Abstract
Background: Transcribed ultraconserved regions (T-UCRs) are a subset of long noncoding RNAs. It has been reported that T-UCRs are dysregulated in cancers and play an important role in the development and progression of malignancies. uc.160 was found to be a suppressive factor of cancer development, but its role has not been fully elucidated. Methods: The uc.160 expression was examined in gastric cancer tissues and established cell lines by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The biological function of gastric cancer cells with uc.160 over-expression were investigated, and the interaction between uc.160 and microRNA miR-155 was examined by dual-luciferase reporter assay. PTEN levels were detected by Western blotting. Anti-tumor effects of uc.160 were further explored in tumor transplantation models. Results: uc.160 expression was significantly down-regulated in gastric cancer tissues and gastric cell lines as compared to adjacent normal tissues and immortalized gastric epithelial cell line (GES-1), respectively. Over-expression of uc.160 in SGC-7901 and AGS gastric cancer cells significantly suppressed their proliferation in vitro and in vivo. Moreover, uc.160 positively regulated the tumor suppressor protein PTEN. Interestingly, uc.160 was inhibited by microRNA miR-155 that is also a negative regulator of gastric cancer. Conclusion: uc.160 is significantly down-regulated in gastric carcinomas and can inhibit the tumor growth both in vitro and in vivo, suggesting that uc.160 may be used as a diagnostic marker and therapeutic target of gastric malignancies.
Keywords: Noncoding RNA, uc.160, tumor suppressor
Introduction
Gastric cancer (GC) is one of the most common malignancies and the fifth most common cause of cancer-related death worldwide [1]. There were almost 500,000 gastric cancer patients died in China in 2015 [2]. Several key genes and related molecular pathways have been identified to be linked to the pathogenesis of gastric cancer, but the specific mechanisms remain to be elucidated.
The transcribed-ultraconserved regions (T-UCRs) are a new group of long noncoding RNAs and highly conservative among human beings, rats, and mice [3,4]. There are 481 ultraconserved DNA regions (UCR) described so far, but the roles of their transcribed products are poorly understood. The high conservation and wide expression of UCRs suggest that they play a critical role in physiological and/or pathological processes. Studies have shown differential expression of T-UCRs in human carcinomas [4,5]. For instance, uc.338 is over-expressed in the hepatocellular carcinoma (HCC) and affects the tumor development [6]. uc.73 is able to increase cell proliferation and reduce apoptosis of colorectal cancer cells [4,7], while uc.160, uc.283+A and uc.346 can undergo CpG island hypermethylation-associated gene silencing [5]. uc.160 is down-regulated in GC tissues and can possibly act as a tumor suppressor [8]. However, the mechanisms are largely unknown.
miRNAs are a group of small non-coding RNAs that can inhibit gene expression at the posttranscriptional level by binding the 3’-untranslated region (3’UTR) of target [9,10]. miR-155 is a typical multifunctional miRNA [14] and has been found over-expressed in hematopoietic disease [15]. Increased miR-155 expression has been reported to be associated with the poor outcome of lung and breast cancer [16]. Although miR-155 is significantly up-regulated and acts as an oncogene in GC [17], Li et al and Ma et al found that miR-155 expression was down-regulated and miR-155 acted as a tumor suppressor in GC [18,19]. Clearly, there is controversy on the role of miR-155 in GC, thus it is necessary to explore the exact function of miR-155 in GC.
In the present study, qRT-PCR was employed to detect the expression of uc.160 and miR-155 in human GC tissues, and the roles of uc.160 and miR-155 in proliferation and apoptosis of GC cells as well as the interaction between miR-155 and uc.160 were further investigated.
Materials and methods
Sample collection
45 paired GC tissues and adjacent normal tissues were collected at Subei People’s Hospital of Jiangsu Province and Affiliated Hospital of Yangzhou University, China. The pathological diagnosis was conducted by independent pathologists. 24 patients were diagnosed with GC at stage I-II, and 21 with GC at stage III-IV. All tissues were obtained during surgery and stored in liquid nitrogen until use. This study was approved by the Ethic Committee of the Institutional Review Board of the Yangzhou University School of Medicine.
Cell lines and cell culture
Three GC cell lines (SGC-7901, MGC-803, AGS) and the immortalized gastric epithelial cell line (GES-1) were obtained from the Department of Integrative Chinese & Western Medicine, Yangzhou University school of Medicine. All cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum (FBS) at 37°C in an environment with 5% CO2. Human embryonic kidney cell line 293T (HEK 293T) was stored in our institute, and grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS.
Plasmids and transfection
Genomic fragments involving uc.160 and human miR-155 were amplified into MSCV-pig retroviral vector by PCR using the following primers designed with Bgl II and Xho I restriction sites: miR-155-Forward: 5’-AGTCGCTAGCCACAAACCAGGAAGGGGAA-3’, miR-155-Reverse: 5’-ATCGGAATTCAAATGATAAAGCCTGAAGTC-3’, uc.160-Forward: 5’-ACACAGATCTAGTAAATGAGGCGAGTGTGC-3’, uc.160-Reverse: 5’-ACACCTCGAGAGCATCCTTAATATTTCCTT-3’.
