Abstract
Alpha-amylases are major digestive enzymes that act in the first step of maltopolysaccharide digestion. In insects, these enzymes have long been studied for applied as well as purely scientific purposes. In many species, amylases are produced by multiple gene copies. Rare species are devoid of Amy gene. They are predominantly secreted in the midgut but salivary expression is also frequent, with extraoral activity. Enzymological parameters are quite variable among insects, with visible trends according to phylogeny: Coleopteran amylases have acidic optimum activity, whereas dipteran amylases have neutral preference and lepidopteran ones have clear alkaline preference. The enzyme structure shows interesting variations shaped by evolutionary convergences, such as the recurrent loss of a loop involved in substrate handling. Many works have focused on the action of plant amylase inhibitors on pest insect amylases, in the frame of crop protection by transgenesis. It appears that sensitivity or resistance to inhibitors is finely tuned and very specific and that amylases and their inhibitors have coevolved. The multicopy feature of insect amylases appears to allow tissue-specific or stage-specific regulation, but also to broaden enzymological abilities, such as pH range, and to overcome plant inhibitory defenses.
Keywords: Amylase inhibitors, multigene families, diet, midgut, salivary gland
Introduction
While conquering virtually all terrestrial and freshwater habitats, insects have evolved various feeding preferences. Many are phytophagous: They may consume seeds (eg, grain pests like weevils), stems (eg, lepidopteran stem borers like Sesamia species), roots (like the corn rootworm Diabrotica virgifera), or leaves (eg, leaf miner moths) or be sap feeders or nectar feeders. Other insects are carnivorous, saprophagous (eg, Drosophila melanogaster), or bloodsuckers. Particularly, phytophagous insects may be polyphagous or more specialized on a single or few host plants. Indeed even in the saprophagous drosophilids, some species are specialized on a single host plant (Drosophila sechellia on Morinda citrifolia fruit,1 Drosophila erecta on Pandanus fruit2). These preliminary remarks are of importance because insects must harbor enzymatic tools devoted to their respective diets, for detoxifying or circumventing the plant defenses, and for metabolizing useful nutriments. Nutritional content is obviously different in plants and in animal flesh, or in blood, and insects have evolved to optimize energetic uptake from their food.
Here I will draw a quick and noncomprehensive picture of insect alpha-amylases. Alpha-amylases (EC 3.2.1.1) are glycosyl hydrolases that break down alpha-1,4 glycosidic bonds inside a maltopolysaccharide linear chain, mainly in starch and glycogen, resulting in maltose, maltotriose, and residual branched maltodextrins as final products. These molecules are in turn hydrolyzed into glucose by alpha-glucosidases. Starch granules may have various structures and composition, which are more or less resistant to amylase: It was shown that the amylases of the weevil Sitophilus oryzae were unable to attack raw starch granules from potato, tapioca, wheat, or amylose-containing corn, but did degrade pea starch.3 In the bruchid Zabrotes subfasciatus, mastication seems a necessary process to damage starch granules and enable them susceptible to enzymatic degradation.4 In the living word, alpha-amylases (hereafter named simply amylases) are almost ubiquitous and are of utmost importance for nutrition of bacteria, plants, fungi, and animals, a lot of which having multiple copies of amylase genes, owing to gene duplications5 or horizontal transfer.6 The question of the evolutionary advantage for an organism to have several, sometimes diverged, amylase gene copies remains raised. Some enlightenment could be gained from insects, most of which rely on polysaccharides for their energy supply, and then depend on amylase activity.
One may acknowledge roughly two types of amylase studies on insects according to their focus: first, basic research dealing, for instance, with enzymology, genetics, evolution, and ecology; second, applied research that seeks to characterize digestive enzymes of insect of economical importance, such as crop pests or disease vectors. Intriguingly, it sometimes seems that these two research communities, ie, basic vs applied, are somewhat ignorant of each other.
