Abstract
Long non-coding RNAs (lncRNAs) have been suggested to serve important roles in the development of a number of human cancer types. An increasing amount of data has indicated that the lncRNA small ubiquitin-like modifier 1 (SUMO1) pseudogene 3 (SUMO1P3) has been involved in various types of human cancer. However, the function SUMO1P3 in the development of pancreatic cancer remains unclear. Firstly, reverse transcription-quantitative polymerase chain reaction was performed to determine the expression of SUMO1P3 in pancreatic cancer tissues and cell lines. Then, cell counting kit-8, wound-healing and transwell assays were conducted to explore the effect of SUMO1P3 on pancreatic cancer cell proliferation, migration and invasion. Finally, the EMT-associated proteins were evaluated by western blotting. The results of the present study revealed that SUMO1P3 expression was elevated in pancreatic tissues compared with the corresponding adjacent normal tissues. Additionally, the data indicated that the increased expression of SUMO1P3 is significantly associated with tumor progression and the poor survival of patients with pancreatic cancer. Furthermore, the present study identified that SUMO1P3 knockdown may suppress the proliferation, migration and invasion of pancreatic cancer cells. Additionally, downregulation of SUMO1P3 suppressed the epithelial-mesenchymal transition (EMT) and increased the expression of epithelial cadherin, and decreased the expression of neuronal cadherin, vimentin and β-catenin. Taken together, the results of the present study demonstrated that SUMO1P3 may participate in EMT and pancreatic cancer progression, thus suggesting that it may be a novel diagnostic and therapeutic biological target for pancreatic cancer.
Keywords: small ubiquitin-like modifier 1 pseudogene 3, pancreatic cancer, prognosis, epithelial-mesenchymal
Introduction
Pancreatic cancer is one of the most common types of cancer that leads to increased mortality (1). Although significant improvements have been made in the diagnosis and treatment of pancreatic cancer in the last few decades, the prognosis of pancreatic cancer remains poor due to its aggressiveness and early systemic dissemination (2). The majority of patients with pancreatic cancer is diagnosed at an advanced stage of disease and is not eligible for curative resection, thus leading to high mortality rate (3). Therefore, it is imperative to investigate the molecular mechanisms underlying the development of pancreatic cancer in order to identify biological markers for the early diagnosis and the development of novel therapeutic agents in pancreatic cancer.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules >200 nucleotides in length that do not encode proteins (4). Accumulating evidence has suggested that lncRNAs exhibit important roles in a number of fundamental cellular processes, including cell growth, apoptosis and differentiation by regulating gene expression at the transcriptional or post-transcriptional level (5,6). Previous studies have demonstrated that several lncRNAs may be dysregulated in various types of human cancer (7,8). Pseudogenes including small ubiquitin-like modifier 1 pseudogene (SUMO1P3) are considered as a separate class of lncRNAs (9).
SUMO1P3 was originally identified as a potential biomarker for the diagnosis of gastric cancer in 2013 (9). Furthermore, increased expression of SUMO1P3 has been reported in bladder cancer tissues, and it is significantly associated with a greater histological grade and advanced tumor-node metastasis (TNM) stage (10). An in vitro study further demonstrated that knockdown of SUMO1P3 inhibited bladder cancer cell proliferation and induced bladder cancer cell apoptosis (10). However, the biological function of SUMO1P3 in pancreatic cancer and its potential molecular mechanisms remain unclear.
In the present study, the expression of SUMO1P3 in pancreatic cancer tissues and the paired adjacent normal pancreatic tissues was evaluated, and its effects on epithelial-mesenchymal transition (EMT). The objective of the present study was to clarify the biological function and potential mechanism of SUMO1P3 in the development of pancreatic cancer.
