Short abstract
Introduction
Electronic health records provide an unparalleled opportunity for the use of patient data that is routinely collected and stored, in order to drive research and develop an epidemiological understanding of disease. Diabetes, in particular, stands to benefit, being a data-rich, chronic-disease state. This article aims to provide an understanding of the extent to which the healthcare sector is using routinely collected and stored data to inform research and epidemiological understanding of diabetes mellitus.
Methods
Narrative literature review of articles, published in both the medical- and engineering-based informatics literature.
Results
There has been a significant increase in the number of papers published, which utilise electronic health records as a direct data source for diabetes research. These articles consider a diverse range of research questions. Internationally, the secondary use of electronic health records, as a research tool, is most prominent in the USA. The barriers most commonly described in research studies include missing values and misclassification, alongside challenges of establishing the generalisability of results.
Discussion
Electronic health record research is an important and expanding area of healthcare research. Much of the research output remains in the form of conference abstracts and proceedings, rather than journal articles. There is enormous opportunity within the United Kingdom to develop these research methodologies, due to national patient identifiers. Such a healthcare context may enable UK researchers to overcome many of the barriers encountered elsewhere and thus to truly unlock the potential of electronic health records.
Keywords: electronic health records, electronic patient records, electronic medical records, informatics, diabetes mellitus
Introduction
The expansion in use of electronic health records (EHRs) provides an unparalleled opportunity for the use of routinely collected patient data to drive research and deliver an epidemiological understanding of the basis of disease. Whilst some challenges exist around legal, ethical and technological issues,1 there is an increasing number of studies using EHRs to deliver meaningful research outputs.2 These studies are somewhat sporadic, in diverse specialist areas including chronic obstructive pulmonary disease3 and heart failure research.4,5 Additionally, there are published opinion pieces highlighting the potential importance the use of EHRs in research.6
An EHR can be defined as ‘a repository of patient data in digital form, stored and exchanged securely, and accessible by multiple authorised users. It contains retrospective, concurrent, and prospective information and its primary purpose is to support continuing, efficient and quality integrated health care’.7 Whilst there is a myriad of different definitions applied to EHRs,8 this represents a widely accepted and comprehensive definition.9 Critical to this paper is that an EHR’s primary purpose is the provision of healthcare. Use of the information routinely collected and stored for the purposes of research is a secondary use of these EHRs. This paper is specifically looking at the direct secondary use of these EHRs as research tools. By direct use, we are looking at instances where research teams have taken data directly from the clinical EHR, rather than indirect uses where multiple EHRs from different sources have been combined together (often with anonymisation or pseudo-anonymisation) into registries or similar repositories. The consideration, specifically of direct secondary uses, is important as it represents an assessment of what immediately can be achieved from data we already have, rather than data requiring third-party processing, complex information governance arrangements and often payment. Neglecting these existing real-life data sources would represent waste of a ready and potentially valuable resource.
The first EHR was created by Loughead Aircraft Manufacturing Company (better known as Lockheed Martin Aerospace) in the 1960s, alongside a small number of other pioneer academic groups. Wider adoption of EHRs only began with commercialisation of systems in the 1990s.10 Use, in the UK, was somewhat delayed; potentially due to the absence of commercial market forces in the National Health Service (NHS), yet the UK now represents the largest EHR market in Europe.11
The wider uptake of high quality EHRs has been fuelled by government and trans-national initiatives, including the $19 billion HiTECH Act in the USA12 and €2 billion Innovative Medicines Initiative in the European Union.13 There is, however, significant geographic variation in the adoption and approach to EHRs, with some countries (notably Denmark and Sweden) utilising national EHRs and other countries (the USA and UK) adopting organisation specific EHRs.14
Diabetes represents a truly data rich pathology with a wealth of routinely collected data, including but not limited to; average blood glucose, foot health, eye health, cardiovascular health and renal health. Diabetes therefore represents a pathology particularly able to benefit from the use of EHRs in research. Diabetes further represents a critical international challenge with a global prevalence that has doubled since 1980 from 4.5 to 7.8% of the adult population.15 Developing and exploiting new data and information-driven research methods is therefore essential to tackling this emerging global burden. At the time of writing this paper, no review has been performed to identify what has been achieved through EHR-based diabetes research, how that progress has been achieved or an appraisal of such studies.
