Abstract
The etiology and pathogenesis of bladder cancer (BCa) is complex. MicroRNA (miRNA) has been implicated in BCa. Targeting of signal transducer and activator of transcription 3 (STAT3) by miR-124 to regulate tumorigenesis has been demonstrated in other types of cancer. In the present study, miR-124 levels were downregulated in the BCa T24 cell line and STAT3 was increased in BCa cell lines. Transfection of miR-124 mimics into T24 cells significantly inhibited STAT3 expression. A luciferase assay confirmed that miR-124 directly targeted the STAT3 3′untranslated region to inhibit STAT expression. Knockdown of STAT3 expression led to increased apoptosis of T24 cells and reduced tumor growth in vitro. The results demonstrated the molecular mechanisms and biological functions of the miR-124/STAT3 signal pathway at the cellular level and indicate the potential of miR-124 as a therapeutic target for BCa.
Keywords: microRNA-124, signal transducer and activator of transcription 3, bladder cancer
Introduction
Bladder cancer (BCa) is one of the most common genitourinary malignancies worldwide that causes an estimated 150,000 mortalities annually globally (1). The incidence of BCa is complex and includes genetic and environmental factors (2). Smoking is an established risk factor (3). Clinical data indicate that BCa has a high mortality rate, due to its propensity to recur and metastasize, and the absence of symptoms in the early stage of the disease (3). Therefore, early diagnosis and appropriate treatment is the key to improving survival in patients with BCa (4,5). With the lack of effective treatments and monitoring strategies, novel diagnostic markers and effective treatment strategies for BCa are imperative.
MicroRNAs (miRNAs) are small (22 nucleotide long) noncoding RNA molecules that are highly conserved in eukaryotes (6). Generally, miRNAs identify their target gene by imperfectly base pairing with their 3′untranslated region (UTR) to exert their biological function (7). In 2007, the expression profiles of miRNA in BCa were examined, and 10 miRNAs were identified as being upregulated (8). Subsequently, the use of the Solexa sequencing technology identified 33 miRNAs that were upregulated and 41 miRNAs that were downregulated in BCa (9). These data indicate that the dysregulation of miRNA is markedly associated with BCa tumorigenesis. miR-124 is a miRNA highly expressed in brain tissue that is the most extensively studied miRNA in the nervous system (10). Previously, miR-124 was identified as a tumor suppressor that downregulates a variety of human tumor types, including gastric, breast and colorectal cancer, and BCa (11–14). Furthermore, signal transducer and activator of transcription 3 (STAT3) has been demonstrated to be an miR-124-downregulated target gene in tumors (15). However, the mechanisms underlying miR-124-mediated inhibition of STAT3 in BCa are unknown.
The present study demonstrated a downregulation of miR-124 in T24 BCa cells that was inversely associated with STAT3 expression. Furthermore, miR-124 inhibited STAT3 expression by directly targeting its 3′UTR. Knockdown of STAT3 significantly promoted apoptosis and suppressed cell cycle progression, migration, proliferation, and colony formation in T24 cells. The collective results suggest that miR-124 may directly inhibit the expression of STAT3 in BCa, and that miR-124 may suppress BCa tumorigenesis by targeting STAT3.
Materials and methods
Cell culture and RNA extraction
Human BCa T24, UM-UC-3, SW780, HT1376, RT4 and J82 cell lines, immortalized human bladder epithelium SV-HUC-1 cells and the 293 cell line were purchased from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured according to the manufacturer's instructions at 37°C in an atmosphere of 5% CO2. Total mRNA and miRNA were independently extracted from the cells using RNAzol RT RNA Isolation Reagent (Molecular Research Center, Inc., Cincinnati, OH, USA) and stored at −20°C for subsequent analysis.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) for miR-124 and STAT3
The expression of miR-124 was quantified using an All-in-one miRNA qPCR kit (GeneCopoeia, Inc., Rockville, USA) and U6 was used as the control. The primers used for the RT-PCR were: miR-124 (forward 5′-GCGGCCGTGTTCACAGCGGACC-3′ and reverse 5′-GTGCAGGGTCCGAGGT-3′) and U6 (forward 5′-CGCTTCGGCAGCACATATACTA-3′ and reverse 5′-CGCTTCACGAATTTGCGTGTCA-3′). To determine the mRNA expression of STAT3, the total RNA in T24 cells was reverse transcribed into cDNA at 37°C using Eastep RT Master Mix kit (Promega Corporation, Madison, WI, USA), then STAT3 mRNA was quantified using SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan) and GAPDH was used as the control. The thermocycling conditions were as follows: 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec, 60°C for 34 sec, then 72°C for 45 sec. The primers used for qPCR were: STAT3 (forward 5′-ATCACGCCTTCTACAGACTGC-3′ and reverse 5′-CATCCTGGAGATTCTCTACCACT-3′) and GAPDH (forward 5′-CCACTCCTCCACCTTTGAC-3′ and reverse 5′-ACCCTGTTGCTGTAGCCA-3′). Relative expression was calculated using the 2−ΔΔCq method (16).