Historically, insect amylases were widely studied early when electrophoresis techniques were developed, for it was easy and cheap. It allowed numerous studies on polymorphism when this unexpected kind of variation at the molecular level was evidenced,7–9 especially in Drosophila, raising questions about the functional or adaptive significance of the observed polymorphism, in terms of fitness and selection,10–15 or at the molecular level, in terms of catalytic activity or regulation. Amylase was then used as a gene model during the controversy between selectionnists and neutralists.12 For instance, researchers wondered if the various “isozymes” of amylases (ie, electrophoretic variants) had similar catalytic activities, similar heat sensitivities, similar tissular or temporal expression profiles?9,16–23 Also, attempts were made to link some characteristics of amylases to the natural diets of their producers, eg, electric charge,24 or catalytic activity.25,26
A Multigene Family in Insects
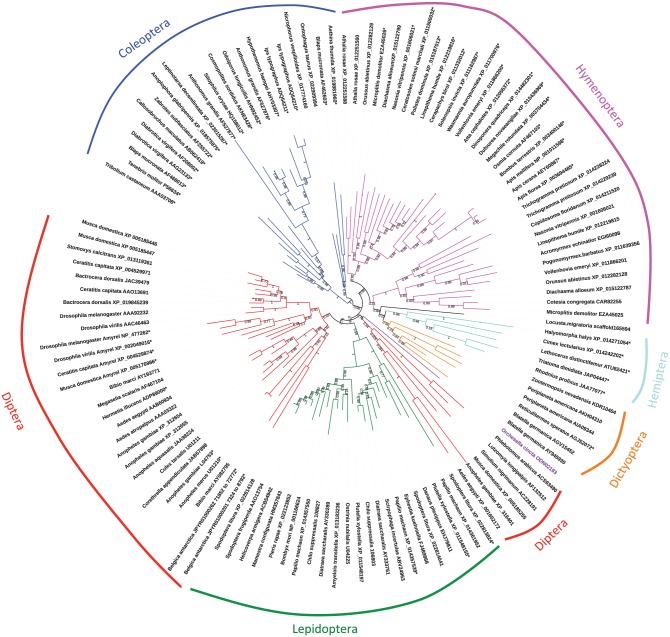
Several amylase gene copies (Amy) were reported in many insect species. Table 1 shows the number of gene copies that were reported in literature or by search in databases. The copy number varies from only 1 (eg, in honeybees) to more that 12 (in some mosquitoes). Indeed, most species harbor several copies. The Amy family was well described in Drosophila, as soon as 1967 for D melanogaster when Bahn28 discovered the Amy gene duplication, but importantly, like in many other insect groups, Amy duplications occurred largely independently in many Drosophila lineages.29–31,50–52 In this only genus, the number of gene copies vary from 1 (eg, Drosophila virilis)53 to 6 (Drosophila ananassae),30 not counting the paralog named Amyrel (Amy-related), a divergent copy (40% in amino acids) which is present throughout the drosophilids and is probably ancestral to Muscomorpha.54,55 Even more profound sequence divergence between Amy copies within genomes exists in most insect orders. Figure 1 shows a tree of selected amylase protein sequences of insects. Deep splitting of clusters is visible within Coleoptera, Lepidoptera, Hymenoptera, and Diptera, showing important divergences between paralogs. It is possible that intraspecific copy number variation occurs in species that have several Amy copies, like D ananassae, but there is no published report to my knowledge in insects, whereas it is well documented in humans59 and dogs.60 Classically, for a multigene family, sequence divergence30 and concerted evolution among copies61,62 were reported. Note the absence of amylase gene in rare genomes, such as the bloodsucker louse Pediculus humanus, the sap-feeding aphid Acyrthosiphon pisum, and other aphidomorphs (Table 1). It is tempting to link this deficiency to their specific, specialized feeding habits. However, amylase activity was detected in some aphids,63 although the genes were not identified, and a purely bloodsucking bug like Rhodnius prolixus does have an amylase gene. Nonetheless, exaptation of such an enzyme to another function linked to hematophagy is a possibility, because in this species an α-glucosidase was recruited for hemozoin formation from the heme of hemoglobin.64 The number of Amy gene copies cannot be clearly related to the diet. For instance, the copy number may vary greatly between related species that share similar diets (D virilis vs D melanogaster; Tenebrio molitor vs Tribolium castaneum; A pisum vs Bemisia tabaci; Table 1). However, it has been proposed that several gene copies may increase dietary flexibility, for instance, in housefly33 or in the soldier bug Podisus maculiventris.48
Table 1.
Number of reported Amy genes in insects from the literature or from genome database searches.
| Order | Species | Number of Amy copies | Reference |
|---|---|---|---|
| Diptera | Drosophila melanogaster | 2 + Amyrel | 28, FlyBase |
| Drosophila ficusphila | 2 + Amyrel | FlyBase | |
| Drosophila eugracilis | 2 + Amyrel | FlyBase | |
| Drosophila biarmipes | 2 + Amyrel | FlyBase | |
| Drosophila takahashii | 2 + Amyrel | FlyBase | |
| Drosophila elegans | 3 + Amyrel | FlyBase | |
| Drosophila rhopaloa | 2 + Amyrel | FlyBase | |
| Drosophila kikkawai | 4 + Amyrel | 29 | |
| Drosophila ananassae | 6 + Amyrel | 30, FlyBase | |
| Drosophila bipectinata | 2 + ψ + Amyrel | FlyBase | |
| Drosophila pseudoobscura | 3 + Amyrel | 31, FlyBase | |
| Drosophila persimilis | 3 + Amyrel | FlyBase | |
| Drosophila miranda | 2 + ψ+ Amyrel | 32 | |
| Drosophila willistoni | 2 + ψ + Amyrel | FlyBase | |
| Drosophila mojavensis | 1 + Amyrel | FlyBase | |
| Drosophila virilis | 1 + Amyrel | FlyBase | |
| Drosophila albomicans | 2 + Amyrel | FlyBase | |