The fragments of uc.160-BS which has the binding sites with the seed sequence of miR-155 and uc.160-NBS which is upstream fragment of uc.160 and does not have the binding sites with the seed sequence of miR-155 were also amplified using following primers with restriction sites of EcoR I and Xba I and engineered to pGL3-BS luciferase reporter vector: uc.160-BS-Forward: 5’-ACACGAATTCATTCTCCCCTGATCTCAGGT-3’, uc.160-BS-Reverse: 5’- ACACTCTAGACAGGATGGGACAGGATATGA-3’, uc.160-NBS-Forward: 5’-ACACGAATTCCAGGAGAATCGCTTGAACCT-3’, uc.160-NBS-Reverse: 5’-ACACTCTAGACCTATGATTGCCTTTGAGGC-3’.
The retroviral vectors were transfected into GC cells and 293T cells independently with Lipofectamine 2000 (Invitrogen). 48 h later, puromycin was used to select the transfected cells for 2-3 days.
Quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells and tissues with TRIzol regent (Invitrogen). 1 μg of RNA was reversely transcribed to cDNA using the PrimerScriptTM RT reagent Kit with gDNA Eraser (Takara Japan), and following strand-specific (allele-specific) reverse transcription primers were used: uc.160: 5’-CTGACAAGGGACTGAAAGAG-3’, β-actin: 5’-CTTTACGGATGTCAACGTCA-3’. For miR-155, 1 μg of RNA was reversely transcribed to cDNA with the Mir-XTM miRNA First-Strand Synthesis Kit (Takara USA) according to the manufacturer’s instructions. qRT-PCR was processed using the TB GreenTM Premix ExTM II (Tli RNaseH Plus) (Takara Japan) and conducted on an Applied Biosystems 7500 Real Time PCR system (Applied Biosystems, White Plains, NY, USA). Relative UCR and miRNA level were calculated based on 2-ΔCt method. β-actin and small RNA U6 were used as internal controls. All the reactions were run in triplicate. PCR primers for uc.160 were: uc.160-Forward: 5’-ATGTACCTGCCTACTTAGCC-3’, uc.160- Reverse: 5’-AATCACAGATGTAGAGTATC-3’. Primers for β-actin were: β-actin-Forward: 5’-GAAGCTGTGCTATGTTGCTC-3’, β-actin-Reverse: 5’-GAATGTAGTTTCATGGATGC-3’. Primers for miR-155 were: miR-155-Forward: 5’-TTAATGCTAATCGTGATAGGGGT-3’. Primers for U6 were: U6-Forward: 5’-CGCTTCGGCAGCACATATAC-3’.
Luciferase assay
As described above, the uc.160-BS and uc.160-NBS were cloned into pGL3-BS luciferase reporter vector. 293T cells in 24-well plates were transfected with 0.4 μg of pGL3-BS reporters, 0.01 μg of Renilla luciferase vector and 0.4 μg of miR-155-MSCV-pig using Lipofectamine 2000 (Invitrogen). 48 hours after transfection, the Dual-Luciferase Reporter (DLR) Assay Kit (Promega) was used to detect the luciferase activity.
Cell proliferation and viability assay
Flow cytometry (FCM, BD Biosciences) was done to detect the green fluorescent protein (GFP) for assessing the cell proliferation. Moreover, cells with stable expression of uc.160, miR-155, and MSCV-pig as control were seeded into 96-well plates (5000 cells/well). After incubation for 24, 48, and 72 h, cell proliferation was evaluated by using a Cell Counting Kit-8 (CCK-8) Kit. Absorbance was measured at 450 nm with the Microplate Reader. Finally, 1 × 105 cells were seeded into 24-well plates and cell counting was conducted at 24, 48, and 72 h. All experiments were conducted in triplicate.
Apoptosis assay
The uc.160 and miR-155 over-expressed cells were washed twice with cold PBS, then treated with Annexin V-APC (BD Biosciences) and Propidium Iodide (PI, BD Biosciences). Cell apoptosis was determined by flow cytometry (FCM, BD Biosciences).
SCID beige mouse experiments
8-week-old SCID mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. Cells were subcutaneously injected into the right flank of SCID mice at the density of 2 × 106 in 200 μl of PBS. Tumor volume was measured using a caliper every 3 days once the tumor was palpable. Tumor volume was calculated using the follow formula: tumor volume = 1/2 × (length × width2). After 23 days, tumors were harvested and weighed.
Western blotting
Protein lysates from SGC-7901 and AGS cells transfected with MSCV-pig and uc.160 were extracted using cell lysis buffer with 1:1000 protease inhibitor mixture (Beyotime institute of Biotechnology). Lysates (10 μg) were resolved by 10% SDS-PAGE gel and transferred onto PVDF membrane (Millipore). Then protein was detected with PTEN antibody (Cell Signaling Technology) and GAPDH antibody (Cell Signaling Technology) for overnight at 4°C. After incubation with horseradish peroxidase (HRP)-conjugated secondary antibody, immunoreactive proteins were detected on FluorChem FC2 Imaging System (Alpha Innotech, San Leandro, CA, USA).