This work represents first a review of the direct use of EHRs as a research tool in diabetes. It considers current applications, barriers and future strategies. It also provides a meaningful benefit by developing an understanding of where gaps currently lie in the research literature, and how we can approach and overcome challenges to EHR research.
Methods
Prospective registration
This review was prospectively registered with the PROSPERO database (registration number: CRD42016038550). PROSPERO is an ‘international database of prospectively registered systematic reviews in health and social care, welfare, public health, education, crime, justice, and international development, where there is a health-related outcome’.16 The system is supported by the University of York and aims to both reduce duplication of research and avoid bias. All registered trials undergo a review process prior to acceptance.
Search strategy
This review is underpinned by a comprehensive search strategy including MEDLINE, Embase and Engineering Village databases. Whilst Medline represents an important source of medicine, nursing and pharmacy literature, it is also important to include Embase in such a study, based on its coverage of pharmacological articles. The OVID search platform allows both Embase and MEDLINE to be searched simultaneously. In addition, informatics and computer systems engineering approaches have been particularly relevant to EHR research, and for a truly comprehensive search strategy, it is therefore necessary to consider a search of the relevant engineering literature in this review. The Engineering Village search platform comprises 12 separate databases including Ei Compendex, Inspec, GEPBASE, GeoFef, US Patents, NTIS, EnCompassLIT, EnCompassPAT, PaperChem, CBNB and Chimica. Combining both the OVID and Engineering Village search platforms adequately and comprehensively covers the relevant medical and engineering literature.
The search included only English language papers, published between 2006 and June 2018. This is appropriate to the time period, for which there has been the existence of meaningful high-quality EHRs within clinical systems. Whilst it could be argued to include a five year cut off, there is a risk this would unintentionally exclude the earliest secondary research uses of EHRs, which could be of significant value and interest. This study focused solely on original research articles, matching our aim to identify more specifically what original research has been performed using EHRs as a direct research data source.
There is an important need to consider the extent to which the grey literature should be incorporated within this study methodology. A natural concern with the application of the grey literature is the variability of study quality and absence of peer review. Given the risks of bias associated with inappropriately dealing with large data-sets contained within EHRs,17 a consideration of the grey literature is not included here. There remains contention as to a formal definition of ‘grey literature’, with some authorities including and others excluding published conference abstracts.18 Conference abstracts can be important to demonstrate early and developing research, as well as research that has not progressed to publication. Such information is therefore valuable to this review, and published conference abstracts indexed in the databases searched are included within the review.
Search terms
‘Electronic health record(s)’ or ‘Electronic Patient Record(s)’ OR ‘Electronic Medical Records’ and ‘diabetes’ were the key search terms employed. ‘Electronic Health Records’ is the relevant NIH MeSH term, however the other search terms are included to ensure a comprehensive search. There was no attempt to distinguish Type 1 from Type 2 diabetes; not only can these be poorly recorded in EHRs19 but also the data variables available in EHRs are usually applicable to both.
Research studies that do not explicitly use EHR data, as a direct research data source, are excluded. In pilot work for this study, the following types of study were identified that would need excluding: studies where a separate research data registry is created, usually through manual inputting of data;20 research where the EHR is used solely for patient recruitment/identification;21 and research where a health record database is created solely for research purposes.22 Whilst excluded from this study, these approaches are themselves interesting and could potentially form the future bases of additional research reviews.
Papers were selected by initially screening article title and article abstract. Those papers identified as relevant were subject to a second stage of screening through review of the whole article. Inclusion criteria (Table 1) represent English language articles, published in the last 10 years, applying an interrogation of EHR data to answer a specific medical research question. Papers specifically considering the design/formatting of EHRs and the use of EHRs for operational management, rather than clinical research purposes, will also be excluded.
Table 1.