Western blot analysis
T24 cells were centrifuged in 1,000 × g for 5 min at 37°C subsequent to the addition of 1 ml ice-cold PBS and washed three times with ice-cold PBS and lysed using a Nuclear and Cytoplasmic Protein Extraction kit (Beyotime Institute of Biotechnology, Haimen, China) for 30 min at 4°C. The cell lysate protein was centrifuged at 15,000 × g at 4°C for 10 min and the protein was quantified using a BCA Protein Assay kit (Beyotime Institute of Biotechnology). A total of 20 µg protein/lane was separated by 10% SDS-PAGE, and the resolved proteins were transferred to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). The membrane was blocked using 5% skim milk powder at 4°C overnight and then hybridized with the primary antibodies targeting the proteins including GADPH (1:2,500; cat no. ab9485), STAT3 (1:1,000; cat no. ab68153), vascular endothelial growth factor receptor (VEGFR; 1:1,000; cat no. ab11939), B-cell lymphoma 2 (Bcl-2; 1:1,000; cat no. ab32124), B-cell lymphoma-extra large (Bcl-xl; 1:1,000; cat no. ab32370), MCL1, Bcl2 family apoptosis regulator (Mcl-1; 1:2,000; cat no. ab32087), Cyclin D1 (1:10,000; cat no. ab134175) and MYC proto-oncogene, BHLH transcription factor (cMyc; 1:1,000; cat no. ab32072) (all from Abcam, Cambridge, UK) at 4°C overnight. Subsequent to washing using PBS with 0.05% Tween-20 three times at 5 min each time, the membrane was incubated with the horseradish peroxidase-conjugated secondary antibody Goat Anti-Rabbit immunoglobulin G H&L (1:2,000; cat no. ab6721; Abcam) conjugated to horseradish peroxidase at 4°C overnight. Results were quantified by the Molecular Imager ChemiDoc XRS+ System 2.0 (Bio-Rad Laboratories, Hercules, CA, USA). GAPDH was used as the control.
Luciferase assay
The whole 3′UTR of the STAT3 gene was cloned and amplified in 293 cells. TargetScan 6.2 bioinformatics prediction software (http://www.targetscan.org/; date of access, 20 February 2017) was used to predict their binding sites in 20 February, 2017. The conserved sites of miR-124 were selected and then target gene STAT3 was selected for sites in its 3′UTR. A mutation in the 3′UTR of the STAT3 gene at the putative binding site of miR-124 was generated with the QuickChange Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc. Santa Clara, CA, USA). The wild and mutant STAT3 genes were cloned into the pmirGlO-vector (Promega Corporation) immediately downstream of the Renilla luciferase gene. Cells were co-transfected with pmirGlO-3′UTR-STAT3 (50 ng), miR-124 (50 nM), or scramble mimic (50 nM) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cell lysates were prepared using Passive Lysis Buffer (Promega Corporation) 48 h after transfection. The luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega Corporation) and normalized to Renilla luciferase activity.
Transfection
T24 (1×105 cells/plate) were plated in 6-well plates at 37°C overnight and transfected with miR-124 mimics or NC mimics (20 nM; GeneCopoeia, Inc.) using Lipofectamine® 2000. The miR-124 mimic sequence used was: Forward, 5′-GCTCTAGAGGCCTCTCTCTCCGTGTTCCACAGCGGACCTTGATTTAAATGTCCATACAATTAAGGCACGCGGTTGAATGCCAAGAATGGGGCTG-3′ and reverse, 5′-CGGGATCCCAGCCCCATTCTTGGCATTCACCGCGTGCCTTAATTGTATGGACATTTAAATCAAGGTCCGCTGTGAACACGGAGAGAGAGGCCT-3′. Following transfection for 6 h at 25°C, the Opti-MEM medium (Gibco; Thermo Fisher Scientific, Inc.) without serum was changed with the fresh medium. Then cells were assayed by RT-qPCR and western blot analysis for each group according to the aforementioned protocols following culture for 48 h.