Statistical analysis
Statistical analysis was performed using SPSS 16.0 software package (SPSS, Chicago, USA). Data from three independent experiments were expressed as mean ± standard deviation (SD). Comparisons were done with Student’s t test. A value of two-tailed P < 0.05 was considered statistically significant.
Results
uc.160 expression decreases in the GC tissues and cell lines
In order to demonstrate the role of uc.160 in GC development, uc.160 expression was detected by qRT-PCR in 45 matched human GC tissues and adjacent normal tissues. In addition, the expression of uc.160 was also measured in three GC cell lines and an immortalized gastric epithelial cell line (GES-1). As shown in Figure 1A, uc.160 expression was significantly down-regulated in GC tissues compared with matched adjacent normal tissues, but it was similar between GC tissues at different clinicopathologic stages (Figure 1B). In addition, lower uc.160 expression was also detected in three GC cell lines (SGC-7901, MGC-803 and AGS) compared to that in the immortalized gastric epithelial cell line (GES-1) (Figure 1C). These findings suggest that uc.160 may play a crucial role in GC development and progression.
Figure 1.
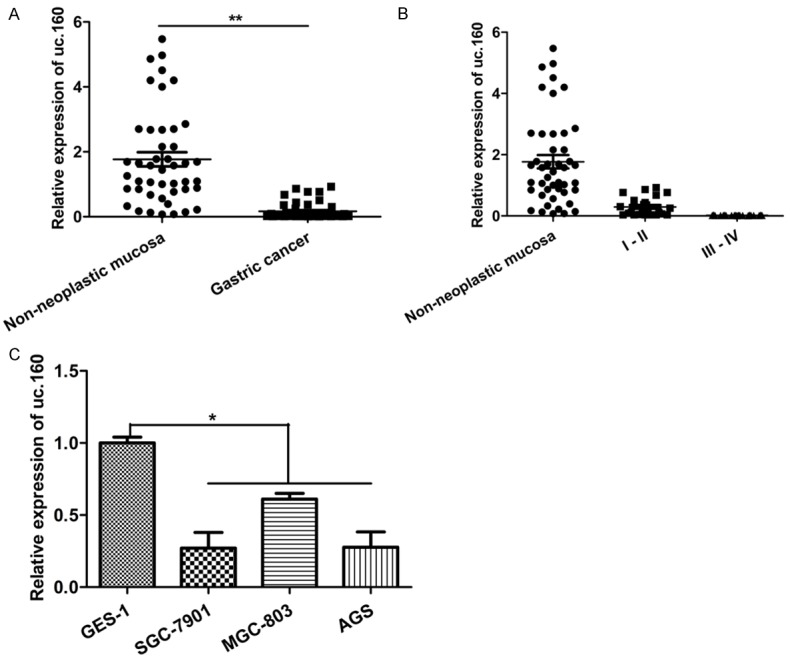
uc.160 expression was down-regulated in GC tissues and GC cell lines. A. Quantitative analysis of uc.160 level in 45 paired human GC tissues compared with their adjacent normal tissues. **P < 0.01 (t-test). The loading control was β-actin. B. Quantitative analysis of uc.160 level in GC tissues at different cliniclpathologcal stages (stage I and II, n = 24; stage III and IV, n = 21), and the relative expression level of uc.160 was similar. Pathological classification was done according to the AJCC Cancer’s TNM Classification system (7th Edition). C. Quantitative analysis of uc.160 level in three GC cell lines (SGC-7901, MGC-803, and AGS) and the immortalized gastric epithelial cell line (GES-1). The Y-axis shows relative expression of uc.160 and the uc.160 level from GES-1 was assigned as a relative value of 1. *P < 0.05 (t-test). The loading control was β-actin.
uc.160 over-expression inhibits the GC cell proliferation and viability
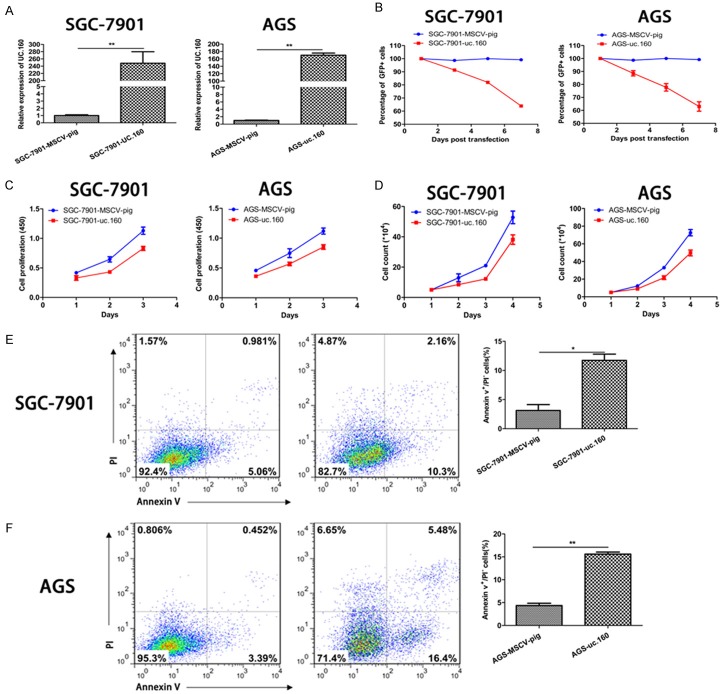
Subsequently, uc.160 was independently transfected into SGC-7901 and AGS cells, and ectopic uc.160 expression was confirmed. Compared with control MSCV-pig, uc.160 expression increased significantly (Figure 2A). Then, by tracking the percentage of GFP+ cells, CCK-8 assay and cell count assay, ectopically expressed uc.160 was found to inhibit the proliferation and viability of two GC cell lines compared with the control (Figure 2B, 2C). Moreover, GC cells with uc.160 over-expression displayed significantly higher apoptosis rate as compared to control GC cells (Figure 2E, 2F). These results indicate that uc.160 acts as a tumor suppressor in GC.
