Abstract
Previous studies have suggested that circular RNAs (circRNAs) serve crucial functions in various human diseases, including cancer. The results of the present study demonstrated that the expression levels of circ-0067934 were significantly increased in non-small cell lung cancer (NSCLC) tissues and NSCLC cells compared with that in adjacent normal tissues and a normal human bronchial epithelium cell line, as assessed using reverse transcription-quantitative polymerase chain reaction. Increased expression levels of circ-0067934 were associated with lymph node metastasis and advanced tumor stage in patients with NSCLC. Increased levels of circ-0067934 were significantly associated with the poor overall survival (OS) time of patients with NSCLC compared with low circ-0067934 expression levels, as assessed using Kaplan-Meier estimator survival curves and log-rank tests. Additionally, circ-0067934 was identified as an independent risk factor for OS time in patients with NSCLC, as assessed using univariate and multivariate Cox proportional hazards regression models. Additionally, cell proliferation and colony formation assays were employed to assess cell proliferation. The results demonstrated that the proliferative ability was significantly inhibited when circ-0067934 was knocked down in NSCLC cells. Therefore, these results indicated that circ-0067934 may be a predictive marker for the prognosis of NSCLC and a target for the treatment of the disease.
Keywords: non-small cell lung cancer, circular RNA, circ-0067934, prognosis, cell proliferation
Introduction
Non-small cell lung cancer (NSCLC) accounts for >80% of lung cancer cases worldwide (1). Tumor proliferation, invasion and metastasis lead to increased mortality, accounting for ~1.4 million cases of mortality per year. Despite advances in surgery and in the development of novel drugs for NSCLC, the outcome for affected patients remains unsatisfactory, with a 5-year overall survival (OS) rate of 15% (2,3). The identification of biological markers for predicting the prognosis of NSCLC and potential targets for the treatment of NSCLC is required.
Circular RNAs (circRNAs) are a class of non-coding RNA molecules that shape a covalently closed continuous loop with no 5′-3′ polarity and a polyadenylated tail (4,5). Recent advances in high-throughput RNA sequencing have contributed to the identification of a number of circRNAs that are involved in tumor progression (6). For example, it has been demonstrated that hsa-circ-0000190 is downregulated in gastric cancer and may be used as a novel non-invasive biomarker for the diagnosis of gastric cancer (7). circRNA plasmacytoma variant translocation 1 was identified as an independent prognostic marker for OS and disease-free survival time in patients with gastric cancer (8). circRNAs have also been associated with progression and metastasis in NSCLC. For example, it was demonstrated that hsa-circ-0013958 promoted cell proliferation and invasion, and inhibited cell apoptosis in lung adenocarcinoma (9). Additionally, circ-0067934 was upregulated in esophageal squamous cell carcinoma and promoted proliferation (10). However, the function of circ-0067934 in NSCLC remains unclear.
The aim of the present study investigated the clinical role and functional significance of circ-0067934. A hypothesis was that circ-0067934 would be significantly increased in NSCLC tissues and cells and that these increased expression levels of circ-0067934 would be associated with prognosis in patients with NSCLC. Additionally, the present study hypothesized that circ-0067934 overexpression affected cell proliferation ability. Thus, the aim of the present study was to investigate whether circ-0067934 could represent a predictive marker for the prognosis of NSCLC and a target for the treatment of the disease.
Patients and methods
Patients
A total of 79 NSCLC tissues and adjacent non-cancerous tissues were obtained from patients (age ranged from 34–80 years, mean 55± 14.22 years) enrolled in the Department of Thoracic Surgery, The First Hospital of Jilin University (Jilin, China) between April 2010 and March 2013. Two experienced pathologists confirmed the diagnosis of NSCLC. The clinical and pathological characteristics of the patients with NSCLC are presented in Table I. The Institutional Review Board of The First Hospital of Jilin University approved the present study and written informed consent was collected from all patients.
Table I.
