed in gliomas and is positively correlated with grades. We then asked, what effect does DNMT3A have on the properties of gliomas? Pearson correlation analysis was implemented using MEV software to identify target genes that were positively associated with DNMT3A expression in CGGA, Rembrandt and GSE4290 databases (r>0.4). In total 375 upregulated genes were identified (Figure 4F). To clarify the associations between these genes, DAVID Web tool (https://david.ncifcrf.gov/tools.jsp) were used for Gene Oncology enrichment analysis and KEGG pathway analysis. The up-regulated genes were mainly enriched in the terms, positive regulation of cell cycle and cell proliferation (Figure 4G).
Figure 4.
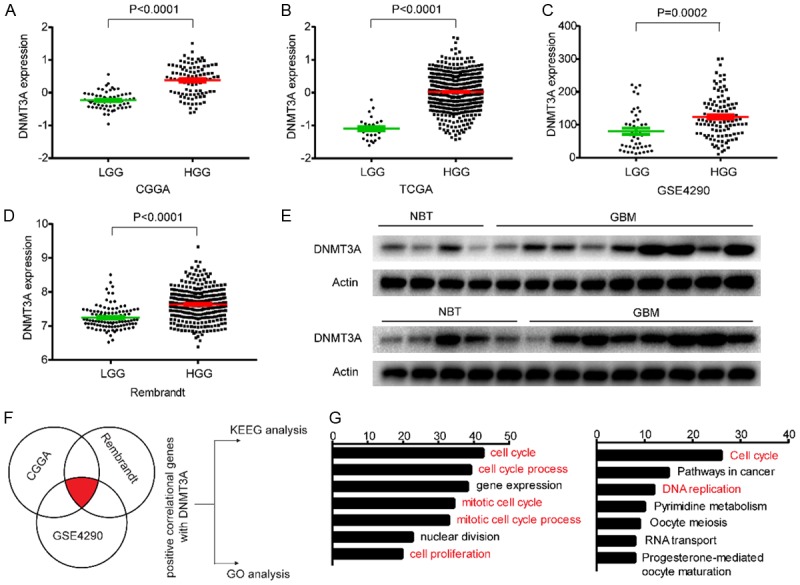
DNMT3A is highly expressed in gliomas and is associated with cell proliferation. A-D. Levels of DNMT3A were analyzed in low-grade glioma (LGG) and high-grade glioma (HGG) in CGGA, TCGA, GSE4290 and Rembrandt databases. E. Western blotting was applied to detect the level of DNMT3A in clinical tissues, β-Actin was used as the loading control. F, G. Gene positively-associated with DNMT3A expression in the CGGA, Rembrandt, and GSE4290 glioblastoma samples were subjected to gene ontology (GO) analysis and KEEG pathway analysis. Enrichment for biological process in the GO database and KEGG pathways analysis are shown. The orders of the different biological processes is based on their enriched number.
Down-regulation of DNMT3A inhibits glioma cell proliferation in vitro
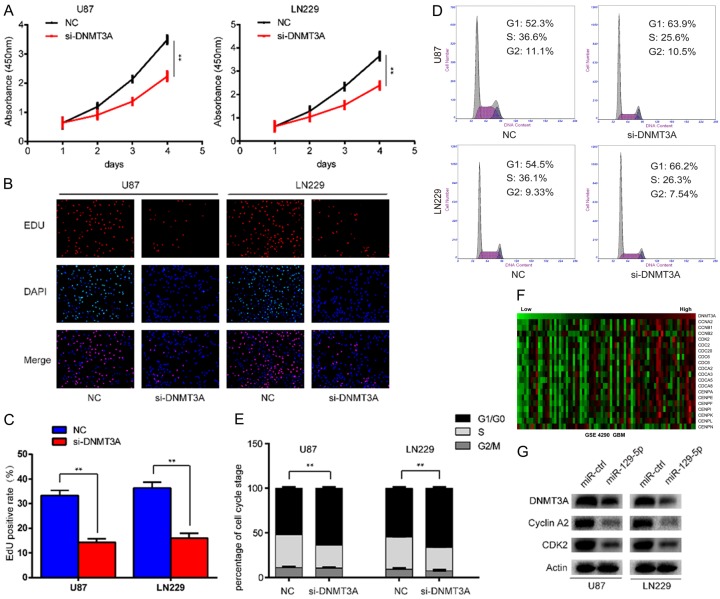
To confirm that DNMT3A affects cell proliferation and the cell cycle in gliomas, we performed in vitro proliferation-related experiments. We planted U87 or LN229 glioma cells into 96-well plates at 1000 cells per well. The following day we transfected the cells with siRNA for DNMT3A. CCK-8 was then used to continuously monitor changes in cell proliferation at 0, 24, 48, 72, and 96 h after transfection. We found that proliferation was significantly reduced after DNMT3A down-regulation (Figure 5A). We validated these findings through EDU experiments (Figure 5B, 5C). We then asked, what effect does DNMT3A have on the cell cycle of gliomas? We detected the distribution of cells in the different stage of the cell cycle after DNMT3A down-regulation by flow cytometry, and found that cells were arrested in the G1 phase (Figure 5D, 5E). Cell cycle progression is regulated by many cyclins and Cyclin-dependent kinases (CDKS). What then is the relationship between DNMT3A and these cycle-related proteins and kinases in gliomas? Through database analysis, we found that 19 cycle-related proteins or kinases were positively correlated with the expression of DNMT3A, as illustrated in a heat map generated using MEV software (Figure 5F). The proteins mainly associated with G1 arrest were cyclinA2 and CDK2. Western blotting showed that the protein levels of DNMT3A, cyclinA2 and CDK2 were decreased after overexpression of miR-129-5p (Figure 5G). In summary, down-regulation of DNMT3A can inhibit the proliferation of glioma cells, and can also affect the cyclinA2 and CDK2, which arrest the cell cycle in G1 phase.