Inclusion and exclusion criteria.
| Inclusion criteria | Exclusion criteria |
|---|---|
| Registry data-based studies | |
| EHRs used solely for patientidentification/recruitment | |
| Research generated database | |
| English language | Non-English language studies |
| Studies from1 Aug 06 onwards | Studies prior to 1 Aug 06 |
| Human | Non-human studies |
| Adult (over 18) | Non-adult studies (under 18) |
Data, extracted from included papers, were structured according to a pre-defined and piloted proforma that incorporated: year of publication, number of unique patient records extracted, country of publication, type of research question, primary/secondary care, single centre/regional/national data source, whether barriers are discussed and whether opportunities for further research are discussed. Under each of these headings further information has been extracted for the narrative of the clinical review. Definitions covering the type of research question are included in Table 2; studies can belong to multiple categories. Meta-analysis was not performed.
Table 2.
Definitions relating to study research questions.
| Term | Definition |
|---|---|
| Epidemiology | Papers relating to the incidence, prevalence or distribution of diabetes included associations with other diseases |
| Prevention | Papers relating to the prevention of Type 2 Diabetes Mellitus (T2DM), including pre-diabetes states |
| Susceptibility | Papers identifying risk factors for the development of diabetes |
| Diagnosis | Papers focusing on the diagnosis of Type 1 Diabetes Mellitus (T1DM) or T2DM |
| Prognosis | Papers focusing on the prognosis of diabetes populations either with or without complications |
| Complications | Papers considering complications of diabetes, their diagnosis, epidemiology and treatment |
| Medication treatment | Papers considering the benefits of medication treatment, or comparing medications for patients with diabetes |
| Medication Side Effect | Papers considering side effects of medications used to treat diabetes or diabetic complications |
| Non-pharmacy intervention | Papers considering non-medication-based intervention for the treatment of diabetes and its complications |
| Insurance based | Papers focusing on health insurance based issues for patients with diabetes |
| Service delivery | Papers focusing on the delivery of care of individuals or populations with diabetes |
Results
Initial search
The search strategy identified 703 research papers meeting the inclusion criteria form the Medline and Embase searches, with 268 papers from Engineering Village. This resulted therefore in a total initial search of 971 papers.
Paper selection
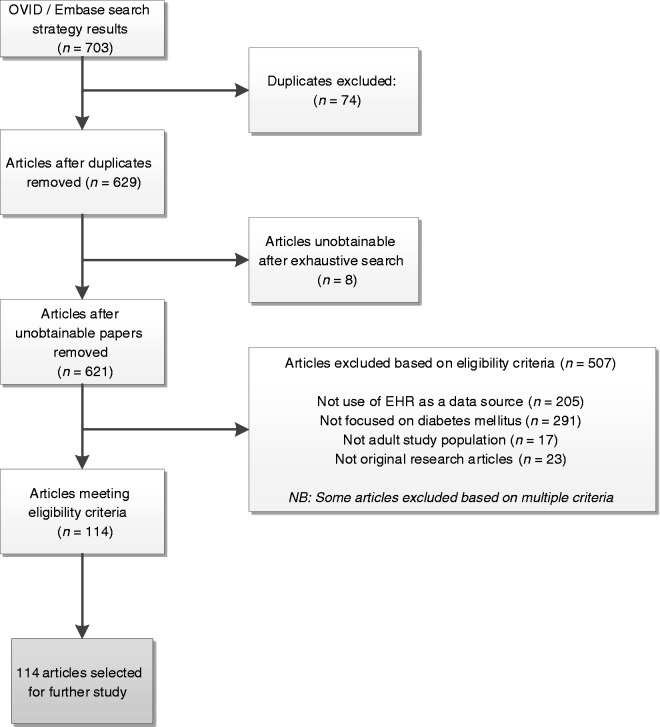
Individual review of articles by title, abstract and full paper resulted in exclusion of 589 articles (84%) from the OVID/Embase search; 114 articles were taken forward for further study and data extraction; 8 articles were unobtainable from the research literature and excluded from the study. The reasoning for papers being excluded is included in the flowchart in Figure 1.
Figure 1.
Flow chart demonstrating assessment of articles for inclusion from OVID/Embase search.