Construction of plasmids
Small interfering RNA Target Finder online design software (Ambion; Thermo Fisher Scientific, Inc.) was used to select a segment (5′-AAGAGTCAAGGAGACATGCAA-3′, 670–690 bp) in the coding region of STAT3 (GenBank serial no. NM139276) as a target sequence. Then, the corresponding DNA template strands containing restriction sites for HindIII and BamH I were designed for the target sequences. The template strand sequences were as follows: Sense, 5′-GATCCGAGTCAAGGAGACCATGCAATTCAAGAGATTGCATGTCTCCTTGACTCTTTTTTGG-3′; and anti-sense, 5′-AGCTTTTCCAAAAAAGAGTCAAGGAGACATGCAATCTCTTGAATTGCATGTCTCCTTGACTCG-3′. The shRNA sequences were obtained by PCR amplification, and vector plasmid pGENESIL (Wuhan Genesil Biotechnology Co., Ltd., Wuhan, China) were digested and connected by HindIII and BamH I enzymes (Thermo Fisher Scientific, Inc.). The recombinant plasmid (pGENESIL-shRNA-STAT3) was amplified and infected (0.8 µg plasmid, at 25°C for 20 min) into the T24 cells. After 6 weeks of G418 (800 µg/ml; Invitrogen; Thermo Fisher Scientific, Inc.) selection, stably-transfected cells (STAT3-shRNA) were used for subsequent experiments.
Cell cycle and apoptosis analysis by flow cytometry
The adherent cells were digested at 37°C with 0.25% trypsin (Bioswamp; Wuhan Myhalic Biotechnological Co., Ltd., Wuhan, China) and centrifuged at 1,000 × g at 25°C for 5 min. The supernatant was discarded and the cells were washed three times with PBS. The cells (1–5×105) were collected and resuspended in 200 µl binding buffer (cat no. C1052-1; Beyotime Institute of Biotechnology) for analysis using the Cell Cycle and Apoptosis Analysis kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol. Analyses were performed using a LSRII Flow Cytometry System equipped with FACSDiva software 4.1 (BD Biosciences, Franklin Lakes, NJ, USA). Data was analyzed with the ModFit LT software package version 4.0 (Verity Software House, Inc., Topsham, ME, USA).
Cell proliferation and colony-forming assays
The WST-1 Cell Proliferation and Cytotoxicity Assay kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol was used to analyze cell viability and cell proliferation at 0, 24, 48, 72 and 96 h. All experiments were performed in triplicate. For the colony formation assay, cells were plated at a density of 200 cells/well in standard culture conditions (5% CO2 and 37°C) for 14 days. The colonies were fixed with 5 ml 4% absolute methanol for 15 min at 25°C, stained at 37°C with 0.1% crystal violet for 20 min and washed three times. Then a light microscope (TS-100F; Nikon, Tokyo, Japan) was used to visualize the results at a magnification of ×40.
Wound healing assay
T24 cells were seeded (1×106 cells/plate) in a 6-well plate. Wounding of the confluent cell growth was performed by scratching with a 10 µl pipette tip 24 h after cell plating. The cells were then cultured for 24 h and wound closure was detected by inverted microscopy (magnification, ×200; Olympus, Tokyo, Japan). The experiment was repeated three times.