Materials and methods
Patient tissue samples
A total of 48 pairs of pancreatic cancer tissues and paired adjacent normal tissues were collected from patients undergoing surgical resection at The Pancreatic Cancer Center, Fenjinting Hospital between January 2009 and October 2013 (mean age 63.3±8.86, age range, 37–71). All of the enrolled patients in the present study underwent surgery during which samples were collected. All specimens were frozen and stored in liquid nitrogen until further use. No patients received preoperative anticancer treatment, including chemotherapy or radiation prior to specimen collection. The present study was conducted with the approval of the Ethics and Research Committees of Fenjinting Hospital (Jiangsu, China) and was performed in accordance with the Declaration of Helsinki. All patients provided written informed consent prior to their participation in the present study. The clinical characteristics of all the patients are summarized in Table I.
Table I.
Association between the expression of SUMO1P3 and the clinicopathological characteristics of patients with pancreatic cancer.
| SUMO1P3 expression, n | ||||
|---|---|---|---|---|
| Clinicopathological parameters | n | High | Low | P-value |
| 48 | 33 | 15 | ||
| Age, years | 0.633 | |||
| <60 | 19 | 12 | 7 | |
| ≥60 | 29 | 21 | 8 | |
| Sex | 0.416 | |||
| Female | 17 | 13 | 4 | |
| Male | 31 | 20 | 11 | |
| Tumor size, cm | 0.019 | |||
| <4 | 30 | 17 | 13 | |
| ≥4 | 18 | 16 | 2 | |
| Histological differentiation | 0.205 | |||
| Well and moderate | 33 | 21 | 12 | |
| Poor | 15 | 12 | 3 | |
| Location | 0.371 | |||
| Head-neck | 27 | 19 | 8 | |
| Body-tail | 21 | 14 | 7 | |
| Lymph node metastasis | 0.028 | |||
| Negative | 35 | 22 | 13 | |
| Positive | 13 | 11 | 2 | |
| TNM stage | 0.034 | |||
| I–II | 41 | 26 | 15 | |
| III | 7 | 7 | 0 | |
TNM, tumor-node metastasis; SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3.
Cell culture
The human pancreatic cancer cell lines BxPC-3, PANC-1, MiaPaCa-2, and ASPC-1 and the normal pancreatic ductal epithelial cell line HPDE6-C7 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 50 U/ml penicillin and 0.1 mg/ml streptomycin. Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2.
Small interfering (si)RNA transfection
siRNAs that targeted SUMO1P3 and a scrambled negative control were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The target sequence of siSUMO1P3 was 5′-TGGCCCTGATGTTCTAGCATGTGAT-3′. The RNAs were introduced into cells at a final concentration of 50 nM. Transfections were performed using the Lipofectamine 3000 kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. The knockdown efficiency was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) 48 h after transfection.
RNA extraction and RT-qPCR
Total RNA from tissues and cells was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using PrimeScript RT Reagent kit (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. qPCR was performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. The primer sequences were as follows: SUMO1P3, 5′-ACTGGGAATGGAGGAAGA-3′ (forward) and 5′-TGAGAAAGGATTGAGGGAAAAG-3′ (reverse); GAPDH, 5′-CGCTCTCTGCTCCTCCTGTTC-3′ (forward) and 5′-ATCCGTTGACTCCGACCTTCAC-3′ (reverse). The thermocycling conditions were as follows: Initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and extension at 60°C for 1 min. Relative expression of PHGDH was normalized to the expression of GAPDH. Relative expression of PHGDH was calculated using the 2−ΔΔCq method (11). All experiments were performed in triplicate. RT-qPCR was performed using the ABI PRISM 7500 PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The median value was the cutoff between low and high SUMO1P3 expression in patients with pancreatic cancer. The median value was included in the low group.
Cell proliferation and colony formation assay
Following transfection with si-SUMO1P3 or si-negative control (NC), BxPC-3 panc-1 cells cell proliferation was accessed using Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol. A total of 2,000 cells were plated into 96-well plates. Following culture for 24 h, 100 µl CCK-8 was added into wells and the viable cells were evaluated at a wavelength of 450 nm. For colony formation assay, 500 cells per plate were seeded in 6-well plates and incubated for two weeks. Then, colonies were fixed with 4% paraformaldehyde at room temperature for 15 min and stained with crystal violet. A total of five fields were randomly selected and cells were counted under a light microscope at low magnification (×100). Colonies that contained >50 cells were designated as survivors.