Association between the expression of circ-0067934 and the clinicopathological features of patients with non-small cell lung cancer.
| Expression of circ-0067934 | ||||
|---|---|---|---|---|
| Variable | Patients (n=79) | Low (n=38) | High (n=41) | P-value |
| Age, years | 0.548 | |||
| ≤60 | 60 | 30 | 30 | |
| >60 | 19 | 8 | 11 | |
| Sex | 0.837 | |||
| Male | 57 | 27 | 30 | |
| Female | 22 | 11 | 11 | |
| Smoking history | 0.745 | |||
| No | 38 | 19 | 19 | |
| Yes | 41 | 19 | 22 | |
| Tumor size, cm | 0.576 | |||
| <5 | 39 | 20 | 19 | |
| >5 | 40 | 18 | 22 | |
| Lymph node metastasis | 0.018 | |||
| Negative | 52 | 30 | 22 | |
| Positive | 27 | 8 | 19 | |
| Tumor differentiation | 0.464 | |||
| Well | 26 | 13 | 13 | |
| Moderately | 25 | 14 | 11 | |
| Poorly | 28 | 11 | 17 | |
| TNM stage | 0.012 | |||
| I–II | 49 | 29 | 20 | |
| III–IV | 30 | 9 | 21 | |
circRNA, circular RNA; TNM, Tumor-Node-Metastasis.
Cell culture
Human NSCLC cell lines (A549, H1299, SK-MES-1, PC-9) and a normal human bronchial epithelial cell line (BEAS-2B) were obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured in RPMI-1640 medium (Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Life Technologies; Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere containing 5% CO2.
Cell transfection
Two specific small interfering RNAs (siRNAs) targeting human circ-0067934 were used. The target sequences of the circ-0067934 siRNAs were as follows: siRNA-1 sense, 5′-UGUUGAUUGGGAUAUGUUAUU-3′ and antisense, 5′-UAACAUAUCCCAAUCAACAUU-3′; and siRNA-2 sense, 5′-CCGAAAUGUUGAUUGGGAUTT-3′ and antisense, 5′-AUCCCAAUCAACAUUUCGGTT-3′. The siRNAs and their matched negative controls (si-NC) were synthesized by Thermo Fisher Scientific, Inc. A549 and SK-MES-1 cells were transfected with siRNA-1 (20 µM), siRNA-2 (20 µM) and si-NC using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following transfection at 48 h, cells were harvested for subsequent experimentation.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from Human NSCLC tissues and adjacent normal tissues or NSCLC cell lines (A549, H1299, SK-MES-1, PC-9) and a normal human bronchial epithelial cell line (BEAS-2B) cells using TRIzol reagent (Thermo Fisher Scientific, Inc.). A total of 1 µg RNA was reverse-transcribed using a Reverse Transcription kit (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was performed using SYBR Premix Ex Taq II kit (Takara Biotechnology Co., Ltd.) on the ABI 7900HT Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The sequences for the primers used were as follows: circ-0067934 forward, 5′-TAGCAGTTCCCCAATCCTTG-3′ and reverse, 5′-CACAAATTCCCATCATTCCC-3′; and GAPDH forward, 5′-GGGAGCCAAAAGGGTCAT-3′ and reverse, 5′-GAGTCCTTCCACGATACCAA-3′. RT-qPCR was conducted at 95°C for 1 min, followed by 40 cycles of 95°C for 30 sec and 60°C for 40 sec. The relative mRNA expression was calculated using the 2−ΔΔCq method (11) and normalized to GAPDH.
Cell proliferation assay
Cells (SK-MES-1 and A549) were seeded in a 96-well plate at a density of 2×103 cells/well. Following overnight incubation at 37°C in a humidified atmosphere containing 5% CO2, the cells were transfected with si-NC, si-circ-0067934-1 (S1) or si-circ-0067934-2 (S2). Following incubation for 24, 48, 72 and 96 h, 10 µl Cell Counting Kit-8 solution (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to each well for 2 h at 37°C. The absorbance was read at 450 nm using a spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Colony formation assay
Cells (SK-MES-1 and A549) were seeded in a 12-well plate at a density of 400 cells/well. Following overnight incubation at 37°C in a humidified atmosphere containing 5% CO2, the cells were transfected with si-NC, si-circ-0067934-1 (S1) or si-circ-0067934-2 (S2) for 14 days. Cell colonies were fixed with 100% methanol at room temperature for 20 min and then stained with 0.1% crystal violet for 20 min at room temperature. Colonies were then counted under a light microscope (Olympus Corporation, Tokyo, Japan).