Statistical analysis
Statistical analysis was conducted using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). Unpaired Student's t-tests were used to analyze the results. Pearson's linear correlation analysis was used to analyze the association between the expression of STAT3 and miR-124. All experiments were performed in triplicate and the data are expressed as mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
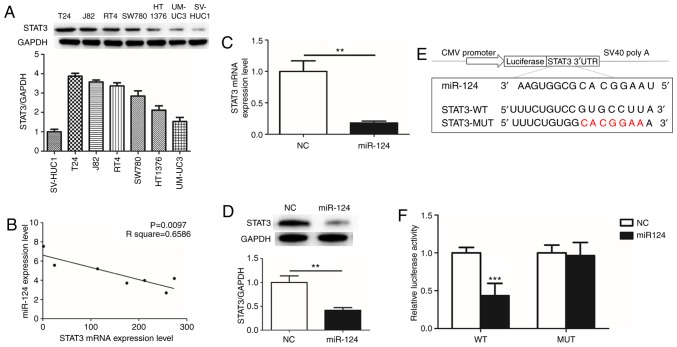
miR-124 and STAT3 are inversely expressed in BCa cell lines
To evaluate the function of STAT3 in human BCa, the expression of STAT3 was detected in 6 human BCa cell lines (T24, UM-UC-3, SW780, HT1376, RT4 and J82) and immortalized human bladder epithelium SV-HUC-1 cells. STAT3 was overexpressed as an oncogene in the BCa cell lines compared with SV-HUC-1 cells. The expression of STAT3 was highest in T24 cells (Fig. 1A). The association between miR-124 and STAT3 in the BCa cell lines was additionally evaluated by RT-qPCR. Notably, there was a significant inverse correlation between miR-124 and STAT3 (Fig. 1B). From these results, it was hypothesized that miR-124 is a potent miRNA regulator of STAT3 expression. T24 cells, which overexpressed STAT3 and exhibited low expression of miR-124, were selected for subsequent experiments.
Figure 1.
Relevance of miR-124 and STAT3 in bladder cancer. (A) The expression of STAT3 protein in bladder cancer cell lines. (B) Relative expression levels of miR-124 and STAT3. (C) Reverse transcription quantitative polymerase chain reaction was used to detect the level of STAT3 mRNA in T24 cell lines following transfection of NC mimics and miR-124 mimics for 48 h. (D) Western blot analysis was used to detect the expression of STAT3 protein in T24 cell lines following transfection of NC mimics and miR-124 mimics for 48 h. (E) Schematic construction of wild-type or mutant miR-124 binding sequences in the STAT3 3′-UTR vector. The mutant binding sequences are in red. (F) Suppressed luciferase activity of wild-type 3′-UTR of STAT3 by pre-miR-124. All data are presented as the mean ± standard deviation of the mean. **P<0.01 and ***P<0.001. miR, microRNA; STAT3, signal transducer and activator of transcription 3; NC, negative control; UTR, untranslated region; CMV promotor, cytomegalovirus promotor; WT, wild type; MUT, mutant.
miR-124 directly targets STAT3 3′UTR
The effects of the transfection of miR-124 mimics on STAT3 mRNA and protein expression levels were determined by RT-qPCR and western blot analysis. There was an ~80% decrease in STAT3 mRNA expression and a 60% decrease in STAT3 protein expression in T24 cells transfected with miR-124 mimics (Fig. 1C and D). This suggested that STAT3 may be downregulated by miR-124. Post-transcriptional regulation by miRNAs involves binding to the 3′UTR of the relevant downstream genes (17). To additionally define the association of miR-124 with STAT3, Target Scan 6.2 bioinformatics prediction software (http://www.targetscan.org/) was used to predict their binding sites. The results revealed that the miR-124 seed-matching sequence in STAT3 3′UTR was ‘CACGGAA’ (Fig. 1E). Luciferase reporter assays demonstrated that the presence of miR-124 resulted in a marked decrease in luciferase activity when the STAT3 plasmid containing wild type 3′UTR was present, while luciferase activity did not change significantly when the mutant 3′UTR was present (Fig. 1F). These data suggested that miR-124 may target STAT3 transcription, potentially contributing to the downregulation of STAT3 expression.