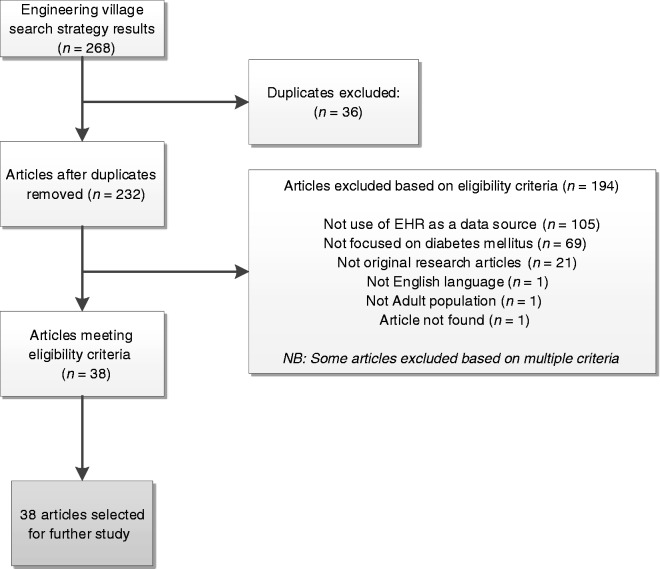
Review of the articles extracted from the Engineering Village search resulted in exclusion of 230 papers (86%), for the reasoning demonstrated in Figure 2. There were 38 papers identified that met the aims/objectives of this study,23,24 however 2 of these had been identified in the OVID/Embase search and were already included within the study. This resulted in a total collection of 150 papers selected for further analysis. It should be noted that, whilst the exclusion rate of papers from the Engineering Village search was high, there was substantial content of interest and relevance to clinical research despite this being a rarely used resource. The articles, whilst not relevant for this study, focused on the design, operation and implementation of EHR systems, or of clinical systems in general and their overall impact on clinical care. We would strongly argue that greater exposure and awareness of this valuable resource is important for future clinical researchers in healthcare research.
Figure 2.
Flow chart demonstrating assessment of articles for inclusion from Engineering Village search.
Data analysis
Publication trends over time
Over the 12-year period of study, 150 original research articles were identified. The distribution of publications over time is demonstrated in Figure 3. It is clear that there is a notable step-change in publication numbers occurring around 2012.
Figure 3.
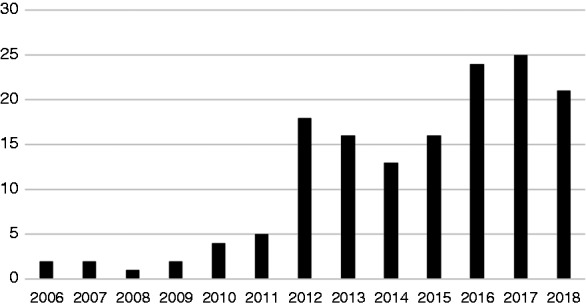
Article publication numbers by year.
Sample size
The largest sample size identified was 4.1 million patients, whilst the smallest was 30 patients. The mean average number of patients per study was 99,757, whilst the median average was 3352. It is important to note, therefore, that there is the potential for these averages to be distorted by outlying values, in particular, large value outliers. There is no clear temporal trend to median sample sizes over time as demonstrated in Figure 4 (all sample sizes included).
Figure 4.
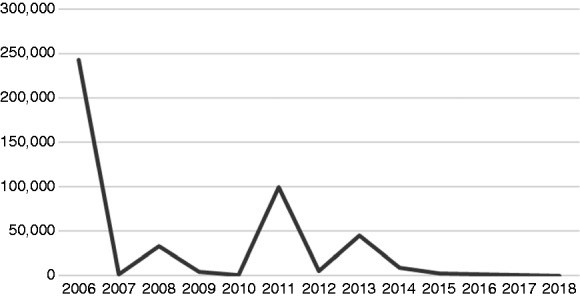
Median sample size by year.