Figure 2.
uc.160 over-expression inhibited the GC cells proliferation and viability and increased their apoptosis. A. Quantitative analysis of uc.160 level in GC cells (SGC-7901 and AGS) transfected with MSCV-pig control and uc.160-MSCV-pig. The relative expression level of uc.160 was increased in GC cells transfected with uc.160-MSCV-pig compared with cells transfected with negative control. The levels from MSCV-pig were assigned an arbitrary value of 1. **P < 0.01 (t-test). The loading control was β-actin. B. The percentage of GFP+ were tracked every 48 h after transfection of uc.160 and MSCV-pig. C. The cell numbers were counted every 24 h after transfection of uc.160. D. Cells were cultured for 24 h, 48 h and 72 h, then measured at 450 nm after adding the CCK-8 reagent. E, F. The apoptosis rate of GC cells was calculated after transfection of uc.160 by flow cytometry (left). The apoptosis rate increased significantly in cells with uc.160 over-expression compared with cells transfected with negative control (right). *P < 0.05, **P < 0.01 (t-test).
miR-155 expression decreases in GC tissues and GC cell lines
uc.160 expression was found to be down-regulated in the chronic lymphocytic leukemia (CLL) compared with the normal control. Meanwhile, a negative interaction was found between miR155 and uc.160 [4]. Thus, miR-155 expression was investigated in GC tissues and cell lines. As shown in Figure 3A, the miR-155 expression was significantly down-regulated in 45 GC tissues compared with the adjacent normal tissues. Meanwhile, miR-155 expression was significantly higher in stage I and II GC than in stage III and IV GC (Figure 3B). Moreover, lower miR-155 expression was also detected in GC cell lines (SGC-7901, MGC-803 and AGS) as compared to the immortalized GES-1 (Figure 3C). These findings suggest that miR-155 might also be a tumor suppressor in GC.
Figure 3.
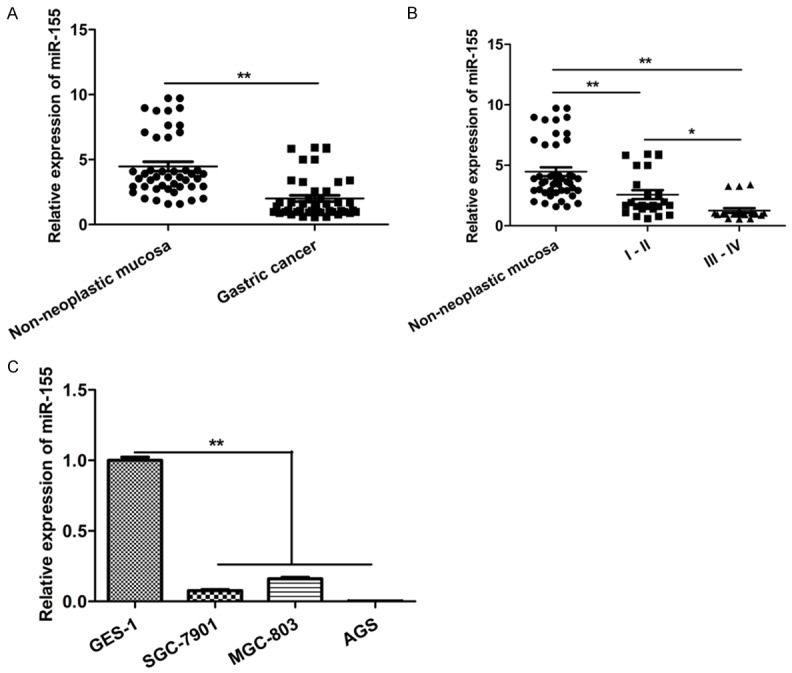
miR-155 expression was down-regulated in GC tissues and GC cell lines. A. Quantitative analysis of miR-155 level in 45 paired human GC tissues compared with their adjacent normal tissues. **P < 0.01 (t-test). The loading control was U6. B. Quantitative analysis of miR-155 level in GC tissues at different cliniclpathologcal stages (stage I and II, n = 24; stage III and IV, n = 21). *P < 0.05, **P < 0.01 (t-test). Pathological classification was done according to the AJCC Cancer,s TNM Classification system (7th Edition). C. Quantitative analysis of miR-155 level in three GC cell lines (SGC-7901, MGC-803, and AGS) and the immortalized gastric epithelial cell line (GES-1). The Y-axis shows fold change of miR-155 with the miR-155 level in GES-1 assigned as a relative value of 1. **P < 0.01 (t-test). The loading control was U6.