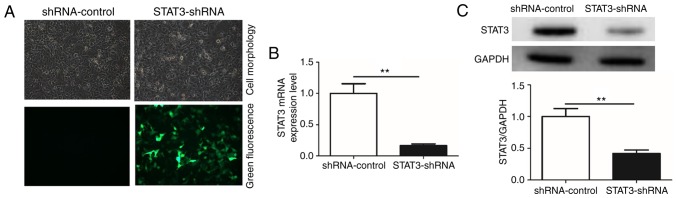
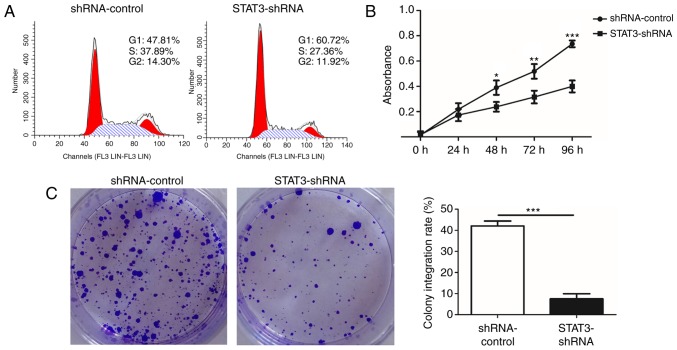
Knockdown of STAT3 inhibits cell cycle progression, proliferation and colony formation of T24 BCa cells in vitro
To additionally investigate the role of miR-124/STAT3 signaling pathway in BCa, a stable cell line with low STAT3 expression (STAT3-shRNA-T24) and a control cell line (shRNA-control-T24) were constructed (Fig. 2A). The result demonstrated that STAT3 mRNA and protein levels in the STAT3-shRNA-T24 were significantly inhibited (Fig. 2B and C). Flow cytometry was conducted to confirm that the knockdown of STAT3 suppressed the growth of T24 BCa cells. A significant increase in numbers of cells in the G1 phase was observed in STAT3-shRNA-T24 cells compared with that in shRNA-control-T24 cells. This increase in G0/G1 cell population was accompanied with a concomitant decrease in cell numbers in S and G2-M phases in the STAT3-shRNA-T24 cells (Fig. 3A). A WST-1 assay was then performed to investigate whether STAT3 exhibited a biological function in the proliferation of T24 cells. A significant inhibition in STAT3-shRNA-T24 was evident (Fig. 3B). Consistent with the result from the WST-1 assay, the colony formation assay revealed markedly fewer and smaller STAT3-shRNA-T24 colonies compared with the shRNA-control-T24 cell line (Fig. 3C).
Figure 2.
Construction and identification of STAT3-shRNA stably-transfected cells. (A) Fluorescence images of shRNA-transfected T24 cells (magnification, ×200). (B) Reverse transcription quantitative polymerase chain reaction was used to detect the level of STAT3 mRNA in STAT3-shRNA-T24 cell lines. (C) Western blot analysis was used to detect the expression of STAT3 protein in STAT3-shRNA-T24 cell lines. All data are presented as the mean ± standard deviation of the mean. **P<0.01. shRNA, short hairpin RNA; STAT3, signal transducer and activator of transcription.
Figure 3.
Knockdown of STAT3 significantly suppresses cell cycle progression, proliferation and colony formation in T24 cells. (A) The effects of knockdown of STAT3 on cell cycle in T24 cells were analyzed by flow cytometry. A total of three independent experiments were performed in each group. (B) Cell viability was examined using an WST-1 assay. (C) Effect of knockdown of STAT3 on the proliferation rate in T24 cells was measured by a colony formation assay. All data are presented as the mean ± standard deviation of the mean. *P<0.05, **P<0.01 and ***P<0.001. shRNA, short hairpin RNA; STAT3, signal transducer and activator of transcription.
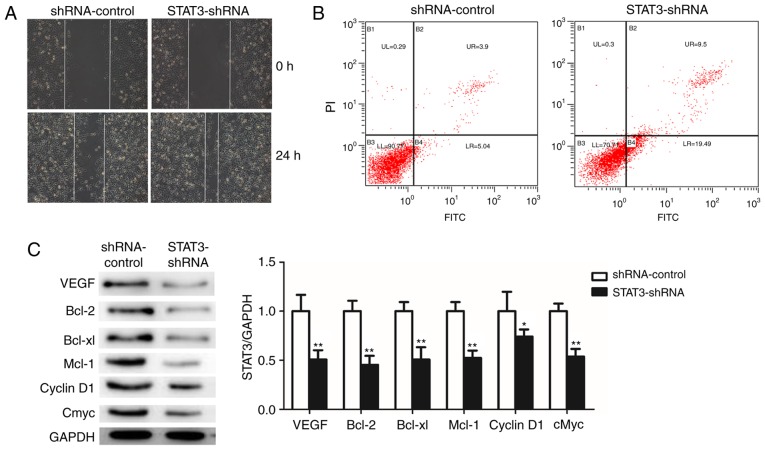
Knockdown of STAT3 promotes apoptosis and suppresses cell migration and STAT3 downstream target genes of T24 BCa cells in vitro
To investigate the effect of STAT3 on the migration of T24 BCa cells, the capacity of STAT3-shRNA-T24 cells to migrate was measured using a wound healing assay. STAT3-shRNA-T24 cells exhibited slower rates of wound closure and decreased numbers of stained cells in the wound healing assay compared with the shRNA-control-T24 cells (Fig. 4A). Furthermore, a marked induction of apoptosis was confirmed in STAT3-shRNA-T24 cells (Fig. 4B). The results demonstrated that the silencing of STAT3 significantly suppressed the migratory capability and induced apoptosis of T24 cells. Finally, the downstream target genes of STAT3 following the knockdown of STAT3 were examined using western blot analysis (Fig. 4C). Notably, cyclin D1, cMyc, Bcl-2, Bcl-xl, Mcl-1 and VEGFR were decreased in the STAT3-shRNA-T24 cells. Taken together, these results demonstrate that STAT3 knockdown repressed cell proliferation, migration, invasion and vasculogenic mimicry, indicating the oncogenic role of STAT3 in BCa.