Location of research
English language, original research articles, which met the inclusion criteria, were identified as originating from 17 different countries. One study represented an international study, utilising electronic health data from both the UK and Canada.25 The largest number of studies originated from the USA (74 studies) with 39 studies from the UK. A full breakdown considering the number of studies per country is demonstrated in Figure 5.
Figure 5.
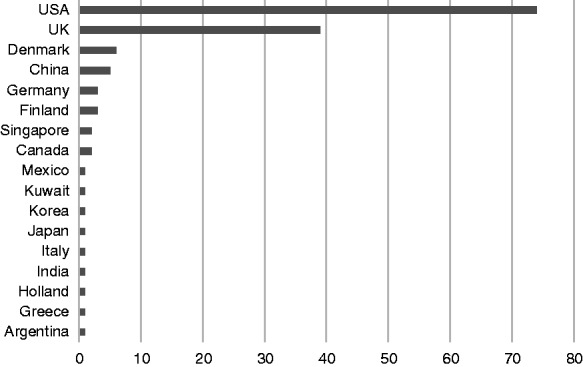
Country of origin of research articles.
Type of publication
Seventy-four articles (49%) extracted from the bibliographic databases were conference proceedings or conference abstracts. For studies originating from the UK 77% were published conference abstracts or proceedings. This is in comparison to only 40% of US studies being published as conference abstracts. This is a finding of some significance and is discussed below.
Nature of articles
Articles were identified for each of the pre-specified study categories: epidemiology, prevention, susceptibility, diagnosis, prognosis, complications, medication treatment, medication side effect, non-pharmacy intervention, service delivery, insurance based. Many articles covered multiple categories. The most common study purposes were to investigate complications (50 articles), epidemiology (34 articles) and diabetes complications (30 articles).
There was considerable variation in the sample sizes used for each of the study types. The median average number of patients, in studies considering medication treatment was 7454, compared to 1861 patients for diabetes complication studies and 12,673 for epidemiology focused studies.
Discussion
Current extent of secondary use of EHRs in diabetes research
This study identifies a number of publications and research outputs that describe the secondary use of EHRs in diabetes research. The number of publications has increased over time, with a step-change in 2012, which we would argue coincides with the increased commercialisation and wider adoption of EHR systems following the US HiTech Act and EU Innovative Medicines Initiative. Since 2012, however, the number of publications has plateaued, perhaps in contrast with medical publication numbers in general, which continue to increase at a near exponential rate. It is clear, therefore, that there are barriers restricting the wider adoption and exploitation of EHR research methodologies, which must be addressed.
The UK’s adoption of EHRs as a research tool in diabetes in particular is embryonic, with the vast majority of publications being conference abstracts. The failure to convert these conference abstracts to full publications could suggest barriers exist to full publication, limitations to existing EHR datasets, or non-specialist researchers experimenting with EHR research.
Internationally, we would argue the potential of these research approaches is evident, with large sample sizes, across multiple centres, tackling a diverse range of research questions. There is the clear ability to adapt sample sizes to the research question under study with epidemiological studies frequently utilising the largest cohort numbers.
A particular challenge to the US studies that currently dominate the published literature are the insurance-based models and data restrictions that exist within such insurance-based healthcare systems and datasets. We might argue that some commercial US healthcare EHRs are designed to have insurance and billing structures,28 with patient care a subsequent (or secondary) addition, and therefore, in effect, making the extraction of data for research purposes a tertiary use.
Barriers to diabetes research
Approximately half of studies reported barriers or limitations, as a result of using EHR data. Many studies reported multiple limitations; many conference abstracts however were brief and did not outline limitations. The most commonly reported limitation was that of missing data values,29 examples include failures to record whether glucose values were fasting or random30 and limited information on diabetes-specific outcome measures such as foot amputation31 or cause of death in the community following hypoglycaemia or diabetic ketoacidosis.32
Limited information on medication compliance was frequently described as a barrier,33–35 this is particularly significant given the high proportion of studies focused specifically on medication treatment in diabetes. Problems with misclassification of diabetes, and difficulty distinguishing between type 1 and type 2 diabetes were also described.36,37 Only two studies reported problems with data extraction, namely the extraction of unstructured data26 and procedural variations in the documentation of information.27 There were, however, concerns regarding a lack of longitudinal data in certain EHRs and fragmentation of patient data across diverse EHRs.30,31 These data fragmentation and longitudinal concerns were more prominent in US studies, rather than UK studies, which would be expected from the nature of NHS records; however, without a single national EHR there will remain problems, despite all patients having a single national identifier number (NHS number).