Cell migration and invasion assay
To measure the migratory ability of pancreatic cancer cells, a wound-healing assay was performed. BXPC-3 and PANC-1 cells were cultured in a 6-well plate until they reached 100% confluence. The monolayer cells were scratched using a 200 µl sterile pipette tip to create the wound. Cells were cultured with DMEM without FBS at 37°C for 24 h. The migration of cells across the gap wound was measured using light microscopy (×100 magnification). The invasive ability of pancreatic cancer cells was assessed using a Matrigel-coated Transwell chamber (BD Biosciences, San Jose, CA, USA). A total of 2×104 cells in 100 µl of serum-free medium were plated in the upper chamber. DMEM medium supplemented with 10% FBS was plated in the lower chamber. Following incubation for 24 h at 37°C in a humidified atmosphere containing 5% CO2. The non-invading cells on the upper surface of the well were scraped off with a cotton swab, and the invading cells on the lower surface were stained with 4% crystal violet at room temperature for 10 min and counted using a light microscope (×200 magnification). Each experiment was performed in triplicate.
Western blot analysis
Cell proteins were extracted with ice-cold lysis buffer (1 mM EDTA, pH 8.0, 50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.1% SDS and 0.5% sodium deoxycholate, pH 7.4). After 30 min on ice, cell debris was removed by centrifugation at 15,000 × g for 15 min at 4°C. A basic protein quantification kit (BioVision, Inc., CA, USA) was used to determine total protein concentration. Equal amounts of the protein (20–40 µg/lane) were separated by SDS-PAGE (10% gels) and transferred onto polyvinylidene difluoride (PVDF) membranes and the blots were blocked for 1 h using 5% fat-free milk at room temperature. The membranes were incubated with the following primary antibodies: Anti-vimentin (5741; Cell Signaling Technology, Inc., Danvers, MA, USA; 1:1,000 dilution); Anti-neuronal (N)-cadherin (13116; Cell Signaling Technology; 1:1,000 dilution); Anti-β-catenin (8480; Cell Signaling Technology; 1:1,000 dilution); Anti-Epithelial (E)-cadherin (3195; Cell Signaling Technology; 1:1,000 dilution); and anti-β-actin antibody (sc-58673, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. Following primary incubation, membranes were incubated with secondary antibodies (cat. no. AB6721; 1:3,000; Abcam) for 1 h at room temperature Finally, protein bands were developed with Amersham ECL Western Blotting Detection reagent (GE Healthcare Life Sciences, Little Chalfont, UK) and visualized using a gel imaging analysis system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and further analyzed using Image Lab software (version 3.0; Bio-Rad Laboratories, Inc.).
Statistical analysis
All results are shown as mean ± standard deviation and were analyzed using GraphPad Prism version 6 (GraphPad Software, Inc., La Jolla, CA, USA) from at least three independent experiments. The χ2 test was performed to explore the associations between SUMO1P3 level and clinicopathological factors. The Kaplan-Meier method was used to calculate the survival curve, and log-rank test to determine statistical significance. Multivariate analysis was applied to determine the independent indicator for overall survival of patients with pancreatic cancer. The differences between groups were analyzed using one-way analysis of variance (ANOVA) followed by the Student-Newman-Keuls test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of SUMO1P3 is upregulated in pancreatic cancer tissues and cells
To explore the role of SUMO1P3 in pancreatic cancer, the relative expression level of SUMO1P3 in 48 pairs of pancreatic cancer tissues and adjacent non-tumor tissues was examined by RT-qPCR analysis. As presented in Fig. 1A, increased expression of SUMO1P3 was identified in pancreatic cancer tissues compared with corresponding adjacent non-tumor tissues. The expression of SUMO1P3 was assessed in 4 human pancreatic cancer cell lines and normal pancreatic ductal epithelial cell line HPDE6-C7. The results revealed that pancreatic cancer cell lines demonstrated a higher expression of SUMO1P3 compared with that in the normal pancreatic ductal epithelial cell (Fig. 1B).