miR-155 over-expression inhibits GC cell proliferation and viability
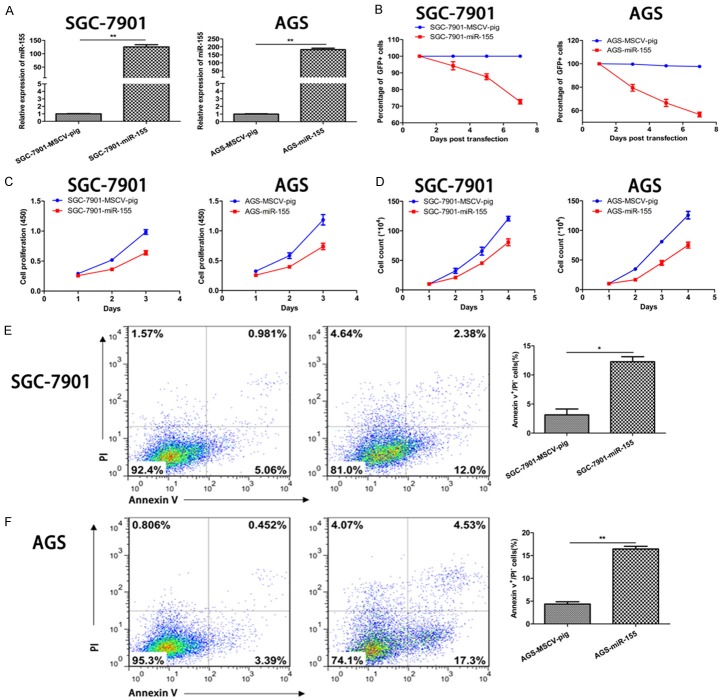
The effects of miR-155 on the proliferation and apoptosis of GC cells were further investigated via over-expressing miR-155 in these cells. Similar to uc.160, miR-155 over-expression significantly inhibited the proliferation and promoted the apoptosis of GC cells (Figure 4A-F). Collectively, these findings indicate that miR-155 plays an inhibitory role in the GC growth.
Figure 4.
miR-155 inhibited the proliferation and viability of GC cells and increased their apoptosis. A. Quantitative analysis of miR-155 level in GC cells (SGC-7901 and AGS) transfected with MSCV-pig control and miR-155-MSCV-pig. The relative expression level of miR-155 was elevated in GC cells transfected with miR-155-MSCV-pig compared with cells transfected with negative control. The levels from MSCV-pig were assigned an arbitrary value of 1. **P < 0.01 (t-test). The loading control was U6. B. The percentage of GFP+ were tracked every 48 h after transfection of miR-155 and MSCV-pig. C. The cell numbers were counted every 24 h after transfection of miR-155. D. Cells were cultured for 24 h, 48 h and 72 h, then measured at 450 nm after adding the CCK-8 reagent. E, F. The apoptosis rate of GC cells was calculated after transfection of miR-155 by flow cytometry (left). The apoptosis rate increased significantly in cells with miR-155 over-expression compared with cells transfected with negative control (right). *P < 0.05, **P < 0.01 (t-test).
miR-155 inhibits the expression of uc.160
Studies have shown that there is an interaction between miR-155 and uc.160 in the CLL but the consequence of this interaction is not clear [4]. Thus, we investigated whether miR-155 and uc.160 were mutually affected. Ectopic expression of miR-155 significantly reduced the expression of uc.160 in GC cells (Figure 5A). On the contrary, ectopic expression of uc.160 did not alter the level of miR-155 (Figure 5B). To confirm whether uc.160 was the direct target of miR-155 in GC cells, RNAhybrid was employed to identify the binding sites between miR-155 and uc.160, and dual luciferase reporter assay was conducted to explore their interaction. As shown in Figure 5C and 5D, co-transfection with miR-155 and uc.160-BS significantly decreased the luciferase activity compared with the negative control, indicating that miR-155 directly binds uc.160.
Figure 5.