Figure 5.
Down-regulation of DNMT3A inhibits glioma cell proliferation in vitro. A. U87-MG and LN229 cells were transfected with DNMT3A siRNA and cultured for in 96 h. Cell proliferation was determined using the CCK-8 assay. Data are presented as the means of triplicate experiments. (**P<0.01). B, C. Proliferating cells were examined using the EDU assay. Representative images are shown (original magnification, 200×). (**P<0.01). D, E. The cell cycle phase of U87-MG and LN229 cells transfected with DNMT3A siRNA or negative control (NC) was analyzed by flow cytometry. (**P<0.01). F. A heat map of relative expression of several DNMT3A-associated cell cycle genes in Rembrandt glioblastoma tissues sorted relative to the level of DNMT3A expression (r>0.4). G. Western blot analysis of DNMT3A, CDK2 and cyclinA2 in U87-MG and LN229 cells after knockdown of DNMT3A.
Overexpression of DNMT3A partially attenuates miR-129-5p inhibited cell growth
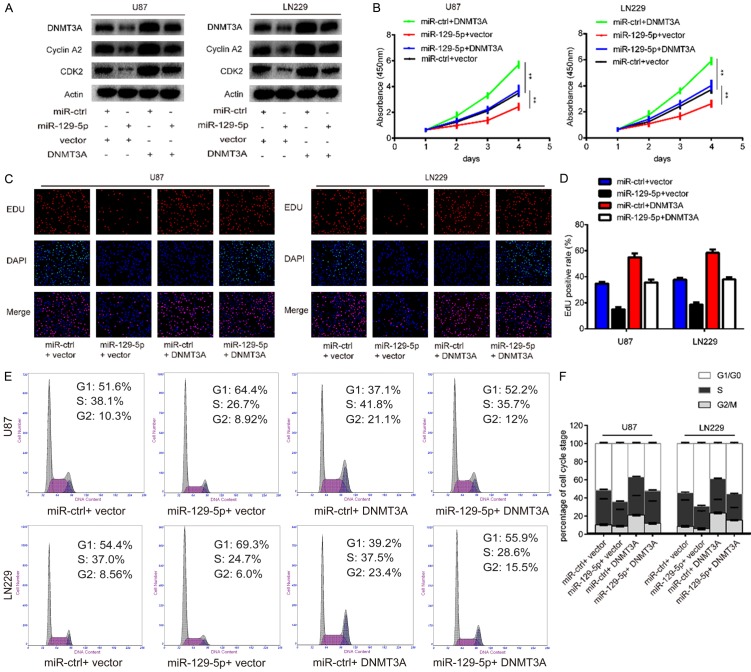
We have shown that DNMT3A is a direct target of miR-129-5p and also a carcinogenic effect in gliomas; therefore, we further investigated whether the role of miR-129-5p in glioma is mediated through DNMT3A. The plasmid pcDNA3.1-DNMT3A was transfected into miR-129-5p-overexpressing U87-MG and LN229 cells to reverse the reduced DNMT3A levels. After transfection, western blot analysis was conducted. As indicated in Figure 6A, the level of DNMT3A was higher in miR-129-5p + DNMT3A cells compared with that in miR-129-5p + vector cells. To further confirm whether DNMT3A is an important target of miR-129-5p with respect to cell proliferation, CCK-8 and EDU assays were performed. Restoration of DNMT3A levels partially rescued miR-129-5p suppression of U87-MG and LN229 proliferation (Figure 6B-D). Furthermore, cell cycle distribution analysis revealed that the increased G1 phase population after miR-129-5p expression was abolished by overexpression of DNMT3A (Figure 6E, 6F). Interestingly, levels of cell cycle-related proteins were altered in a similar way to DNMT3A; that is, decreased levels of CDK2 and cyclin A2 resulting from miR-129-5p overexpression could be rescued by up-regulation of DNMT3A (Figure 6A). Collectively, these findings indicate that DNMT3A acts as a downstream effector of miR-129-5p in the regulation of glioma cell proliferation in vitro.