It is important to note the high proportion of extracted articles that were conference proceedings, rather than journal articles, and to consider this as a barrier in itself. This is despite a wide range of important topics and meaningful findings discussed within these articles. This could represent barriers such as a lack of funding available to develop these research projects into substantial pieces of work sufficient for peer-reviewed journal publication, or a lack of suitable journals accepting such articles for publication. Whatever the reason for a failure to translate such research into full papers represents a barrier to the wider adoption of usefulness of EHR research, it is interesting that this was a particular barrier in the UK, and suggests that we continue not to utilise fully at a system level, the important information held within our EHRs.
Future opportunities for EHRs in diabetes research
There are clear opportunities that could overcome some of the challenges described above. Excitingly, the UK has now moved on from the failed NHS National Programme for IT, becoming the largest EHR market in Europe.11 This offers the exciting potential to overcome key barriers, most particularly that of generalisability. Many of the current EER-based studies are limited by being only single-centre studies or based on small regions. The UK healthcare system has the significant advantage of every member of the population having a unique patient identifier, which enables larger-sized studies and helps avoid some of the barriers generated by missing patient values or misclassifications as patients move between providers or re-present.
Limitations of this study and further work
This study has a number of limitations. First, it represents a narrative review without formal independent two-author article identification, extraction and analysis. Whilst a meta-analysis would be inappropriate given the diversity of the study designs and methodologies, a more systematic two-author approach to article selection and data extraction could be argued to improve the quality of the study. Importantly though, this study did undergo prospective PROSPERO registration, with a pre-defined and piloted data collection tool. In the context of the first review of its kind, the study still has significant potential to add learning to the research literature and should be considered as an exploratory review in an underexplored area on which future research can build.
Restrictions also limited this paper to considering only English language journals, it is certainly likely that EHR datasets internationally have been adopted for research purposes published in other languages – in particular from Asia, South America and Northern Europe. This review excluded the grey literature. It is evident from the articles extracted and references provided that a number of consultancy firms and charities have utilised EHR data and may not have published their work in academic journals. Finally, restrictions were placed on the definition of secondary use of EHRs, excluding registries and the use of EHRs for recruitment to clinical studies. Both these areas represent important research areas, and further study to understand the contribution of EHRs to these fields would be beneficial.
Further research is needed to look beyond simply diabetes and compare the approaches taken in other clinical specialities. The understandings developed for diabetes here might not be generalisable across disease processes. Indeed, in the increasing trend for medical research to occur within speciality ‘siloes’ there is the exciting potential for EHR-based research to cross and unite research teams.
Conclusions
There is clearly an established body of research that utilises EHRs as a data source for diabetes research. This research covers a broad range of research questions. The published studies often include large data sets but are limited by missing values (many specifically required for diabetes related research) and challenges of generalisability. The small number of journal articles published using UK data suggests research of this nature is only in its infancy in the UK. The UK however represents an exciting and almost unique environment for such research, with national unique patient identifiers allowing for large multi-centre sample sizes overcoming challenges of generalisability and maximising the clinical usefulness of results.
Contributorship
All authors have made a substantial contribution to the research article as described by the SAGE Digital Health Journal Guidelines
Declaration of Conflicting Interests
The author(s) declare(s) that there is no conflict of interest.
Ethical approval
Given the nature of this review article, no ethical review was required and no ethical issues were raised.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors
Guarantor
TR
Peer review
This manuscript was reviewed by John Ainsworth, University of Manchester and Zhijun Yin, Vanderbilt University Department of Biomedical Informatics.