| Drosophila grimshawi | 1 + Amyrel | FlyBase | |
| Drosophila hydei | 1 + Amyrel | FlyBase | |
| Ceratitis capitata | 2 + Amyrel | GenBank | |
| Musca domestica | 5 + Amyrel | 33, FlyBase | |
| Glossina morsitans | 1 + ψ | FlyBase | |
| Culex quinquefasciatus | 12 | FlyBase | |
| Aedes albopictus | 13 | GenBank | |
| Aedes aegypti | 9 | GenBank | |
| Anopheles darlingi | 5 | FlyBase | |
| Anopheles gambiae | 5 | FlyBase | |
| Mayetiola destructor | 1 | FlyBase | |
| Rhagoletis zephyria | 3-4 ? | GenBank | |
| Bactrocera oleae | 4 | GenBank | |
| Lucilia cuprina | 4 | GenBank | |
| Zeugodacus cucurbitae | 4 + ψ? | GenBank | |
| Lepidoptera | Bombyx mori | 3 | 34, FlyBase |
| Danaus plexippus | 4 | FlyBase | |
| Papilio machaon | 4 | GenBank | |
| Papilio polytes | 4 | GenBank | |
| Papilio xuthus | 4 | GenBank | |
| Ephestia kuehniella | 3 | 35 | |
| Spodoptera litura | 4 | GenBank | |
| Spodoptera frugiperda | 2 | 5 | |
| Helicoverpa armigera | 5 | 22,36, GenBank | |
| Plutella xylostella | 4 | GenBank | |
| Chilo suppressalis | 2 | Da Lage, unpubl. | |
| Glyphodes pyloalis | 2 | 37 | |
| Bicyclus anynana | 3 | GenBank | |
| Pieris rapae | 3 | GenBank | |
| Coleoptera | Tribolium castaneum | 7-9 | Hickey, unpublished, 38, GenBank |
| Hypothenemus hampei | 2 ? | 39 | |
| Blaps mucronata | 2 | 5 | |
| Sitophilus oryzae | 2 | 38,40 | |
| Tenebrio molitor | 1 | 38 | |
| Diabrotica virgifera | 2 | 41 | |
| Anthonomus grandis | 2 | 42 | |
| Ips typographus | 2 | 43 | |
| Acanthoscelides obtectus | 2 | 44 | |
| Zabrotes subfasciatus | 3 “isoforms” | 45 | |
| Aethina tumida | 3 | GenBank | |
| Anoplophora glabripennis | 4 | GenBank | |
| Agrilus planipennis | 3 | GenBank | |
| Onthophagus taurus | 4 | GenBank | |
| Hymenoptera | Apis mellifera | 1 | 46, Hymenoptera genome database |
| Apis florea | 1 | Hymenoptera genome database | |
| Apis dorsata | 1 | Hymenoptera genome database | |
| Bombus terrestris | 1 | Hymenoptera genome database | |
| Bombus impatiens | 1 | Hymenoptera genome database | |
| Megachile rotundata | 1 | Hymenoptera genome database | |
| Melipona quadrifasciata | 1 | Hymenoptera genome database | |
| Nasonia vitripennis | 5 | Hymenoptera genome database | |
| Acromyrmex echinatior | 2 | Hymenoptera genome database | |
| Atta cephalotes | 2 | Hymenoptera genome database | |
| Solenopsis invicta | 2 | Hymenoptera genome database | |
| Camponotus floridanus | 2 | Hymenoptera genome database | |
| Blattodea | Blattella germanica | 3 | GenBank |
| Periplaneta americana | 1 | 47 | |
| Zootermopsis nevadensis | 2 | GenBank | |
| Cryptotermes secundus | 3 | GenBank | |
| Phasmatodea | Timema cristinae | 2 | InsectBase |
| Sipyloidea sipylus | 1 | InsectBase | |
| Meraudoidea extradentata | 1 | InsectBase | |
| Extatosoma tiaratum | 1 | InsectBase | |
| Aretaon asperrimus | 1 | InsectBase | |
| Orthoptera | Locusta migratoria | 3 | InsectBase |
| Hemiptera | Rhodnius prolixus | 1 | FlyBase |
| Cimex lectularius | 1 | GenBank | |
| Podisus maculiventris | 3 “isoforms” | 48 | |
| Lygus lineolaris | 2 | 49 | |
| Halyomorpha halys | 2 | GenBank | |
| Nilaparvata lugens | 0 | GenBank | |
| Myzus persicae | 0* | GenBank | |
| Diuraphis noxia | 0* | GenBank | |
| Acyrthosiphon pisum | 0* | FlyBase | |
| Bemisia tabaci | 5 | GenBank | |
| Phthiraptera | Pediculus humanus | 0** | FlyBase |
| Siphonaptera | Archaeopsylla erinacei | 1 | InsectBase |
| Trichoptera | Limnephilus lunatus | 8 | InsectBase |
| Thysanoptera | Frankliniella occidentalis | 1 | InsectBase |
| Odonata | Ladona fulva | 2? | InsectBase |
BLAST search was performed on genome data in the indicated databases in June 2018, by BLASTP or TBLASTN27 using Drosophila melanogaster Amy sequence BAB32511 as query. ψ: pseudogene. *: no amylase gene sequence found in Aphidomorphs, but amylase activity was reported in Aphididae species Aphis fabae and Aphis gossypii. **: the “putative amylase” with accession EEB15075 is an amino acid transport protein. FlyBase: flybase.org; GenBank: ncbi.nlm.nih.gov/protein/; Hymenoptera genome database: hymenopteragenome.org; InsectBase: insect-genome.com.
Figure 1.
Unrooted tree of amylase protein sequences of insects. Sequences were aligned with MUSCLE,56 and a Maximum Likelihood tree was built using the online server phylogeny.fr57 with GBLOCKS curation and default parameters. The tree was drawn with iTOL.58 Numbers along branches indicate posterior probabilities. Asterisks indicate the loss of the GHGA motif in proteins. Red: Diptera; green: Lepidoptera; dark blue: Coleoptera; pink: Hymenoptera; orange: Dictyoptera; light blue: Hemiptera; black: Orthoptera. Purple branch and label is a Collembola. Accession numbers or references follow the taxa names of the sequences.