Figure 1.
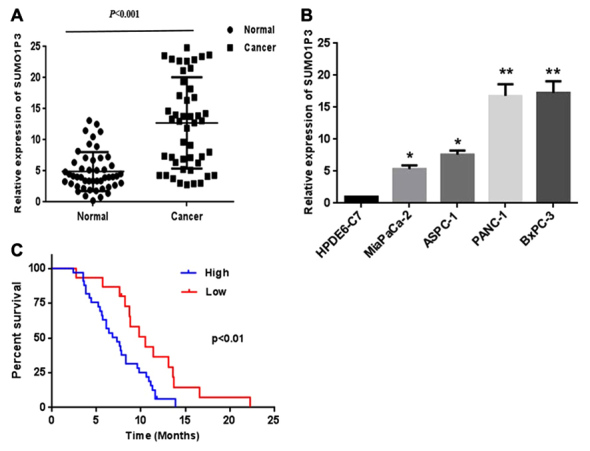
Relative expression of SUMO1P3 in pancreatic cancer tissues and cells. (A) Relative expression of SUMO1P3 in 48 pairs of pancreatic cancer tissues and adjacent non-tumor tissues by reverse transcription-quantitative polymerase chain reaction analysis. (B) SUMO103 is upregulated in pancreatic cancer cells compared with normal pancreatic ductal epithelial cell. (C) Kaplan-Meier curves for overall survival of two groups defined by low and high expression of SUMO1P3 in patients with pancreatic cancer. The median expression of SUMO1P3 is 12.34±8.76 *P<0.05, **P<0.01. SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3.
Increased expression of SUMO1P3 is associated with the progression and poor prognosis of patients with pancreatic cancer
The present study further investigated the association between SUMO1P3 expression and clinicopathological factors in 48 patients with pancreatic cancer (Table I). The results revealed that the increased expression of SUMO1P3 was significantly associated with tumor size (P=0.019), lymph node metastasis (P=0.028) and TNM stage (P=0.034). However, there was no significant association between SUMO1P3 expression and age, sex, histological differentiation or tumor location. Kaplan-Meier survival curves (Fig. 1C) demonstrated that the survival of patients with pancreatic cancer with a lower expression of SUMO1P3 is significantly improved compared with that of the higher expression group (the median value was the cutoff between low and high SUMO1P3 expression). Furthermore, multivariate analysis indicated that increased expression of SUMO1P3 was an independent indicator for overall survival of patients with pancreatic cancer (Table II).
Table II.
Cox multivariate regression analysis of the association of prognostic factors in pancreatic cancer.
| 95% CI | |||
|---|---|---|---|
| Factors | P-value | Lower | Upper |
| SUMO1P3 expression, high/low | 0.027 | 0.371 | 0.814 |
| Age, years | 0.092 | 0.315 | 1.224 |
| Sex | 0.831 | 0.272 | 1.283 |
| Tumor size, cm | 0.063 | 0.329 | 1.072 |
| Histological differentiation | 0.744 | 0.295 | 1.316 |
| Location | 0.083 | 0.255 | 1.097 |
| Lymph node metastasis | 0.062 | 0.216 | 1.013 |
| TNM stage | 0.012 | 0.217 | 0.711 |
TNM, tumor-node metastasis; CI, confidence interval; SUMO1P3, small ubiquitin-like modifier 1 pseudogene.
Knockdown of SUMO1P3 impairs proliferation of BXPC-3 and PANC-1 cells in vitro
In order to explore the potential biological function of SUMO1P3 in the development of pancreatic cancer, SUMO1P3 expression was silenced in BXPC-3 and PANC-1 cells. As presented in Fig. 2A, mRNA expression level of SUMO1P3 was significantly decreased in cells transfected with SUMO1P3 siRNA compared with the control group. Knockdown of SUMO1P3 significantly suppressed the proliferative ability of BXPC-3 and PANC-1 cells (Fig. 2B), as assessed using a CCK-8 assay. Furthermore, a colony formation assay was also used to evaluate the cell proliferation ability. Knockdown of SUMO1P3 significantly decreased the colony formation ability of BXPC-3 and PANC-1 cells (Fig. 2C).