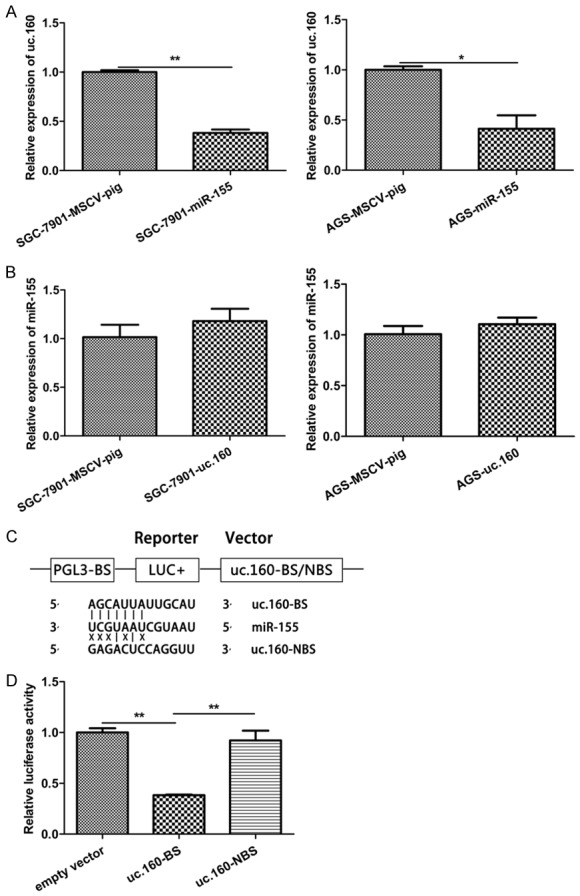
uc.160 was a direct target of miR-155 in GC. A. Quantitative analysis of uc.160 level in GC cells (SGC-7901 and AGS) transfected with miR-155 compared with cells transfected with negative control. *P < 0.05, **P < 0.01 (t-test). The loading control was β-actin. B. Quantitative analysis of uc.160 level in GC cells (SGC-7901 and AGS) transfected with uc.160 and cells transfected with negative control. The loading control was U6. C. Luciferase structures containing the potential binding sites of uc.160 with the miR-155 seed sequence and no binding sites between the upstream of uc.160 sequence and the miR-155 seed sequence. D. Dual-luciferase reporter assay 293T cells co-transfected with negative control or uc.160-BS and miR-155. Luciferase activities were determined 48 h post transfection. Levels from the empty vector were assigned an arbitrary value of 1. **P < 0.01 (t-test).
Both uc.160 and miR-155 inhibit the growth of GC in vivo
The SGC-7901 xenograft was established in SCID mice. The uc.160 and miR-155 over-expressing cells were subcutaneously injected into 8-week-old SCID mice and tumor volume was evaluated. After 12 days, tumors were found in the control groups (SGC-7901 and SGC-7901-MSCV-pig), but not in uc.160 and miR-155 retrovirus producing groups. In addition, the tumors in uc.160 and miR-155 groups grew much slower than in control groups (Figure 6A). At 23 days after injection, tumors were collected and weighed. Consistent with the in vitro findings, the tumors in vivo were much smaller in both uc.160 and miR-155 groups than the control groups (Figure 6B, 6C), demonstrating that both uc.160 and miR-155 are GC suppressive non-coding RNAs.
Figure 6.
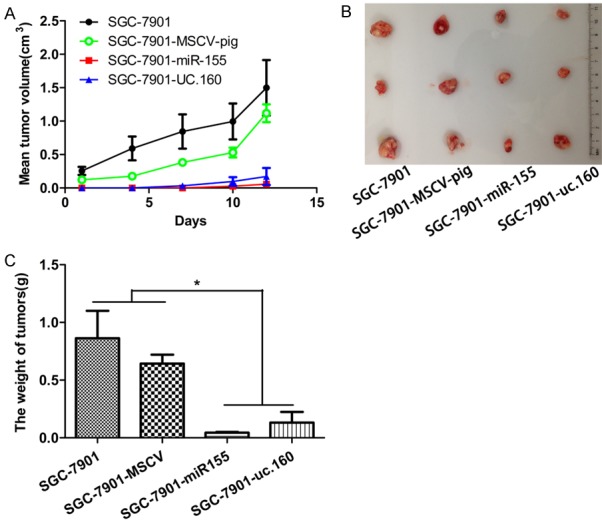
Over-expression of uc.160 and miR-155 inhibited the progression of GC in vivo. A. Analysis of tumor volume in the SCID mice. The tumor grew more slowly in the group of uc.160 and miR-155 compared with negative control group. B, C. Tumor volume at the end of study. The tumor volume and weight reduced significantly in the group of uc.160 and miR-155 compared with negative control group. A representative experiment was displayed (n = 3 mice for each group). *P < 0.05 (t-test).
uc.160 elevated PTEN expression
Because Honma et al. [8] had demonstrated that uc.160 inhibited PTEN expression in MKN-1 and MKN-45 cells, we next determined whether PTEN expression was influenced by uc.160 in SGC-7901 and AGS cells. We first stably increased the expression of uc.160 in SGC-7901 and AGS cells, and confirmed that uc.160 increased PTEN levels (Figure 7A, 7B). Our data indicate that uc.160 also can inhibit the growth of SGC-7901 and AGS cells though elevating the PTEN levels.
Figure 7.