Sequence and Enzymatic Characterization of Insect Amylases
Irrespective of the copy number, it is logically believed that phytophagous insects must have more active amylolytic enzymes than carnivorous insects65 and that various vegetal diet may regulate amylase levels differentially (see below). In line with the fact that insect amylase studies are often devoted to crop pests, most enzymatic characterization of insect amylases are from such insects, like seed-feeding beetles, or lepidopteran stem borers. However, model insects like drosophilids were intensively investigated as well. Using purified recombinant amylases, Commin et al25 attempted to evidence enzymological differences between amylases of the generalist D melanogaster and two specialists, D sechellia and D erecta. But more general and accurate comparisons between the specific activities (kcat) of insect amylases from species differing in their diet are still wanting.
The D melanogaster Amy sequence was published in 1986.66 Innumerable insect amylase sequences were published since then. Figure 1 is a small subset of what is available at GenBank and in various genome databases. These sequences allow comparative studies about the gene structure and protein evolution, regarding conserved or divergent parts of the protein. All insect amylases have about the same size, ie, coding sequences around 1500 nucleotides, corresponding to a mature protein weight around 50 to 55 kDa after removing the signal peptide, as amylase is secreted (Table 2). An exception is in some mosquitoes, where a long N-terminal domain of unknown function occurs in some copies.71 Accordingly, it is surprising that some amylase protein sizes reported in the literature are very different from this value.69,72 This may be in most cases due to migration artifacts arising from abnormal sodium dodecyl sulfate binding on proteins with very basic or acidic isoelectronic point or on glycosylated proteins, resulting in migration defects and false mass estimations. In such cases, amylase band excision from the gel and mass spectrometry analysis should provide more accurate results. The intron content of Amy genes is quite variable in insects, from no intron in D melanogaster to at least 6 in Lepidoptera73 so that gene lengths may vary a lot. At least one case of alternative splicing was reported, in the beetle Ips typographus.43 Importantly, insect amylases are overall quite similar to other animal amylases. They have been assigned to the GH13_15 subfamily of glycosyl hydrolases,74 with other invertebrate amylases, whereas vertebrate amylases belong to GH13_24, a somewhat artificial division. All animal amylases (and beyond) are made of 3 major domains, named A, B and C and the structure requires a calcium ion.75 The catalytic apparatus, in domain A, is conserved, but some interesting facts are to be noticed: An amino acid stretch named “flexible loop” with the motif GHGA, protruding near the catalytic cleft,75 which is an ancestral feature, is missing in many insect amylase sequences.5 Figure 1 indicates the sequences lacking this motif. For example, the GHGA motif is deleted and the flexible loop is much shortened in most coleopteran amylase sequences, except, intriguingly in two of them. This is surprising because, otherwise, it would have been obvious that the GHGA motif was lost in the coleopteran ancestor. In Hymenoptera, two types exist, one gene group with the flexible loop, another group lacking the loop. This suggests that the two types have been coexisting ancestrally. In Muscomorpha flies, the Amyrel paralog also lacks the GHGA motif.54,55 These observations suggest recurrent losses of the flexible loop in the course of evolution (convergences), due to selective constraints that remain to elucidate (see below). Another interesting feature is the substitution of a conserved arginine into a glutamine in some unrelated amylases, ie, another convergence. This arginine is involved in the fixation of an activating chloride ion which changes the protein conformation and without which a detrimental salt bridge interaction would form. The glutamine is found in all Lepidopteran amylases (studied in details by Pytelková et al35) and in the Amyrel protein of a part of drosophilids, eg, D virilis, but not D melanogaster.76 Those glutamine-bearing amylases cannot bind the chloride ion but are nonetheless active, chloride independent,35,76 probably due to compensating mutations, because simply mutating to a glutamine when an arginine is normally present almost abolishes enzymatic activity.76 It was proposed that the chloride independence would be an adaptation to an alkaline pH in the midgut.35
Table 2.
Estimates of amylase molecular weights in some insects.