Figure 2.
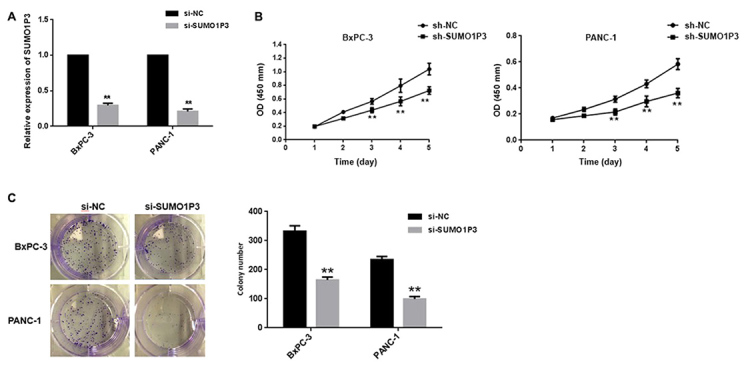
Effects of downregulation of SUMO1P3 on cell proliferation in pancreatic cancer. (A) The expression level of SUMO1P3 in BxPC-3 and PANC-1 cells was significantly decreased in response to si-SUMO1P3 compared with the si-NC group. (B) Knockdown of SUMO1P3 significantly suppressed cell proliferation of BxPC-3 and PANC-1 cells. (C) Colony-formation assay demonstrated that knockdown of SUMO1P3 significantly suppressed the colony-formation ability of BxPC-3 and PANC-1 cells compared with the negative control. **P<0.01. NC, negative control; si, small interfering; OD, optical density; SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3.
Knockdown of SUMO1P3 inhibits cell migration and invasion
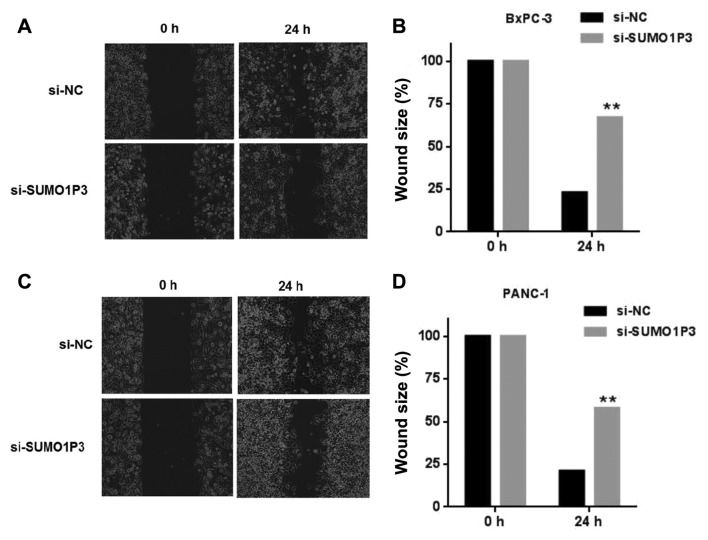
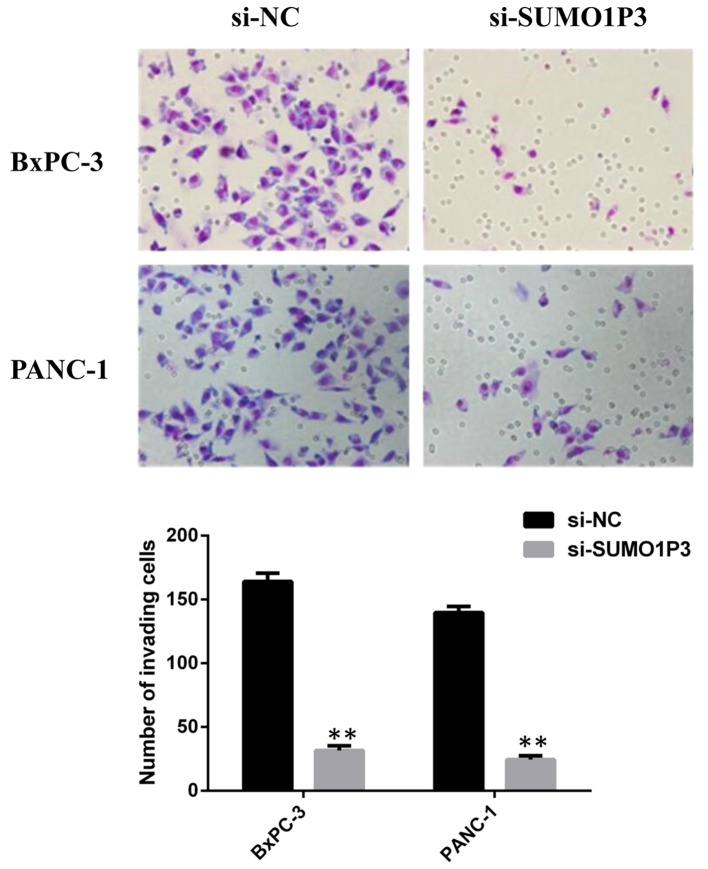
Wound healing and Transwell assays were performed in order to measure the migratory and invasive ability of pancreatic cancer BxPC-3 cells and PANC-1 cells. Knockdown of SUMO1P3 suppressed the migration ability of BxPC-3 (Fig. 3A and B) and PANC-1 (Fig. 3C and D) cells. Knockdown of SUMO1P3 led a significant inhibition of the invasive ability of BxPC-3 and PANC-1 cells compared with that in the si-NC group (Fig. 4).
Figure 3.