Figure 4.
Knockdown of STAT3 significantly promotes apoptosis and suppresses cell migration and STAT3 downstream target genes in T24 cells. (A) The effects of knockdown of STAT3 on migration in T24 were analyzed using wound healing assays with a 24 h recovery period. (B) Apoptosis was determined by fluorescence-activated cell sorting. Cells in early apoptosis are in the bottom right quadrant, while cells in late apoptosis are in the top right quadrant. (C) Western blot analysis was used to detect the expression of STAT3 downstream target proteins. All data are presented as the mean ± standard deviation of the mean. *P<0.05 and **P<0.01. shRNA, short hairpin RNA; STAT3, signal transducer and activator of transcription; PI, propidium iodide; FITC, fluorescein isothiocyanate; VEGFR, vascular endothelial growth factor receptor; Bcl-2, B-cell lymphoma 2; Bcl-xl, B-cell lymphoma-extra large; Mcl-1, MCL1, Bcl2 family apoptosis regulator; cMyc, MYC proto-oncogene, BHLH transcription factor.
Discussion
miRNA has been studied intensively since it was suggest to be associated with cancer in 2002 (18). In previous years, a number of miRNAs, including miR-124, have also been identified to be dysregulated in BCa (19). miR-124 inhibits the expression of its downstream target genes, which include ubiquitin like with PHD and ring finger domains 1, cyclin dependent kinase 4 and Rho-associated protein kinase, to suppress BCa tumorigenesis by competitive binding to their 3′UTRs (20–22). The STAT3 oncogene has been demonstrated as a downstream target gene of miR-124 in other types of cancer (23). In the present study, a significant downregulation of miR-124 was evident in BCa cell lines. The results are consistent with the majority of other similar studies. Next, STAT3 expression was detected in BCa cell lines, and it was determined that it was negatively correlated with miR-124. The ectopic overexpression of miR-124 inhibited STAT3 expression. In addition, miR-124 directly bound to the 3′UTR of STAT3. These data suggest a targeted association between STAT3 and miR-124 in BCa.
STAT3 is a recognized oncogene that belongs to the STAT family. In tumors, it is generally always stimulated by a variety of extracellular signals that include growth factors and cytokines (24–26). Its aberrant activation causes abnormal cell proliferation and malignant transformation (27). Previous studies (28,29) have indicated that increased STAT3 activity is closely associated with BCa tumor growth and survival (30). The use of small interfering RNA technology in the present study revealed that the knockdown of STAT3 expression in T24 BCa cells substantially inhibited tumor cell cycle progression, migration, proliferation, colony formation and in vitro invasiveness. This is consistent with one previous study (28). The STAT3 protein ranges between 750 and 795 amino acids in length and contains 6 functional domains: The amino-terminal domain (SH2), coiled-coil domain, DNA binding domain, linker domain, SH2 domain and transactivation domain (31). Under the effects of external stimuli, STAT3 is activated by tyrosine phosphorylation. The activated STAT3 monomer forms a homodimer with the tyrosine phosphorylated SH2 domain of STAT3 (32). The homodimer translocates into the nucleus and binds to the specific DNA response element to regulate the transcriptional activity of downstream target genes involved in the regulation of cell cycle, proliferation and apoptosis (33–35). In human cancer, 7 downstream target genes of STAT3 have been identified: Cell cycle-associated genes (cyclin D1, cMyc); apoptosis-associated genes (Bcl-2, Bcl-xl and Mcl-1) and an angiogenesis-associated gene (VEGFR). The present study demonstrated that knockdown of STAT3 expression significantly suppressed the protein expression of these genes. These observations suggest that STAT3 is a key regulatory factor regulated by miR-124 in BCa, and that targeting the inhibition of its signaling pathway will effectively suppress tumorigenesis through a number of mechanisms.