References
- 1.Botsis T, Hartvigsen G, Chen F, et al. Secondary use of EHR: data quality issues and informatics opportunities. AMIA Summits Transl Sci Proc 2010; 2010: 1–5. [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao D and, Weng C. Combining PubMed knowledge and EHR data to develop a weighted bayesian network for pancreatic cancer prediction. J Biomed Inform 2011; 44(5): 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothnie KJ, Müllerová H, Hurst JR, et al. Validation of the recording of acute exacerbations of COPD in UK primary care electronic healthcare records. PLoS One 2016; 11(3): e0151357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijayakrishnan R, Steinhubl SR, Ng K, et al. Prevalence of heart failure signs and symptoms in a large primary care population identified through the use of text and data mining of the electronic health record. J Cardiac Fail 2014; 20(7): 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panahiazar M, Taslimitehrani V, Pereira N, et al. Using EHRs and machine learning for heart failure survival analysis. Stud Health Technol Inform 2015; 216: 40. [PMC free article] [PubMed] [Google Scholar]
- 6.Manion FJ, Harris MR, Buyuktur AG, et al. Leveraging EHR data for outcomes and comparative effectiveness research in oncology. Curr Oncol Rep 2012; 14(6), 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Häyrinen K, Saranto K and, Nykänen P. Definition, structure, content, use and impacts of electronic health records: a review of the research literature. Int J Med Inform 2008; 77(5): 291–304. [DOI] [PubMed] [Google Scholar]
- 8.Van Der Meijden MJ, Tange HJ, Troost J, et al. Determinants of success of inpatient clinical information systems: a literature review. J Am Med Inform Assoc 2003; 10(3): 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schloeffel P. Electronic Health Record Definition, Scope and Context. ISO. TC. 2002; ▪: 215. [Google Scholar]
- 10.Utterback J. Utterback, James, Mastering the Dynamics of Innovation: How Companies Can Seize Opportunities in the Face of Technological Change (1994). University of Illinois at Urbana-Champaign's Academy for Entrepreneurial Leadership Historical Research Reference in Entrepreneurship. 1994; Harvard Business Review Press. Book ISBN 0875843425.
- 11.Accenture. Getting EMR Back in the Fast Lane. https://www.accenture.com/_acnmedia/PDF-4/Accenture-Getting-EMR-Back-Fast-Lane.pdf#zoom=50 (2015, accessed 19 Sept 2016)
- 12.Blumenthal D. Launching hitech. N Engl J Med 2010; 2010(362): 382–385. [DOI] [PubMed] [Google Scholar]
- 13.Hunter AJ. The Innovative Medicines Initiative: a pre-competitive initiative to enhance the biomedical science base of Europe to expedite the development of new medicines for patients. Drug Discov Today 2008; 13(9): 371–373. [DOI] [PubMed] [Google Scholar]
- 14.Jha AK. Meaningful use of electronic health records: the road ahead. JAMA 2010; 304(15): 1709–1710. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization. Global Report on Diabetes. Geneva: World Health Organization, 2016. [Google Scholar]
- 16.Booth A, Clarke M, Dooley G, et al. The nuts and bolts of PROSPERO: an international prospective register of systematic reviews. Syst Rev 2012; 1(1): 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eysenbach G. Tackling publication bias and selective reporting in health informatics research: register your eHealth trials in the International eHealth Studies Registry. J Med Internet Res 2004; 6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopewell S, Clarke M, Mallett S. Grey literature and systematic reviews In Rothstein HR, Sutton AJ, Borenstein M. (eds) Publication Bias in Meta-analysis: Prevention, assessment and adjustments. Chichester, UK: Wiley, 2005, pp.48–72. [Google Scholar]
- 19.Nirantharakumar K, Chen YF, Marshall T, et al. Clinical decision support systems in the care of inpatients with diabetes in non‐critical care setting: systematic review. Diabetic Med 2012; 29(6): 698–708. [DOI] [PubMed] [Google Scholar]
- 20.Karter AJ, Ackerson LM, Darbinian JA, et al. Self-monitoring of blood glucose levels and glycemic control: the Northern California Kaiser Permanente Diabetes registry. Am J Med 2001; 111(1), 1–9. [DOI] [PubMed] [Google Scholar]