Statistical analysis
Data were analyzed using SPSS software (version 20.0; IBM Corp., Armonk, NY, USA). Data are expressed as the mean ± standard deviation. Results were analyzed using Student's t-test when only 2 groups were compared, or using one-way analysis of variance with Fishers's least significant difference post-hoc test when >2 groups were compared. A χ2 test was used to analyze the association between circ_0067934 expression and clinicopathological factors. Kaplan-Meier survival analysis and log rank test was used to estimate overall survival of the patients with NSCLC. Univariate and multivariate Cox proportional hazard regression analysis was used to evaluate the risk factors of NSCLC survival. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of circ-0067934 is increased in NSCLC tissues and cells
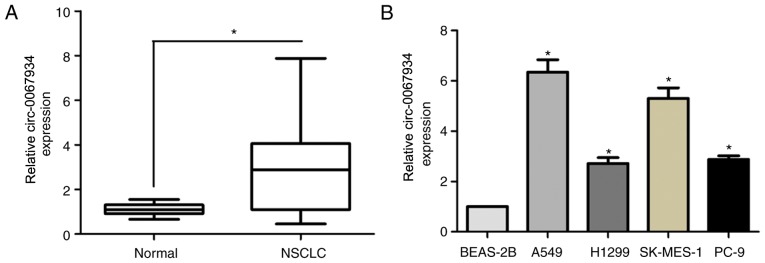
The expression of circ-0067934 in 79 pairs of tumor tissues and adjacent non-cancerous tissues was analyzed using RT-qPCR. The results demonstrated that the expression of circ-0067934 in NSCLC tissues was significantly increased compared with that in adjacent normal tissues (Fig. 1A). Furthermore, RT-qPCR was performed to analyze the expression of circ-0067934 in human NSCLC cell lines (A549, H1299, SK-MES-1 and PC-9) and in a normal human bronchial epithelium cell line, BEAS-2B. As presented in Fig. 1B, the expression of circ-0067934 was significantly increased in the NSCLC cells compared with that in the BEAS-2B cells. These results indicated that the expression of circ-0067934 was increased in NSCLC.
Figure 1.
Expression of circ-0067934 is increased in NSCLC tissues and cells. (A) The expression of circ-0067934 was detected in 79 pairs of tumor tissues and adjacent non-cancerous tissues using RT-qPCR. (B) The expression of circ-0067934 was detected in NSCLC cell lines (A549, H1299, SK-MES-1 and PC-9) and in the normal human bronchial epithelial BEAS-2B cell line using RT-qPCR, BEAS-2B cells represented to control group, the circ-0067934 expression of which was set at a value of 1. *P<0.05 vs. BEAS-2B or as indicated. circRNA, circular RNA; NSCLC; non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Expression of circ-0067934 is associated with prognosis of patients with NSCLC
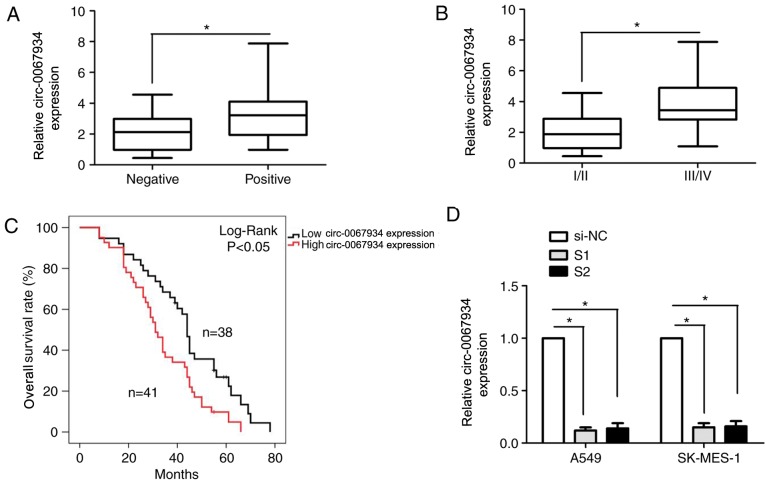
The association between the expression of circ-0067934 and clinicopathological features was evaluated. Based on the median expression of circ-0067934 (2.95 fold) in the tumor tissues, the patients were divided into a high circ-0067934-expression group and low circ-0067934-expression group (Table I). circ-0067934 was markedly associated with positive lymph node metastasis (Fig. 2A) and advanced tumor-node-metastasis (TNM) stage (12) (Fig. 2B) in patients with NSCLC. However, the expression of circ-0067934 was not associated with other clinicopathological factors, including age, sex and tumor size (Table I). Kaplan-Meier estimator survival curves and log-rank tests demonstrated that increased expression of circ-0067934 was significantly associated with a poor OS time in patients with NSCLC compared with low circ-0067934 expression (Fig. 2C). Univariate and multivariate Cox proportional hazards regression models demonstrated that lymph node metastasis [relative risk (RR), 1.882; 95% confidence interval (CI), 1.344–2.603; P<0.05], advanced TNM stage (RR, 2.008; 95% CI, 1.322–3.113; P<0.05) and high circ-0067934 expression (RR, 2.133; 95% CI, 1.677–3.251; P<0.05) were independent risk factors of OS time in patients with NSCLC (Table II). Overall, the results indicated that the expression of circ-0067934 was an independent risk factor for OS time in patients with NSCLC.
Figure 2.
Expression of circ-0067934 is associated with the prognosis of patients with NSCLC. The association between circ-0067934 expression and (A) lymph node metastasis and (B) Tumor-Node-Metastasis stage in patients with NSCLC. (C) The association between circ-0067934 expression and overall survival time in patients with NSCLC was analyzed using Kaplan-Meier analysis and the log-rank test. (D) The expression of circ-0067934 was detected using reverse transcription-quantitative polymerase chain reaction following treatment with si-NC, S1 or S2 in A549 and SK-MES-1 cells. *P<0.05. circRNA, circular RNA; NSCLC; non-small cell lung cancer; si/siRNA, small interfering RNA; NC, negative control; S1, si-circ-0067934-1; S2, si-circ-0067934-2; positive, patients with NSCLC with lymph node metastasis; negative, patients with NSCLC without lymph node metastasis.
Table II.
Univariate and multivariate Cox analysis of overall survival time in patients with non-small cell lung cancer.
| Univariate cox analysis | Multivariate cox analysis | |||
|---|---|---|---|---|
| Clinicopathological characteristics | RR (95% CI) | P-value | RR (95% CI) | P-value |
| Age | 0.645 (0.345–1.522) | 0.876 | ||
| Sex | 1.064 (0.466–1.822) | 0.487 | ||
| Smoking history | 1.224 (0.688–1.769) | 0.285 | ||
| Tumor size | 1.031 (0.525–1.822) | 0.502 | ||
| Lymph node metastasis | 1.968 (1.422–2.912) | 0.003 | 1.882 (1.344–2.603) | 0.007 |
| Tumor differentiation | 1.288 (1.088–2.112) | 0.202 | ||
| TNM stage | 2.448 (1.556–3.554) | 0.001 | 2.008 (1.322–3.113) | 0.001 |
| circ-0067934 | 2.677 (1.891–3.998) | 0.001 | 2.133 (1.677–3.251) | 0.001 |
circRNA, circular RNA; TNM, Tumor-Node-Metastasis; NSCLC, non-small cell lung cancer; RR, relative risk; CI, confidence interval.