| Order | Species | Molecular weight | Method | Reference |
|---|---|---|---|---|
| Hemiptera | Eurygaster integriceps | 49 kDa, 52 kDa | SDS-PAGE | 67 |
| Coleoptera | Anthonomus grandis | 50870 Da, 52680 Da | Protein sequence |
AF527876 AF527877 |
| Coleoptera | Sitophilus oryzae | 51318 Da 53 kDa 56 kDa |
Protein sequence Sedimentation equilibrium centrifugation SDS-PAGE |
HQ158012 38 |
| Coleoptera | Tenebrio molitor | 51240 Da 56 kDa |
Protein sequence SDS-PAGE |
P56634 38 |
| Coleoptera | Tribolium castaneum | 51568 Da 56 kDa |
Protein sequence SDS-PAGE |
AAA03708 38 |
| Coleoptera | Callosobruchus maculatus | 51768 Da | Protein sequence | AB062419 |
| Coleoptera | Zabrotes subfasciatus | 51438 Da | Protein sequence | AF255722 |
| Coleoptera | Diabrotica virgifera | 50910 Da 50517 Da |
Protein sequence |
AAG23133 AF208002 |
| Coleoptera | Morimus funereus | 31 kDa | FPLC column | 68 |
| Coleoptera | Hypothenemus hampei | 51243 Da | Protein sequence | AHY03307 |
| Diptera | Drosophila melanogaster | 51915 Da | Protein sequence | AAA92232 |
| Diptera | Ceratitis capitata | 53010 Da | Protein sequence | XP_004529971 |
| Diptera | Aedes aegypti | 52386 Da | Protein sequence | AAB60934 |
| Diptera | Lutzomyia longipalpis | 54019 Da | Protein sequence | AF132512 |
| Hymenoptera | Apis mellifera | 53870 Da | Protein sequence | NP_001011598 |
| Blattodea | Blattella germanica | 53701 Da 53140 Da |
Protein sequence |
AGV15452 AY945930 |
| Orthoptera | Dociostaurus maroccanus | 73 kDa | SDS-PAGE | 69 |
| Lepidoptera | Ostrinia nubilalis | 54292 Da | Protein sequence | U04225 |
| Lepidoptera | Pieris brassicae | 88 kDa | SDS-PAGE | 70 |
| Lepidoptera | Ephestia kuehniella | 54442 Da | Protein sequence | FJ489868 |
| Lepidoptera | Helicoverpa armigera | 54043 Da | Protein sequence | ACB54942 |
| Lepidoptera | Bombyx mori | 54644 Da | Protein sequence | NP_001166624 |
| Lepidoptera | Chilo suppressalis | 55019 Da 54811 Da |
Protein sequence | 108827 106803 |
| Lepidoptera | Spodoptera frugiperda | 54007 Da | Protein sequence | AAO13754 |
Estimates through protein sequences used the accession numbers indicated in Figure 1, removing the peptide signal. Other estimates used biochemical methods and are given with literature references numbered in italics as in text. FPLC = fast protein liquid chromatography; SDS-PAGE = sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
The optimum pH of amylases generally corresponds to the pH values in the midgut lumen.72 The optimum pH of insect amylases varies greatly depending on the species. Table 3 shows the optimum pH reported for some insect species. Coleoptera show mostly acidic optimum pH for amylase activity, whereas Lepidopteran amylases generally have alkaline preferences. Dipteran amylases have more neutral preference. Therefore, the hypothesis that chloride independence is adapted to high pH values does not hold in the case of Amyrel working at a neutral pH. In some species, several amylases are produced, with different pH optima, due to different tissue specificities37,108 or stage specificities.104 Dow119 suggested that a high gut pH in insects, such as in Lepidoptera, could be an adaptation to feeding on tannin-rich plants, because high pH decreases the binding of tannins to nutritious proteins and thus enhances digestibility.
Table 3.
Optimum pH of insect amylases, from published studies, and inhibition or noninhibition by plant amylase inhibitors or other plant extracts.
| Order | Species | Optimum pH | Active inhibitors | Inactive inhibitors | Reference |
|---|---|---|---|---|---|
| Hemiptera | Leptoglossus zonatus | 5.6 | 79 | ||
| Podisus maculiventris | 6.0 | 48 | |||
| Graphosoma lineatum | 6.0 | αAI1 | 80 | ||
| Eurygaster maura | 6-7 | 81 | |||
| Eurygaster integriceps | 6.5 | 67 | |||
| Aphis fabae | 7 | 63 | |||
| Coleoptera | Acanthoscelides obtectus | 7-7.5 | 0.19 WI, 0.53 WI, HI | αAI1, αAI2 | 80,82-85 |
| Anthonomus grandis | ? | chimeric AI from αAI1 and αAI2, BIII from rye | αAI1, αAI2 0.19 WI, 0.53 WI |
42,86 | |
| Prostephanus truncatus | 6.0 | Amaranth ΑΙ | Maize AI Phaseolus acutifolius AI |
87 | |
| Cryptolestes ferrugineus | 5.0-5.5 | αAI1 | 80 | ||
| Oryzaephilus surinamensis | 4.5-5.0 | αAI1 | 80 | ||