- 21.Embi PJ, Jain A, Clark J, et al. Development of an electronic health record-based Clinical Trial Alert system to enhance recruitment at the point of care. In: AMIA Annual Symposium Proceedings 2005, Hilton Washington & Towers, Washington DC (Vol. 2005, p. 231). American Medical Informatics Association. Published in AMIA Annual Symposium Proceedings Archive as an independent pubmed entry. Held in Hilton Washington & Towers, Washington DC. [PMC free article] [PubMed]
- 22.Mulnier HE, Seaman HE, Raleigh VS, et al. Risk of stroke in people with type 2 diabetes in the UK: a study using the General Practice Research Database. Diabetologia 2006; 49(12): 2859–2865. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Cuevas R, Doubova SV, Suarez-Ortega M, et al. Evaluating quality of care for patients with type 2 diabetes using electronic health record information in Mexico. BMC Med Inform Decis Mak 2012; 12(1): 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pivovarov R, Albers DJ, Hripcsak G, et al. Temporal trends of hemoglobin A1c testing. J Am Med Inform Ass 2014; 21(6): 1038–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamblyn R, Abrahamowicz M, Beauchamp ME. International pharmacosurveillance: Modeling exposure to diabetic drugs in relationship to cardiovascular effects. Pharmacoepidemiol Drug Saf 2013; 22: 182. [Google Scholar]
- 26.Pantalone KM, Hobbs TM, Wells BJ, et al. Clinical characteristics, complications, comorbidities and treatment patterns among patients with type 2 diabetes mellitus in a large integrated health system. BMJ Open Diabetes Res Care 2015; 3(1): e000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollard C, Bailey KA, Petitte T, et al. Electronic patient registries improve diabetes care and clinical outcomes in rural community health centers. J Rural Health 2009; 25(1): 77–84. [DOI] [PubMed] [Google Scholar]
- 28.Menachemi N, Collum TH. Benefits and drawbacks of electronic health record systems. Risk Manag Healthc Policy 2011; 4: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barkhuysen P, de Grauw W, Akkermans R, et al. Is the quality of data in an electronic medical record sufficient for assessing the quality of primary care? J Am Med Inform Ass 2013; 21(4): 692–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Ass 2011; 19(2): 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messenger G, Masoetsa R, Hussain I, et al. Diabetic foot ulcer outcomes from a podiatry led tertiary service in Kuwait. Diabet Foot Ankle 2018; 9(1): 1471927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibb FW, Teoh WL, Graham J, et al. Risk of death following admission to a UK hospital with diabetic ketoacidosis. Diabetologia 2016; 59(10): 2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pantalone KM, Kattan MW, Yu C, et al. Increase in overall mortality risk in patients with type 2 diabetes receiving glipizide, glyburide or glimepiride monotherapy versus metformin: a retrospective analysis. Diabetes Obes Metab 2012; 14(9): 803–809. [DOI] [PubMed] [Google Scholar]
- 34.Egan BM, Shaftman SR, Wagner CS, et al. Demographic differences in the treatment and control of glucose in type 2 diabetic patients: implications for health care practice. Ethn Dis 2012; 22(1): 29–37. [PubMed] [Google Scholar]
- 35.Pantalone KM, Kattan MW, Yu C, et al. The risk of overall mortality in patients with type 2 diabetes receiving glipizide, glyburide, or glimepiride monotherapy: a retrospective analysis. Diabetes Care 2010. Mar 8; 33(6) 1224–1229. [DOI] [PMC free article] [PubMed]
- 36.Klompas M, Eggleston E, McVetta J, et al. Automated detection and classification of type 1 versus type 2 diabetes using electronic health record data. Diabetes Care 2012:DC_120964; 36(4) 914–921. [DOI] [PMC free article] [PubMed]
- 37.Esteban S, Tablado MR, Peper FE, et al. Development and validation of various phenotyping algorithms for Diabetes Mellitus using data from electronic health records. Comput Methods Programs Biomed 2017; 152: 53–70. [DOI] [PubMed] [Google Scholar]