Figure 6.
Reintroduction of DNMT3A reverses the inhibitory effect of miR-129-5p. A. A rescue experiment was performed by introducing pcDNA3.1-DNMT3A or pcDNA3.1 in the presence or absence of ectopic miR-129-5p or miR-ctrl expression in U87-MG and LN229 cells. Western blot assays were performed to detect the levels of DNMT3A, CDK2 and cyclinA2 in the indicated cells. Actin was used as the loading control. B. Cell viability of glioma cells transfected with pcDNA3.1-DNMT3A and miR-129-5p separately or together was detected using the CCK-8 assay. C, D. Cell proliferative potential was evaluated using the EDU assay 48 h after co-transfection of pcDNA3.1-DNMT3A and miR-129-5p. E, F. Cell cycle distribution of glioma cells was measured using flow cytometry.
Discussion
Glioma is the most common malignant tumor in the adult human brain. It has features in common with other malignant tumors: rapid proliferation, infiltrating growth, intracranial metastases, frequent recurrence after surgery and insensitivity to traditional radiotherapy and chemotherapy. Novel therapeutic strategies, for example, using gene therapy, are therefore urgently needed. However, any new approach requires a thorough understanding of the molecular mechanisms of glioma development to enable suitable targets to be found.
MiRNAs have extensive roles in the development of cancer, especially in decreasing the mRNA levels of target genes and in post-transcriptional regulation of gene expression [42,43]. Numerous miRNAs, such as miR-147 [44], miR-146a [45], miR-138 [46,47], miR-340 [48], miR-221 [49] and miR-96 [50], have been found to play critical roles in tumors. The miR-129 family includes three mature members: miR-129-5p, miR-129-1-3p, and miR-129-2-3p. MiR-129-5p is also downregulated in gastric cancer [51], bladder cancer [52] and colorectal cancer [53]. In these tumors, miR-129-5p inhibits growth consistent with miR-129-5p being a tumor suppressor miRNA [54].
Our pre-experimental study found that miR-129-5p was expressed at low levels in the glioma database. We then showed that miR-129-5p was also downregulated in clinical specimens of glioma. Furthermore, the expression level was negatively correlated with the grade of glioma. We then analyzed the expression of miR-129-5p in the existing glioma cell lines U118, LN229, H4, A172, U87 and U251 and in NHAs. The expression of miR-129-5p in glioma cell lines was indeed reduced, and it was particularly evident in LN229 and U87 cells.
So what are the roles of miR-129-5p in glioma? We overexpressed miR-129-5p in both LN229 and U87 cell lines. We confirmed the overexpression of miR-129-5p by RT-PCR, and then detected changes in cell proliferation by CCK-8 and EDU assays, which showed that the proliferation of glioma cells were inhibited after overexpression of miR-129-5p. Proliferation and the cell cycle are closely related. So what was the effect of miR-129-5p on the cell cycle of glioma? We found that the cell cycle was arrested in G1 phase. This partly explained the effect of miR-129-5p on the proliferation of glioma cells.
It is well known that miRNAs function by inducing degradation or preventing translation of specific mRNA targets. As miR-129-5p can influence the proliferation of glioma cells, what is the mechanism for this effect? Among the target genes predicted by Targetscan, DNMT3A attracted our interest. DNMT3A is a 130 kDa protein that is highly conserved in vertebrates, with 98% homology between humans and mice [55,56]. The gene is encoded by 23 exons on human chromosome 2p23 [57]. As a de novo DNA methyltransferase, DNMT3A is traditionally considered to play an important role in the establishment of methylation patterns during primordial germ cell development and early embryogenesis [32], and behaving as an oncogene in human cancers [58]. However, several inactivating mutations of DNMT3A in myeloid malignancies [37,59] and loss of DNMT3A activity at advanced tumor stages 24 were recently identified.