| Sitophilus granarius | 4.5 | WI, HI, Thymus vulgaris extract | αAI1 | 80,82,88 | |
| Sitophilus oryzae | 5.0 | WRP25, ΗΙ | Corn AI | 3,38,82 | |
| Sitophilus zeamais | 5-7 | WΙ | 89 | ||
| Tenebrio molitor | 5.4 | WRP25, Corn AI, 0.28 WI, HI | Amaranth AI (weak effect) 0.19 WI, 0.53 WI | 38,82,90,91 | |
| Tribolium castaneum | 4.5-5.0 | αAI1, WRP25, HI, 0.28 WI, Corn AI Amaranth AI, Withania somnifera AI |
38,80,82,92-94 | ||
| Bruchus pisorum | 5.5 | αAI1, αAI2 | 86,95 | ||
| Rhyzopertha dominica | 7 | WI | Thymus vulgaris extract, Punica granatum extract | 82,88,96 | |
| Callosobruchus maculatus | 5.0 | αΑΙ, Amaranth ΑΙ, 0.19 WI, 0.53 WI, HI, Punica granatum extract, Achyranthes aspera AI | Vigna unguiculata AI | 82,83,85,88,92,97-99 | |
| Zabrotes subfasciatus | 6-7 | αAI2, WRP25, 0.19 WI, 0.53 WI | αAI1, WRP26 | 83,84,86,100,101 | |
| Diabrotica virgifera | 5.7 | αAI1, wheat AI | 41 | ||
| Morimus funereus | 5.2 | 68 | |||
| Hypothenemus hampei | 4.5-5.2 | αAI1, Amaranth AI, Ph. coccineus AI | αAI2 | 38,97,102,103 | |
| Plagiodera versicolora | 41, 82 | 104 | |||
| Alphitobius diaperinus | 5.0 | αAI1 | 105 | ||
| Diptera | Drosophila melanogaster | 7.4** | αAI1, WI | 9,80 | |
| Ceratitis capitata | 8.0 | 106 | |||
| Sarcophaga bullata | 7.0 | αAI1 | 80 | ||
| Aedes aegypti | 7.0 | αAI1 | 80 | ||
| Lutzomyia longipalpis | 8.5 | 107 | |||
| Hymenoptera | Monomorium pharaonis | 5.0-5.5 | αAI1 | 80 | |
| Apis mellifera | 5.0-5.5 | αAI1 | 80 | ||
| Venturia canescens | 5.0-5.5 | αAI1 | 80 | ||
| Blattodea | Blattella germanica | 6.0 | αAI1 | 80 | |
| Orthoptera | Acheta domesticus | 5.5-6.5 | αAI1 | 80 | |
| Dociostaurus maroccanus | 6.0 | αAI1, αAI2 | 108 | ||
| Calliptamus italicus | 8 | 108 | |||
| Gryllodes sigillatus | 6.6-7.0 | 69 | |||
| Lepidoptera | Manduca sexta | 10 | αAI1 | 80 | |
| Ostrinia nubilalis | 11 | αAI1 | 80 | ||
| Pieris brassicae | 8 | 70 | |||
| Ephestia kuehniella | 9 | WI1, WI3* | αAI1 | 35 | |
| Helicoverpa armigera | 9-11* | WI, Achyranthes aspera AI | Amaranth AI | 36,92,99 | |
| Bombyx mori | 6.83-9.24 | 108 | |||
| Chilo suppressalis | 9 | 109 | |||
| Acherontia atropos | 12 | 110 | |||
| Lasiocampa quercus | 10.8 | 110 | |||
| Lichnoptera felina | 10.8 | 110 | |||
| Antheraea mylitta | 9.5 | 111 | |||
| Spodoptera littoralis | 9.5 | 112 | |||
| Mamestra brassicae | 9.5 | 113 | |||
| Erinnyis ello | 9.8 | 114 | |||
| Tecia solanivora | 9.0 | Amaranth AI | 115 | ||
| Glyphodes pyloalis | 95-104 | 37 | |||
| Naranga aenescens | 10 | 116 | |||
| Spodoptera frugiperda | 8.5-9.5 | wheat tetrameric inhibitor | 117 | ||
| Tuta absoluta | 8.0 | Amaranth AI, WI | 118 |
AI: amylase inhibitor; 0.19, 0.28, 0.53 WI, WRP25, WRP26: wheat inhibitors; HI: barley inhibitor; *depending on the amylase paralog; **no amylase activity in the acidic mid-midgut77; 1: larval amylase; 2: adult amylase; 3: hemolymph amylase; 4: digestive amylase; 5: salivary amylase. Another table of insect target/amylase inhibitor has been published elsewhere.78
The optimum temperatures reported are typical of mesophilic amylases for the insects studied to date. Note however that results may vary significantly according to purification and assay conditions. Indeed, the temperature for maximal activity is strongly dependent on the assay duration, because long incubation at high temperature accelerates the enzyme denaturation; raw extracts contain proteases that are also activated by increased temperature and therefore degrade proteins in the sample, including amylases. This results in a lower apparent optimal temperature. For instance, optimum temperature for amylases of D melanogaster, D sechellia, and D erecta was estimated 37°C on raw extracts,26 but rather 57°C to 60°C using purified enzymes produced in vitro.25,120 To avoid this drawback, addition of commercially available protease inhibitor cocktails to crude extracts is a good practice. It is supposed that species that experience sun exposure in open fields should have more thermal-resistant amylases than species living in cold areas. But therefore it is not easy to compare optimum temperatures among amylases from insects that have contrasted thermal preferences without using standardized enzyme purification and assay protocols.