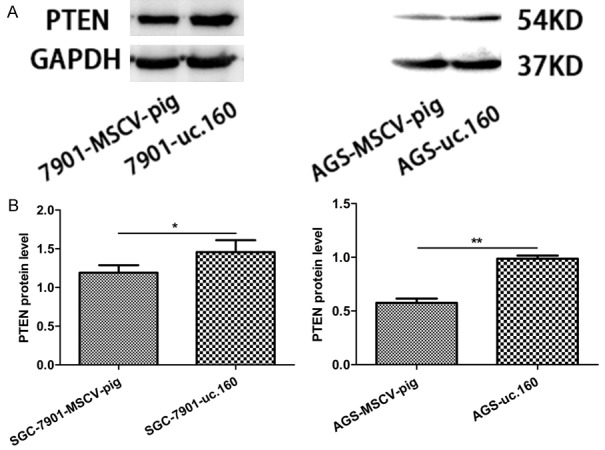
uc.160 promoted PTEN expression. A. Whole cell lysates were prepared from cells stably expressing MSCV-pig or uc.160 for Western blotting. Ectopic expression of uc.160 increased PTEN protein levels. GAPDH was used as a loading control. B. Quantitative image analysis of PTEN protein expression in SGC-7901 and AGS cells transfected with MSCV-pig and uc.160. *P < 0.05, **P < 0.01 (t-test).
Discussion
T-UCRs are a class of non-coding RNA and can regulate the expression of mRNAs. In recent years, T-UCRs have been proven to act as the oncogenes and tumor suppressor genes in human tumorigenesis. For example, up-regulated uc.73+A expression in colorectal cancer was found to promote the cell proliferation [4,7]. In breast cancer, uc.63 not only promotes the cell proliferation, but is also related to the prognosis [20]. In this study, we focused on the effect of uc.160 on the GC growth. uc.160 expression was down-regulated in the gastric tumors compared with non-neoplastic mucosa tissues of the stomach [8], and it was associated with the hypermethylation of CpG island [5,8]. In the current study, uc.160 expression was shown to be inhibited in human GC tissues and GC cell lines. Moreover, over-expression of uc.160 inhibited the proliferation of GC cells and promoted their apoptosis. Our data also demonstrates that over-expression of uc.160 elevates the PTEN levels which result in silenting the MAPK signaling. These observations support the hypothesis that uc.160 is a suppressor in gastric tumorigenesis.
miR-155 is mainly expressed in immunocytes and inflammatory tissues [21]. Currently, the role of miR-155 in GC is poorly elucidated and there is still controversy on the role of miR-155 in GC [17-19]. Our data clearly show that the level of miR-155 is significantly reduced in GC cells and over-expression of miR-155 strongly suppresses the growth of GC in both in vitro and in vivo assays, demonstrating miR-155 as a tumor suppressive miRNA in GC.
Although uc.160 is found to inhibit the phosphorylation of Akt by up-regulating PTEN in GC [8], the specific mechanism underlying the role of uc.160 in GC remains elucidated. Calin et al. reveal the negative relationship between uc.160 and miR-155 in CLL [4]. Moreover, it is reported that uc.416+A is a direct target of miR-153 in GC, and there is an inverse correlation between them [22]. In our study, luciferase reporter assay confirmed the direct interaction between uc.160 and miR-155. qRT-PCR showed that miR155 down-regulated the expression of uc.160. Of note, both uc.160 and miR-155 were down-regulated in GC tissues and GC cell lines and both uc.160 and miR-155 could inhibit the GC growth. These apparently conflicted findings highlight the complicated gene regulatory mechanism in GC development. In fact, the activity and levels of majority of gene products must be tightly regulated for proper cellular functions. For instance, acetylation enhances the function of transcription factor Gata1 in erythroid cells but at the same time signals its ubiquitination, suggesting that transcription factor activation and degradation are directly linked [23]. We assume that in response to physiological signaling miR-155 restrains the level of uc.160 to prevent the accelerated cell cycle arrest and apoptosis and maintain the homeostasis in gastric epithelial cells. Further investigation should be conducted for the precise reasons why uc.160 needs to be repressed by miR-155 under pathological conditions.
In summary, our study shows that both uc.160 and miR-155 may function as tumor suppressors in human GC. Interestingly, uc.160 is directly repressed by miR-155 in GC. Our data provides strong evidence on the crucial role of T-UCR in GC though the exact mechanism of how miR-155 coordinates uc.160 in the pathogenesis of GC is still unclear. Future studies are needed to elucidate the specific gene regulatory mechanisms involved in uc.160 and miR-155 in GC.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (Grant No. 81670186, D.Y.), grants from the Priority Academic Program Development of Jiangsu Higher Education Institution (Veterinary Medicine, D.Y.) and the science and technology project fund of Yangzhou City [No. YZ 2016052].
Disclosure of conflict of interest
None.