Downregulation of circ-0067934 suppresses the proliferation of NSCLC cells
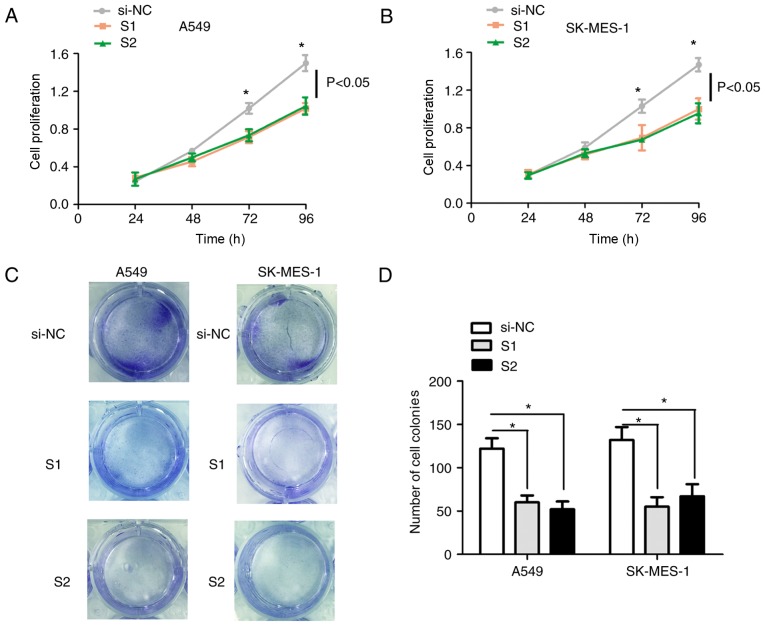
To assess the effects of circ-0067934 on cellular proliferation, loss-of-function experiments were performed in A549 and SK-MES-1 cells (Fig. 2D). Downregulation of circ-0067934 significantly decreased cellular proliferation in A549 (Fig. 3A) or SK-MES-1 (Fig. 3B) cells at the 72 and 96 h time intervals compared with that in the control group, as assessed using a CCK-8 assay. Additionally, the number of cell colonies was markedly decreased when the expression of circ-0067934 was knocked down in A549 (Fig. 3C) and in SK-MES-1 (Fig. 3D) compared with that in the control group, as assessed using a cell colony formation assay (Fig. 3C and D). These results demonstrated that the downregulation of circ-0067934 suppressed the proliferative ability of the NSCLC cells.
Figure 3.
Downregulation of circ-0067934 suppresses the proliferation of NSCLC cells. Cell proliferation was analyzed in (A) A549 and (B) SK-MES-1 cells that were treated with si-NC, S1 or S2 for 24, 48, 72 and 96 h. *P<0.05 vs. si-NC group. (C) Representative images and (D) numbers of colonies of A549 and SK-MES-1 cells that were treated with si-NC, S1 or S2. *P<0.05. circRNA, circular RNA; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; si/siRNA, small interfering RNA; NC, negative control; S1, si-circ-0067934-1; S2, si-circ-0067934-2.
Discussion
Recent studies have revealed that circRNAs serve as key regulators in the development of human diseases, including cancer (13,14). Comprehensive profiling of circRNAs revealed the regulatory function of the circRNA-100338/miR-141-3p signaling pathway in hepatitis B-associated hepatocellular carcinoma (15). A previous study demonstrated that hsa-circRNA-0020397 regulated cell viability, apoptosis and invasion by promoting the expression of miR-138 targets, including telomerase reverse transcriptase (TERT) and programmed death ligand 1 (PD-L1), in colorectal cancer (16). It has been identified that increased levels of circRNA ubiquitin-associated protein 2 act as a sponge for miR-143 to promote the progression of osteosarcoma (17). These findings indicate that circRNAs may be involved in tumor progression.