Localization, Secretion, and Regulation of Amylases in Insects
In most species studied, amylase is secreted at least in the midgut. It seems that in a number of insect species, the enzyme is partly recovered from the residual undigested food, through endo-ectoperitrophic circulation.72,121 In Drosophila, Amy tissue-specific expression was studied in details; no clear expression was found outside larval or adult midgut, as can be seen in RNAseq data at FlyBase. In D melanogaster, compartmentalization was found along the midgut, with no expression in the acidic mid-midgut, and 3 areas in the anterior midgut and 2 areas in the posterior midgut, with various combinations depending on genotypes and diet.122 This tissue-specific expression was controlled by a putative trans-acting factor named “map” (midgut activity pattern),77 located 2 cM downstream of the structural genes. However, the map gene was never identified until now in genome annotation. Similar complex midgut expression was found in other Drosophila species in larvae and adults.17,123 In D ananassae, different gene copies were expressed in different parts of the midgut.17 Whereas extraoral amylase activity was recognized in Drosophila,124,125 leading to a “social digestion,”126 the enzyme is produced by the midgut and regurgitated, but not by the salivary glands. A fine picture of amylase secretion in adult D melanogaster was also published more recently.127 In other Diptera, amylase expression may take place in salivary glands, as in the adult sand fly Lutzomyia longipalpis,107 where it is downregulated after a blood meal. In Aedes aegypti, an amylase gene is specifically expressed in adult female salivary glands,71 showing that the occurrence of several Amy copies may serve fine regulation. A general review of midgut amylase secretion in insects was given in a rich review on digestive enzymes of insects by Terra and Ferreira,72 who studied the precise localization and secretion process, whether apocrine secretion (Lepidoptera, Coleoptera) or exocytosis (eg, Diptera). It appeared that the enzyme may be produced in the midgut and be moved forth to the foregut, where the first step of digestion, that involves cutting long polysaccharides by amylase, occurs. This is the case in Coleoptera, Dictyoptera, and Orthoptera.72,128,129
In bugs, seed-feeding species have exclusively midgut-produced amylases, contrary to predatory species.79 The predatory spined soldier bug P maculiventris injects a salivary amylase into its prey, performing an extraoral digestion,48 like the Miridae Lygus lineolaris.49 In the omnivorous Hemiptera Apolygus lucorum, amylase is produced mainly in salivary glands, and to a lesser extent in the midgut.130 In Coleoptera, expression may be limited to the midgut like in T molitor121 but may take place also (or alternatively) in the foregut and hindgut,22 or in the head in I typographus. In this species, the head-specific amylase is an unusually smaller protein due to alternative splicing.43 Lepidoptera often produce amylase in their salivary glands in addition to midgut, which may be excreted through the mouth, eg, in Sesamia nonagrioides (Noctuidae) (Da Lage, unpublished), in the mulberry moth Glyphodes pyloalis (Pyralidae) (possibly produced by different gene copies),37 in Chilo suppressalis (Pyralidae) with a tissular differentiation of the electromorphs,109 or in Helicoverpa armigera.22 In Bombyx mori, amylase activity was also reported in hemolymph, although at a much lower level than in the digestive tract.108 In the Tasar silkworm Antheraea mylitta, there is also a hemolymph activity.111 In Hymenoptera, the ant Acromyrmex subterraneus shows amylase activity mostly in the midgut but also in labial glands.131 In the honeybee Apis mellifera, amylase activity is important in the hypopharyngeal gland of foragers, but not nurses.46 Indeed, amylase is a component of honey. In Blattella germanica (Dictyoptera), an amylase named BGTG1 is active in the tergal gland and could play a nondigestive role by processing phagostimulating sugars that function as nuptial feeding stimulants.132,133
Many studies have focused on the regulation of amylase secretion by food. At the genetic and molecular level, Drosophila has been the main model. D melanogaster larvae adapt amylase excretion to the hardness of food.125 In the fruit fly, mostly downregulation by glucose or other sugars was reported,134-136 and also induction by starch, especially in larvae.136 Glucose repression was largely dependent on the strain, therefore on the genotype.135 Chng et al127 have demonstrated the involvement of the transforming growth factor β /activin signaling pathway in this repression. Such regulation is classically interpreted by sparing resource when amylase is not necessary.16 In a selection experiment, it was shown that genotypes favoring low amylase activity were favored in glucose-rich environments, and that natural populations of D melanogaster were adapted to a sugar-rich (but variable) environment.16 This is not the case in housefly, which seems insensitive to dietary glucose, and may secrete amylase constitutively due to its polyphagous diet.33 In the omnivorous bug A lucorum, amylase production is induced by vegetal food, whereas proteases are induced by animal food.130 In the moth H armigera, amylase expression depends on food richness in starch and saccharose. Higher levels of sugars occurring in the natural host plant lower H. armigera amylase gene expression.22 In the Western corn rootworm D virgifera (Chrysomelidae), there is much more amylase produced on maize seedlings than on an artificial diet.41 Interestingly, amylase secretion may be upregulated in the presence of an inhibitor in Ephestia kuehniella as a compensation for loss of activity.35 Also, amylase is upregulated upon insecticidal treatment in the cockroach Periplaneta americana.129
Amylase regulation is also stage specific. Larvae and adults may have very different feeding habits; in some species adults do not feed at all. In Lepidoptera and Coleoptera, most studies were done on larvae, the stages which damage crops. Amy genes may be differentially expressed in larvae or in adults, in the sense that not only the same gene may be differentially expressed,137 but also different gene copies.80 In Muscomorpha, Amyrel is expressed only in larvae.54 In D ananassae, which has six Amy copies, some are active in larvae but not in adults.17 In D. serrata and D. lebanonensis, larvae also express different amylase variants from those of adult flies.136 In the bug P maculiventris, the 3 isoforms show specific temporal expression.48
Insect Amylases and Their Inhibitors