References
- 1.Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe C, Hamadeh RR, Moore A, Werdecker A, Gessner BD, Te Ao B, McMahon B, Karimkhani C, Yu C, Cooke GS, Schwebel DC, Carpenter DO, Pereira DM, Nash D, Kazi DS, De Leo D, Plass D, Ukwaja KN, Thurston GD, Yun Jin K, Simard EP, Mills E, Park EK, Catala-Lopez F, deVeber G, Gotay C, Khan G, Hosgood HD 3rd, Santos IS, Leasher JL, Singh J, Leigh J, Jonas JB, Sanabria J, Beardsley J, Jacobsen KH, Takahashi K, Franklin RC, Ronfani L, Montico M, Naldi L, Tonelli M, Geleijnse J, Petzold M, Shrime MG, Younis M, Yonemoto N, Breitborde N, Yip P, Pourmalek F, Lotufo PA, Esteghamati A, Hankey GJ, Ali R, Lunevicius R, Malekzadeh R, Dellavalle R, Weintraub R, Lucas R, Hay R, Rojas-Rueda D, Westerman R, Sepanlou SG, Nolte S, Patten S, Weichenthal S, Abera SF, Fereshtehnejad SM, Shiue I, Driscoll T, Vasankari T, Alsharif U, Rahimi-Movaghar V, Vlassov VV, Marcenes WS, Mekonnen W, Melaku YA, Yano Y, Artaman A, Campos I, MacLachlan J, Mueller U, Kim D, Trillini M, Eshrati B, Williams HC, Shibuya K, Dandona R, Murthy K, Cowie B, Amare AT, Antonio CA, Castaneda-Orjuela C, van Gool CH, Violante F, Oh IH, Deribe K, Soreide K, Knibbs L, Kereselidze M, Green M, Cardenas R, Roy N, Tillmann T, Li Y, Krueger H, Monasta L, Dey S, Sheikhbahaei S, Hafezi-Nejad N, Kumar GA, Sreeramareddy CT, Dandona L, Wang H, Vollset SE, Mokdad A, Salomon JA, Lozano R, Vos T, Forouzanfar M, Lopez A, Murray C, Naghavi M. The global burden of cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 5.Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, Croce CM, Calin GA, Esteller M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, Terracciano L, Croce CM, Patel T. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2011;108:786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sana J, Hankeova S, Svoboda M, Kiss I, Vyzula R, Slaby O. Expression levels of transcribed ultraconserved regions uc.73 and uc.388 are altered in colorectal cancer. Oncology. 2012;82:114–118. doi: 10.1159/000336479. [DOI] [PubMed] [Google Scholar]
- 8.Honma R, Goto K, Sakamoto N, Sekino Y, Sentani K, Oue N, Yasui W. Expression and function of Uc.160+, a transcribed ultraconserved region, in gastric cancer. Gastric Cancer. 2017;20:960–969. doi: 10.1007/s10120-017-0714-9. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han S, Yang S, Cai Z, Pan D, Li Z, Huang Z, Zhang P, Zhu H, Lei L, Wang W. Anti-Warburg effect of rosmarinic acid via miR-155 in gastric cancer cells. Drug Des Devel Ther. 2015;9:2695–2703. doi: 10.2147/DDDT.S82342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci (Lond) 2011;121:141–158. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 14.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Ma T, Huang C, Hu T, Li J. The pivotal role of microRNA-155 in the control of cancer. J Cell Physiol. 2014;229:545–550. doi: 10.1002/jcp.24492. [DOI] [PubMed] [Google Scholar]
- 16.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY, Cheng JQ. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim BH, Hong SW, Kim A, Choi SH, Yoon SO. Prognostic implications for high expression of oncogenic microRNAs in advanced gastric carcinoma. J Surg Oncol. 2013;107:505–510. doi: 10.1002/jso.23271. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Xie S, Liu M, Chen Z, Liu X, Wang L, Li D, Zhou Y. The clinical significance of downregulation of mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric cancer tumorigenesis. Int J Oncol. 2014;45:197–208. doi: 10.3892/ijo.2014.2415. [DOI] [PubMed] [Google Scholar]
- 19.Ma Z, Ma Y, Xia Q, Li Y, Li R, Chang W, Chen J, Leng Z, Tao K. MicroRNA-155 expression inversely correlates with pathologic stage of gastric cancer and it inhibits gastric cancer cell growth by targeting cyclin D1. J Cancer Res Clin Oncol. 2016;142:1201–1212. doi: 10.1007/s00432-016-2139-y. [DOI] [PubMed] [Google Scholar]
- 20.Marini A, Lena AM, Panatta E, Ivan C, Han L, Liang H, Annicchiarico-Petruzzelli M, Di Daniele N, Calin GA, Candi E, Melino G. Ultraconserved long non-coding RNA uc.63 in breast cancer. Oncotarget. 2017;8:35669–35680. doi: 10.18632/oncotarget.10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Goto K, Ishikawa S, Honma R, Tanimoto K, Sakamoto N, Sentani K, Oue N, Teishima J, Matsubara A, Yasui W. The transcribed-ultraconserved regions in prostate and gastric cancer: DNA hypermethylation and microRNAassociated regulation. Oncogene. 2016;35:3598–3606. doi: 10.1038/onc.2015.445. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez-Hernandez A, Ray P, Litos G, Ciro M, Ottolenghi S, Beug H, Boyes J. Acetylation and MAPK phosphorylation cooperate to regulate the degradation of active GATA-1. EMBO J. 2006;25:3264–3274. doi: 10.1038/sj.emboj.7601228. [DOI] [PMC free article] [PubMed] [Google Scholar]