We analyzed the expression profile of DNMT3A in the commonly used glioma databases. DNMT3A was highly expressed in gliomas and was positively correlated with glioma grade. We then determined the expression of DNMT3A in clinical tissue samples by western blotting and the results were consistent with the analysis of the databases. What was the effect of DNMT3A on the characteristics of glioma? We screened the glioma databases for genes that are positively related to the expression of DNMT3A. Through GO and Pathway analysis, we found that DNMT3A mainly affected proliferation and cell cycle in glioma. To verify the results of analysis, we down-regulated the expression of DNMT3A in LN229 and U87 cells and showed that the proliferation of glioma cells was inhibited. In addition, we also found that the cell cycle was arrested in G1 phase. Therefore, we propose that DNMT3A promotes the proliferation of glioma cells.
What is the relationship between the expression of miR-129-5p and DNMT3A in clinical glioma samples? Spearman correlation analysis showed that miR-129-5p was negatively correlated with DNMT3A, which further indicated that DNMT3A is a target gene of miR-129-5p. Luciferase reporter assays and rescue experiments confirmed that DNMT3A was indeed a functional target of miR-129-5p and that miR-129-5p affected the proliferation of glioma cells by targeting DNMT3A.
In summary, our study adds our knowledge of the roles that miR-129-5p plays in glioma proliferation and provides evidence of the complex signaling network in glioma development and cell cycle. Both DNMT3A and miR-129-5p might serve as prognosis or predictive factors and as potential therapeutic targets in glioma diagnosis and treatment.
Acknowledgements
We thank Prof. Jian Chen (Nantong University), Dr. Huangcheng Song (Suzhou University) and the other members of our laboratory. We thank Dr. Dongjing Song (Jilin University) for English writing assistance. We thank Jeremy Allen, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript. We also thank the neurosurgery department of Affiliated Hospital of Nantong University for supporting glioma specimens. This work is funded by the Haimen People’s Hospital scientific research project (017) (No. 2017N001) and the Haimen People’s Hospital Research Mutual Fund (No. HM2017051).
Disclosure of conflict of interest
None.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM, Wrensch MR, Barnholtz-Sloan JS. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis FG, Freels S, Grutsch J, Barlas S, Brem S. Survival rates in patients with primary malignant brain tumors stratified by patient age and tumor histological type: an analysis based on surveillance, epidemiology, and end results (SEER) data, 1973-1991. J Neurosurg. 1998;88:1–10. doi: 10.3171/jns.1998.88.1.0001. [DOI] [PubMed] [Google Scholar]
- 4.Penas-Prado M, Gilbert MR. Molecularly targeted therapies for malignant gliomas: advances and challenges. Expert Rev Anticancer Ther. 2007;7:641–661. doi: 10.1586/14737140.7.5.641. [DOI] [PubMed] [Google Scholar]
- 5.van den Bent M, Chinot OL, Cairncross JG. Recent developments in the molecular characterization and treatment of oligodendroglial tumors. Neuro Oncol. 2003;5:128–138. doi: 10.1215/S1522-8517-02-00028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalpathy-Cramer J, Gerstner ER, Emblem KE, Andronesi O, Rosen B. Advanced magnetic resonance imaging of the physical processes in human glioblastoma. Cancer Res. 2014;74:4622–4637. doi: 10.1158/0008-5472.CAN-14-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen AL, Colman H. Glioma biology and molecular markers. Cancer Treat Res. 2015;163:15–30. doi: 10.1007/978-3-319-12048-5_2. [DOI] [PubMed] [Google Scholar]
- 8.Li SD, Zhang JR, Wang YQ, Wan XP. The role of microRNAs in ovarian cancer initiation and progression. J Cell Mol Med. 2010;14:2240–2249. doi: 10.1111/j.1582-4934.2010.01058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song JL, Nigam P, Tektas SS, Selva E. microRNA regulation of Wnt signaling pathways in development and disease. Cell Signal. 2015;27:1380–1391. doi: 10.1016/j.cellsig.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sirotkin AV, Laukova M, Ovcharenko D, Brenaut P, Mlyncek M. Identification of microRNAs controlling human ovarian cell proliferation and apoptosis. J Cell Physiol. 2010;223:49–56. doi: 10.1002/jcp.21999. [DOI] [PubMed] [Google Scholar]