A previous study revealed that the expression level of hsa-circRNA-100876 was significantly elevated in NSCLC tissues. Additionally, patients with increased levels of hsa-circRNA-100876 exhibited a significantly decreased survival time compared with the low hsa-circRNA-100876-expression group (18). In the present study, it was demonstrated that the expression levels of circ-0067934 were significantly increased in NSCLC tissues compared with those in adjacent normal tissues. Increased expression levels of circ-0067934 were associated with lymph node metastasis, advanced tumor stage and decreased OS time. Univariate and multivariate analysis demonstrated that circ-0067934 was an independent risk factor for OS time. These results indicated that circ-0067934 may act as a prognostic marker of NSCLC.
A number of circRNAs have been identified to regulate cell proliferation in hepatocellular carcinoma. It was revealed that mirRNA-200b suppressed the invasion and migration of hepatocellular carcinoma by downregulating Rho A and hsa-circRNA-000839 (19). circRNA oiled-coil domain-containing protein 66 controlled multiple pathological processes, including cell proliferation, migration, invasion and anchorage-independent proliferation, in colon cancer (20). A previous study demonstrated that hsa-circRNA-104916 was downregulated in gastric cancer and suppressed the migration and invasion of gastric cancer cells (21). In the present study, the effects of circ-0067934 on the cell proliferation ability were evaluated using cell proliferation and colony formation assays. The results demonstrated that the proliferative ability was significantly inhibited following circ-0067934-knockdown in NSCLC cells.
In conclusion, the results of the present study demonstrated that the expression of circ-0067934 was increased in NSCLC and indicated a poor OS time for affected patients. Additionally, downregulation of circ-0067934 decreased the proliferative ability of NSCLC cells. These results indicated that circ-0067934 may be a predictive marker for the prognosis of NSCLC and a target for the treatment of the disease.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed in the present study are available from the corresponding author upon reasonable request.
Authors' contributions
QZ, TW and GL performed the experiments. LZ, BL, SS and BL performed the data interpretation. QZ and TW drafted the manuscript and are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. QZ coordinated and supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Institutional Review Board of The First Hospital of Jilin University (Jilin, China) approved the present study and written informed consent was collected from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Raungrut P, Wongkotsila A, Lirdprapamongkol K, Svasti J, Geater SL, Phukaoloun M, Suwiwat S, Thongsuksai P. Prognostic significance of 14-3-3γ overexpression in advanced non-small cell lung cancer. Asian Pac J Cancer Prev. 2014;15:3513–3518. doi: 10.7314/APJCP.2014.15.8.3513. [DOI] [PubMed] [Google Scholar]
- 3.Grinberg-Rashi H, Ofek E, Perelman M, Skarda J, Yaron P, Hajdúch M, Jacob-Hirsch J, Amariglio N, Krupsky M, Simansky DA, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res. 2009;15:1755–1761. doi: 10.1158/1078-0432.CCR-08-2124. [DOI] [PubMed] [Google Scholar]
- 4.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: A novel type of biomarker for cancer. Breast Cancer. 2017;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 5.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, Han S, Wu G. Hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. Febs J. 2017;284:2170–2182. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 10.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, Yin R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Sobin LH, Gospodarowicz MK, Wittekind CH. TNM classification of malignant tumours. 7th. Wiley-Blackwell; Hoboken, NJ: 2009. [Google Scholar]
- 13.Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, Finn SP. Circular RNAs: Biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett SP, Salzman J. Circular RNAs: Analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, Zhou J, Tang ZY, Huang XY. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis, and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41:1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W, Tong D, Wu D, Li C, Wei Q, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8:61687–61697. doi: 10.18632/oncotarget.18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, Nan KJ. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Wang BG, Li JS, Liu YF, Xu Q. MicroRNA-200b suppresses the invasion and migration of hepatocellular carcinoma by downregulating RhoA and circRNA_000839. Tumour Biol. 2017;39:1010428317719577. doi: 10.1177/1010428317719577. [DOI] [PubMed] [Google Scholar]
- 20.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Zhen L, Zhang Y, Zhao L, Liu H, Cai D, Chen H, Yu J, Qi X, Li G. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther. 2017;10:3521–3529. doi: 10.2147/OTT.S136347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed in the present study are available from the corresponding author upon reasonable request.