In conclusion, the data of the present study indicate a novel role of the miR-124/STAT3 signaling pathway in BCa and demonstrate the potential to use miR-124 or STAT3 as a diagnostic marker or therapeutic tool for human BCa. However, there were several limitations in the present study. The study was validated in only one bladder cancer cell line¸ T24, therefore the results of this study suggest that this may be cell-type specific. Therefore, the association between miR-124 and STAT3 in vitro requires additional exploration.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from Project of Hainan Natural Science Foundation of China (grant no. 20168304) and Project capital of Hainan Provincial Department of health (grant no. 2013 self raising-10).
Availability of data and materials
All datasets used during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SW was responsible for the project and performed the experiments. PL lead experimental work and conducted immunohistochemistry work; GW actualized the fluorescence detection. YH conducted the fluorescence detection and specimen treatment. The immunohistochemistry experiments were performed by PS, JC and YW. JY implemented autofluorescence acquisition and software system processing.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malats N, Real FX. Epidemiology of bladder cancer. Hematol Oncol Clin North Am. 2015;29:177–189, vii. doi: 10.1016/j.hoc.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C, Shariat S, Lotan Y. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63:234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 4.Käsmann L, Manig L, Janssen S, Rades D. Chemoradiation including paclitaxel for locally recurrent muscle-invasive bladder cancer in elderly patients. In Vivo. 2017;31:239–241. doi: 10.21873/invivo.11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ide H, Inoue S, Miyamoto H. Histopathological and prognostic significance of the expression of sex hormone receptors in bladder cancer: A meta-analysis of immunohistochemical studies. PLoS One. 2017;12:e0174746. doi: 10.1371/journal.pone.0174746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilczynska A, Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33. doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 9.Chen YH, Wang SQ, Wu XL, Shen M, Chen ZG, Chen XG, Liu YX, Zhu XL, Guo F, Duan XZ, et al. Characterization of microRNAs expression profiling in one group of Chinese urothelial cell carcinoma identified by Solexa sequencing. Urol Oncol. 2013;31:219–227. doi: 10.1016/j.urolonc.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Xia H, Cheung WK, Ng SS, Jiang X, Jiang S, Sze J, Leung GK, Lu G, Chan DT, Bian XW, et al. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu T, Suzuki H, Nojima M, Kitamura H, Yamamoto E, Maruyama R, Ashida M, Hatahira T, Kai M, Masumori N, et al. Methylation of a panel of microRNA genes is a novel biomarker for detection of bladder cancer. Eur Urol. 2013;63:1091–1100. doi: 10.1016/j.eururo.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 12.Hu CB, Li QL, Hu JF, Zhang Q, Xie JP, Deng L. miR-124 inhibits growth and invasion of gastric cancer by targeting ROCK1. Asian Pac J Cancer Prev. 2014;15:6543–6546. doi: 10.7314/APJCP.2014.15.16.6543. [DOI] [PubMed] [Google Scholar]
- 13.Feng T, Xu D, Tu C, Li W, Ning Y, Ding J, Wang S, Yuan L, Xu N, Qian K, et al. MiR-124 inhibits cell proliferation in breast cancer through downregulation of CDK4. Tumour Biol. 2015;36:5987–5997. doi: 10.1007/s13277-015-3275-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhou L, Xu Z, Ren X, Chen K, Xin S. MicroRNA-124 (MiR-124) inhibits cell proliferation, metastasis and invasion in colorectal cancer by downregulating Rho-associated protein kinase 1(ROCK1) Cell Physiol Biochem. 2016;38:1785–1795. doi: 10.1159/000443117. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, Yao R, Cui L. The tumor suppressor miR-124 inhibits cell proliferation by targeting STAT3 and functions as a prognostic marker for postoperative NSCLC patients. Int J Oncol. 2015;46:798–808. doi: 10.3892/ijo.2014.2786. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Banaei-Esfahani A, Moazzeni H, Nosar PN, Amin S, Arefian E, Soleimani M, Yazdani S, Elahi E. MicroRNAs that target RGS5. Iran J Basic Med Sci. 2015;18:108–114. [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia; Proc Natl Acad Sci USA; 2002; pp. 