- 11.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F, Bao G, Kong H, Ge C, Zhang F, Yu T, Li J, He X, Yao M. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol Cancer. 2014;13:166. doi: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 16.Xia M, Li H, Wang JJ, Zeng HJ, Wang SH. MiR-99a suppress proliferation, migration and invasion through regulating insulin-like growth factor 1 receptor in breast cancer. Eur Rev Med Pharmacol Sci. 2016;20:1755–1763. [PubMed] [Google Scholar]
- 17.Wu D, Niu X, Pan H, Zhou Y, Qu P, Zhou J. MicroRNA-335 is downregulated in bladder cancer and inhibits cell growth, migration and invasion via targeting ROCK1. Mol Med Rep. 2016;13:4379–4385. doi: 10.3892/mmr.2016.5055. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Bo L, Lu W, Zhou G, Chen Q. MicroRNA-148b targets Rho-associated protein kinase 1 to inhibit cell proliferation, migration and invasion in hepatocellular carcinoma. Mol Med Rep. 2016;13:477–482. doi: 10.3892/mmr.2015.4500. [DOI] [PubMed] [Google Scholar]
- 19.Xiao G, Li X, Li G, Zhang B, Xu C, Qin S, Du N, Wang J, Tang SC, Zhang J, Ren H, Chen K, Sun X. MiR-129 blocks estrogen induction of NOTCH signaling activity in breast cancer stem-like cells. Oncotarget. 2017;8:103261–103273. doi: 10.18632/oncotarget.21143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han H, Li W, Shen H, Zhang J, Zhu Y, Li Y. microRNA-129-5p, a c-Myc negative target, affects hepatocellular carcinoma progression by blocking the Warburg effect. J Mol Cell Biol. 2016 doi: 10.1093/jmcb/mjw010. [DOI] [PubMed] [Google Scholar]
- 21.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, Gemma A, Kudoh S, Croce CM, Harris CC. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng A, Yin J, Li Y, Li R, Wang Z, Zhou X, Jin X, Shen F, Yan W, You Y. miR-129-5p targets Wnt5a to block PKC/ERK/NF-kappaB and JNK pathways in glioblastoma. Cell Death Dis. 2018;9:394. doi: 10.1038/s41419-018-0343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutz BM, Bekker A, Tao YX. Noncoding RNAs: new players in chronic pain. Anesthesiology. 2014;121:409–417. doi: 10.1097/ALN.0000000000000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang L, Lutz BM, Bekker A, Tao YX. Epigenetic regulation of chronic pain. Epigenomics. 2015;7:235–245. doi: 10.2217/epi.14.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 26.Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2007;96(Suppl):R26–30. [PubMed] [Google Scholar]
- 27.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010;17:2141–2151. doi: 10.2174/092986710791299966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 31.Pradhan S, Esteve PO. Mammalian DNA (cytosine-5) methyltransferases and their expression. Clin Immunol. 2003;109:6–16. doi: 10.1016/s1521-6616(03)00204-3. [DOI] [PubMed] [Google Scholar]
- 32.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 33.Lin H, Yamada Y, Nguyen S, Linhart H, Jackson-Grusby L, Meissner A, Meletis K, Lo G, Jaenisch R. Suppression of intestinal neoplasia by deletion of Dnmt3b. Mol Cell Biol. 2006;26:2976–2983. doi: 10.1128/MCB.26.8.2976-2983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Alteration of DNA methyltransferases contributes to 5’CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55:205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, Meissner A, Jaenisch R. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, Baylin SB, Vogelstein B. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 37.Singh RR, Bains A, Patel KP, Rahimi H, Barkoh BA, Paladugu A, Bisrat T, Ravandi-Kashani F, Cortes JE, Kantarjian HM, Medeiros LJ, Luthra R. Detection of high-frequency and novel DNMT3A mutations in acute myeloid leukemia by high-resolution melting curve analysis. J Mol Diagn. 2012;14:336–345. doi: 10.1016/j.jmoldx.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metzeler KH, Walker A, Geyer S, Garzon R, Klisovic RB, Bloomfield CD, Blum W, Marcucci G. DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia. 2012;26:1106–1107. doi: 10.1038/leu.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Q, Steine EJ, Barrasa MI, Hockemeyer D, Pawlak M, Fu D, Reddy S, Bell GW, Jaenisch R. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc Natl Acad Sci U S A. 2011;108:18061–18066. doi: 10.1073/pnas.1114946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi H, Gong H, Cao K, Zou S, Zhu B, Bao H, Wu Y, Gao Y, Tang Y, Yu R. Nrdp1-mediated ErbB3 degradation inhibits glioma cell migration and invasion by reducing cytoplasmic localization of p27(Kip1) J Neurooncol. 2015;124:357–364. doi: 10.1007/s11060-015-1851-9. [DOI] [PubMed] [Google Scholar]