Effects of downregulation of SUMO1P3 on migration of pancreatic cancer cells. (A) Representative images and (B) quantification of wound healing in response to si-NC and si-SUMO1P3 in BxPC-3 cells. (C) Representative images and (D) quantification of wound healing in response to si-NC and si-SUMO1P3 in PANC-1 cells. (magnification, ×200) **P<0.01. SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3; NC, negative control; si, small interfering.
Figure 4.
Effects of downregulation of SUMO1P3 on invasive ability of pancreatic cancer cells. Representative images and quantification of cell invasion in response to si-NC and si-SUMO1P3 in BxPC-3 and PANC-1 cells. **P<0.01 vs. si-NC. SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3; NC, negative control; si, small interfering.
SUMO1P3 is associated with EMT
Since EMT is essential for tumor progression and metastasis (12), the effects of SUMO1P3 on EMT were analyzed by western blotting. The results showed that downregulation of SUMO1P3 resulted in upregulation of epithelial markers (E-cadherin) and downregulation of mesenchymal markers (N-cadherin, vimentin and β-catenin) (Fig. 5). Taken together, these findings reveal that SUMO1P3 may be a positive regulator of EMT, with an important biological function in the development of pancreatic cancer.
Figure 5.
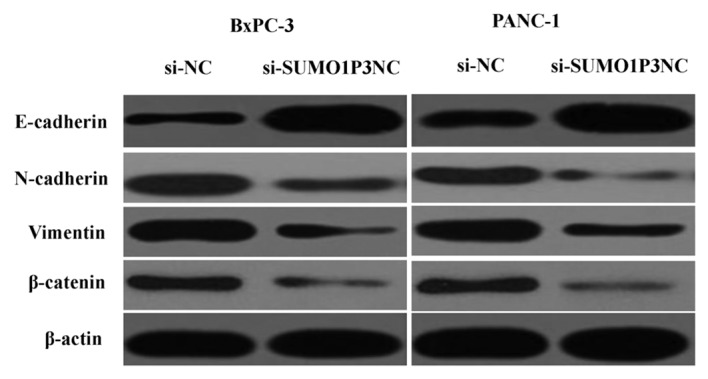
Effects of downregulation of SUMO1P3 on the expression of E-cadherin, N-cadherin, vimentin and β-catenin. E, epithelial; N, neuronal; SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3; NC, negative control; si, small interfering.