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong F, Xu T, Shen Y, Zhong S, Chen S, Ding Q, Shen Z. Dysregulation of miRNAs in bladder cancer: Altered expression with aberrant biogenesis procedure. Oncotarget. 2017;8:27547–27568. doi: 10.18632/oncotarget.15173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang T, Wang J, Zhai X, Li H, Li C, Chang J. MiR-124 retards bladder cancer growth by directly targeting CDK4. Acta Biochim Biophys Sin (Shanghai) 2014;46:1072–1079. doi: 10.1093/abbs/gmu105. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Wu Q, Xu B, Wang P, Fan W, Cai Y, Gu X, Meng F. MiR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1. FEBS J. 2015;282:4376–4388. doi: 10.1111/febs.13502. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Li S, Lin Y, Chen H, Hu Z, Mao Y, Xu X, Wu J, Zhu Y, Zheng X, et al. MicroRNA-124-3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J Transl Med. 2013;11:276. doi: 10.1186/1479-5876-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Y, Li Y, Nian Y, Liu D, Dai F, Zhang J. STAT3 is involved in miR-124-mediated suppressive effects on esophageal cancer cells. BMC Cancer. 2015;15:306. doi: 10.1186/s12885-015-1303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan J, Zhao XF, Vojtek A, Goldman D. Retinal injury, growth factors, and cytokines converge on β-catenin and pStat3 signaling to stimulate retina regeneration. Cell Rep. 2014;9:285–297. doi: 10.1016/j.celrep.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Simone V, Franzè E, Ronchetti G, Colantoni A, Fantini MC, Di Fusco D, Sica GS, Sileri P, MacDonald TT, Pallone F, et al. Th17-type cytokines, IL-6 and TNF-α synergistically activate STAT3 and NF-kB to promote colorectal cancer cell growth. Oncogene. 2015;34:3493–3503. doi: 10.1038/onc.2014.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Reilly S, Ciechomska M, Cant R, van Laar JM. Interleukin-6 (IL-6) trans signaling drives a STAT3-dependent pathway that leads to hyperactive transforming growth factor-β (TGF-β) signaling promoting SMAD3 activation and fibrosis via Gremlin protein. J Biol Chem. 2014;289:9952–9960. doi: 10.1074/jbc.M113.545822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siveen KS, Sikka S, Surana R, Dai X, Zhang J, Kumar AP, Tan BK, Sethi G, Bishayee A. Targeting the STAT3 signaling pathway in cancer: Role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014;1845:136–154. doi: 10.1016/j.bbcan.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Huang SY, Chang SF, Liao KF, Chiu SC. Tanshinone IIA inhibits epithelial-mesenchymal transition in bladder cancer cells via modulation of STAT3-CCL2 signaling. Int J Mol Sci. 2017;18(pii):E1616. doi: 10.3390/ijms18081616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Ren Y, Lou ZG, Wan X, Weng GB, Cen D. Paeoniflorin inhibits the growth of bladder carcinoma via deactivation of STAT3. Acta Pharm. 2018;68:211–222. doi: 10.2478/acph-2018-0013. [DOI] [PubMed] [Google Scholar]
- 30.Ho PL, Lay EJ, Jian W, Parra D, Chan KS. Stat3 activation in urothelial stem cells leads to direct progression to invasive bladder cancer. Cancer Res. 2012;72:3135–3142. doi: 10.1158/0008-5472.CAN-11-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T, Kee WH, Seow KT, Fung W, Cao X. The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Mol Cell Biol. 2000;20:7132–7139. doi: 10.1128/MCB.20.19.7132-7139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang TT, Su JC, Liu CY, Shiau CW, Chen KF. Alteration of SHP-1/p-STAT3 signaling: A potential target for anticancer therapy. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18061234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamran MZ, Patil P, Gude RP. Role of STAT3 in cancer metastasis and translational advances. Biomed Res Int. 2013;2013:421821. doi: 10.1155/2013/421821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter RL, Lo HW. STAT3 target genes relevant to human cancers. Cancers (Basel) 2014;6:897–925. doi: 10.3390/cancers6020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used during the current study are available from the corresponding author on reasonable request.