- 42.Wang F, Li T, Zhang B, Li H, Wu Q, Yang L, Nie Y, Wu K, Shi Y, Fan D. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem Biophys Res Commun. 2013;434:688–694. doi: 10.1016/j.bbrc.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 43.Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ, Yang AG, Zhang R. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting beta-catenin. Biochem Biophys Res Commun. 2012;420:787–792. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Zhang HE, Liu Z. MicroRNA-147 suppresses proliferation, invasion and migration through the AKT/mTOR signaling pathway in breast cancer. Oncol Lett. 2016;11:405–410. doi: 10.3892/ol.2015.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornett AL, Lutz CS. Regulation of COX-2 expression by miR-146a in lung cancer cells. RNA. 2014;20:1419–1430. doi: 10.1261/rna.044149.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang R, Liu M, Liang H, Guo S, Guo X, Yuan M, Lian H, Yan X, Zhang S, Chen X, Fang F, Guo H, Zhang C. miR-138-5p contributes to cell proliferation and invasion by targeting survivin in bladder cancer cells. Mol Cancer. 2016;15:82. doi: 10.1186/s12943-016-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei J, Nduom EK, Kong LY, Hashimoto Y, Xu S, Gabrusiewicz K, Ling X, Huang N, Qiao W, Zhou S, Ivan C, Fuller GN, Gilbert MR, Overwijk W, Calin GA, Heimberger AB. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol. 2016;18:639–648. doi: 10.1093/neuonc/nov292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohammadi-Yeganeh S, Paryan M, Arefian E, Vasei M, Ghanbarian H, Mahdian R, Karimipoor M, Soleimani M. MicroRNA-340 inhibits the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway. Tumour Biol. 2016;37:8993–9000. doi: 10.1007/s13277-015-4513-9. [DOI] [PubMed] [Google Scholar]
- 49.Roscigno G, Quintavalle C, Donnarumma E, Puoti I, Diaz-Lagares A, Iaboni M, Fiore D, Russo V, Todaro M, Romano G, Thomas R, Cortino G, Gaggianesi M, Esteller M, Croce CM, Condorelli G. MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget. 2016;7:580–592. doi: 10.18632/oncotarget.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, Liu L, Xu Q, Wu P, Zuo X, Ji A. MicroRNA as a novel drug target for cancer therapy. Expert Opin Biol Ther. 2012;12:573–580. doi: 10.1517/14712598.2012.671293. [DOI] [PubMed] [Google Scholar]
- 51.Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, Kao HW, Fang WL, Huang KH, Chan WC, Lin WC. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer. 2011;129:2600–2610. doi: 10.1002/ijc.25919. [DOI] [PubMed] [Google Scholar]
- 52.Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, Kauppinen S, Ulhoi BP, Kjems J, Borre M, Orntoft TF. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 53.Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 54.Wu J, Qian J, Li C, Kwok L, Cheng F, Liu P, Perdomo C, Kotton D, Vaziri C, Anderlind C, Spira A, Cardoso WV, Lu J. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle. 2010;9:1809–1818. doi: 10.4161/cc.9.9.11535. [DOI] [PubMed] [Google Scholar]
- 55.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 56.Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 57.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 58.Fernandez AF, Huidobro C, Fraga MF. De novo DNA methyltransferases: oncogenes, tumor suppressors, or both? Trends Genet. 2012;28:474–479. doi: 10.1016/j.tig.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 59.Wang S, Zhang YX, Huang T, Sui JN, Lu J, Chen XJ, Wang KK, Xi XD, Li JM, Huang JY, Chen B. Mutation profile and associated clinical features in Chinese patients with cytogenetically normal acute myeloid leukemia. Int J Lab Hematol. 2018;40:408–418. doi: 10.1111/ijlh.12802. [DOI] [PubMed] [Google Scholar]