Discussion
Numerous studies have demonstrated that several lncRNAs serve a critical function in the development and progression of various types of cancer, including pancreatic cancer (13). For example, lncRNA-plasmacytoma variant translocation 1 functions as an endogenous sponge by competing with microRNA (miR)-488 to regulate Serpin Family E Member 1 mRNA Binding Protein 1 and therefore promote cell proliferation and migration in pancreatic cancer (14). LncRNA-taurine-upregulated gene 1 may enhance the cell proliferation and migration in pancreatic cancer by regulating EMT (15). Linc00673 may regulate non-small cell lung cancer proliferation, migration, invasion and EMT by functioning as an endogenous sponge by competing with miR-150-5p (16).
LncRNA SUMO1P3 has been previously reported to be upregulated and may serve as a potential therapeutic target in several types of cancer (9–10). In the present study, SUMO1P3 was demonstrated to be upregulated in pancreatic cancer tissues compared with paired adjacent non-tumor tissue and increased expression of SUMO1P3 was significantly associated with tumor size, lymph node metastasis and TNM stage. Furthermore, the survival of patients with pancreatic cancer with a lower expression of SUMO1P3 was significantly improved compared with that of the higher expression group. Furthermore, knockdown of SUMO1P3 inhibited cell proliferation, migration and invasion. These results suggested that SUMO1P3 may serve an important role in the development of pancreatic cancer.
EMT is a vital pathological progress in the development and progression in numerous types of human cancer, including pancreatic cancer (15,16). During this process, cancer cells lose cell polarity and cell-cell adhesion and acquire mesenchymal characteristics, including motility and invasiveness (17,18). Cadherin switch (loss E-cadherin and gain of N-cadherin expression) represents an important characteristic in EMT (19). E-cadherin, a canonical epithelial marker, has been widely accepted as a critical suppressor of motility and invasiveness of epithelial cells in numerous types of cancer (20). Transcriptional downregulation or genomic deletion of E-cadherin expression may result in key pathological changes in tumor cells (21). However, N-cadherin may promote invasion and distal metastasis in various types of human cancer (22). Vimentin and β-catenin are well-established markers for EMT (23,24).
Therefore, the present study further assessed whether the biological function of SUMO1P3 on pancreatic cancer cells was via EMT induction. The results demonstrated that downregulation of SUMO1P3 led to upregulation of epithelial markers (E-cadherin) and downregulation of mesenchymal markers (N-cadherin, vimentin and β-catenin). These results revealed that SUMO1P3 may function as an oncogene in pancreatic cancer by regulating EMT.
In conclusion, the expression of SUMO1P3 was increased in pancreatic cancer tissues and cell lines, and increased expression of SUMO1P3 was associated with the malignant status and poor prognosis in patients with pancreatic cancer. Knockdown of SUMO1P3 suppressed cell proliferation, migration and invasion in pancreatic cancer by regulating EMT in vitro.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Fund for 333 Engineering Project in Jiangsu Province (grant no. BRA2017266).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on request.
Authors' contributions
CT and YJ performed the majority of the research and were the major contributors in writing the manuscript. CT, YJ and SS prepared experimental materials and reviewed the article. CT and SS made substantial contributions to the design of the work, drafting the manuscript and revising it critically for important intellectual content. SS gave final approval of the version to be published.
Ethics approval and consent to participate
The present study was conducted with the approval of the Ethics and Research Committees of Fenjinting Hospital (Jiangsu, China) and was performed in accordance with the Declaration of Helsinki. All patients provided written informed consent prior to their participation in the present study.
Patient consent for publication
All patients provided written informed consent for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Xu W, Xu B, Yao Y, Yu X, Cao H, Zhang J, Liu J, Sheng H. Overexpression and biological function of IQGAP3 in human pancreatic cancer. Am J Transl Res. 2016;8:5421–5432. [PMC free article] [PubMed] [Google Scholar]
- 3.Yao W, Ji S, Qin Y, Yang J, Xu J, Zhang B, Xu W, Liu J, Shi S, Liu L, et al. Profilin-1 suppresses tumorigenicity in pancreatic cancer through regulation of the SIRT3-HIF1α axis. Mol Cancer. 2014;13:187. doi: 10.1186/1476-4598-13-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 7.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: Progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 9.Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, Ding X, Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709. doi: 10.1007/s12032-013-0709-2. [DOI] [PubMed] [Google Scholar]
- 10.Zhan Y, Liu Y, Wang C, Lin J, Chen M, Chen X, Zhuang C, Liu L, Xu W, Zhou Q, et al. Increased expression of SUMO1P3 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Oncotarget. 2016;7:16038–16048. doi: 10.18632/oncotarget.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin C, Zhao GC, Xu YD, Wang DS, Jin DY, Ji Y, Lou WH, Wu WC. Increased expression of αTubulin is associated with poor prognosis in patients with pancreatic cancer after surgical resection. Oncotarget. 2016;7:60657–60664. doi: 10.18632/oncotarget.10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: The dark matter matters. Curr Opin Genet Dev. 2018;48:8–15. doi: 10.1016/j.gde.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol. 2018;233:4044–4055. doi: 10.1002/jcp.26072. [DOI] [PubMed] [Google Scholar]
- 15.Qin CF, Zhao FL. Long non-coding RNA TUG1 can promote proliferation and migration of pancreatic cancer via EMT pathway. Eur Rev Med Pharmacol Sci. 2017;21:2377–2384. [PubMed] [Google Scholar]
- 16.Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H, Kong J, Ding K, Shen HM, Wu H, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16:118. doi: 10.1186/s12943-017-0716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C, Xie D, Cui J, Li Q, Gao Y, Xie K. FOXM1c promotes pancreatic cancer epithelial-to-mesenchymal transition and metastasis via upregulation of expression of the urokinase plasminogen activator system. Clin Cancer Res. 2014;20:1477–1488. doi: 10.1158/1078-0432.CCR-13-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma R, Chen J, Jiang S, Lin S, Zhang X, Liang X. Up regulation of NAT10 promotes metastasis of hepatocellular carcinoma cells through epithelial-to-mesenchymal transition. Am J Transl Res. 2016;8:4215–4223. [PMC free article] [PubMed] [Google Scholar]
- 19.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu JF, Mao L, Bu LL, Ma SR, Huang CF, Zhang WF, Sun ZJ. C4.4A as a biomarker of head and neck squamous cell carcinoma and correlated with epithelial mesenchymal transition. Am J Cancer Res. 2015;5:3505–3515. [PMC free article] [PubMed] [Google Scholar]
- 21.Guo F, Kerrigan Parker BC, Yang D, Hu L, Shmulevich I, Sood AK, Xue F, Zhang W. Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. J Hematol Oncol. 2014;7:19. doi: 10.1186/1756-8722-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan ZZ, Yan XB, Yan LL, Tian Y, Meng QC, Qiu WW, Zhang Z, Jin ZM. Overexpression of Tbx3 is correlated with Epithelial-Mesenchymal Transition phenotype and predicts poor prognosis of colorectal cancer. Am J Cancer Res. 2014;5:344–353. [PMC free article] [PubMed] [Google Scholar]
- 23.Dai C, Cao J, Zeng Y, Xu S, Jia X, Xu P. E-cadherin expression as a prognostic factor in patients with ovarian cancer: A meta-analysis. Oncotarget. 2017;8:81052–81061. doi: 10.18632/oncotarget.18898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Liu G, Kang Y, Dong Z, Qian Q, Ma X. N-cadherin expression is associated with acquisition of EMT phenotype and with enhanced invasion in erlotinib-resistant lung cancer cell lines. PLoS One. 2013;8:e57692. doi: 10.1371/journal.pone.0057692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on request.