One of the most fascinating aspects of insect amylases is their relationships with plant defenses directed toward them, ie, amylase inhibitors. Phytophagous insects face inhibitory molecules produced by plants as defenses against their feeding on them. Many studies were devoted to the sensitivity or resistance to plant extracts, mainly proteinaceous inhibitors, which are abundant in cereals and leguminosae, with the goal of making transgenic plants resistant to their own pests. As shown in Table 3, a given insect amylase may be insensitive to a plant inhibitor, and sensitive to an inhibitor from another plant, and reciprocally, a given inhibitor may inhibit strongly one insect amylase but have no action on a related species. Kluh et al80 have compared various insect species for their amylase sensitivity toward the αAI1 inhibitor from Phaseolus vulgaris, the most studied proteinaceous inhibitor. Importantly, there is a pH dependence in amylase/inhibitor interaction, so that the study must be done at the relevant biological pH at which interaction forms.80,101 There was a general trend among insect orders regarding their sensitivities (in fact more between legume feeders and the others), and at a lower taxonomic level, there were contrasted results too. For instance, Acanthoscelides obtectus amylase was tolerant to αAI1, but the one of T castaneum was very sensitive. It is related to the fact that A. obtectus feeds on legume seeds. Experiments were carried out using 1% αAI1 in the food, a realistic value. Also, amylase paralogs in a species may have contrasted sensitivities toward inhibitors, and this is another adaptive response to overcome plant defenses.35,80,138 The pea weevil Bruchus pisorum is sensitive to the bean inhibitor αAI1, but not to the pea inhibitor. This lead to design transgenic peas expressing the bean inhibitor in their developing seeds, yielding a high larval mortality (93%) in the pests.95 Unfortunately, immunogenicity was reported for transgenic amylase inhibitors likely because of minor changes in molecular architecture of the transferred protein.139 Similar experiments were done on another grain legume culture, the cowpea Vigna unguiculata, transgenized with the same αAI1. It became resistant to its pests Callosobruchus maculatus and C. chinensis.140 Coffee plants were also transformed with αAI1 and became resistant to the coffee berry borer Hypothenemus hampei.141 αAI1 has a paralog in some wild accessions of P vulgaris, named αAI2. They share 78% identity86 but have different inhibitory properties on insects83 (Table 3). None of them are able to inhibit the Anthonomus grandis amylase, but chimeric proteins made from pieces of both inhibitors were able to show inhibition.86 Note that another way to overcome plant inhibitors is to produce a lot of amylase.84
At the molecular level, interactions between amylases and their inhibitors were studied, in part to elucidate why closely related amylases may exhibit so contrasted sensitivities. The tridimensional structure of the T molitor amylase (TMA) was published, in interaction with different inhibitors.91,142 Compared with mammal amylases, a striking feature of TMA is the lack of the flexible loop. It was supposed to explain differences in sensitivities between mammals and insects toward some inhibitors, because in porcine amylase the existing loop is pushed away in the presence of inhibitor instead of moving toward the saccharide.94 However, many insects have the loop, and this may not be the reason.143,144 Loops protruding from the inhibitors interact with the catalytic cleft of the enzyme through ionic and hydrogen bonds98 and may also block the sugar-binding “subsites”91; the formation of the complex depends on a large number of amino acids at the interface of the two proteins.102 The sensitivity or resistance to an inhibitor depends rather from multiple incompatible structural changes rather than a single crucial mutation.80 Importantly, the efficiency of inhibitors also depends on their own natural resistance to the insect proteases encountered in the gut.84,102
Concluding Remarks
Most insects are strongly dependent on their amylases for development and survival. In this review, I have shown that the presence of several gene copies can be of interest in different ways: for more enzyme production, for fine developmental and tissue-specific expression, for broadening pH and substrate range, for overcoming the natural defenses of plants. The coevolution between insect amylases and proteinaceous plant inhibitors is a passionating adaptation paradigm and would deserve more basic studies. In this respect, more structures of insect amylases would be needed, but only the one of T molitor is publicly available to date.
Acknowledgments
I am grateful to Dr P.-A. Calatayud for inviting me to write this review.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: This work was supported by regular funding from the CNRS to the author.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: JLDL designed the review, analyzed online data and wrote the article.
References
- 1. R’Kha S, Capy P, David JR. Host-plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetical analysis. Proc Natl Acad Sci U S A. 1991;88:1835–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rio B, Couturier G, Lemeunier F, Lachaise D. Evolution d’une spécialisation saisonnière chez Drosophila erecta (Dipt., Drosophilidae). Ann Soc Entomol Fr (NS). 1983;19:235–248. [Google Scholar]
- 3. Celinska E, Bialas W, Borkowska M, Grajek W. Cloning, expression, and purification of insect (Sitophilus oryzae) alpha-amylase, able to digest granular starch, in Yarrowia lipolytica host. Appl Microbiol Biotech. 2015;99:2727–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Silva CP, Terra WR, Xavier-Filho J, et al. Digestion of legume starch granules by larvae of Zabrotes subfasciatus (Coleoptera: Bruchidae) and the induction of alpha-amylases in response to different diets. Insect Biochem Mol Biol. 2001;31:41–50. [DOI] [PubMed] [Google Scholar]
- 5. Da Lage J-L, van Wormhoudt A, Cariou M-L. Diversity and evolution of the alpha-amylase genes in animals. Biol Bratisl. 2002;57:181–189. [Google Scholar]
- 6. Da Lage J-L, Danchin EGJ, Casane D. Where do animal α-amylases come from? An interkingdom trip. FEBS Lett. 2007;581:3927–3935. [DOI] [PubMed] [Google Scholar]
- 7. Kikkawa H. An electrophoretic study on amylase in Drosophila melanogaster. Jpn J Genet. 1964;39:401–411. [Google Scholar]
- 8. Doane WW. Quantitation of amylases in Drosophila separated by acrylamide gel electrophoresis. J Exp Zool. 1967;164:363–378. [DOI] [PubMed] [Google Scholar]
- 9. Doane WW. Amylase variants in Drosophila melanogaster: linkage studies and characterization of enzyme extracts. J Exp Zool. 1969;171:321–342. [DOI] [PubMed] [Google Scholar]
- 10. Yardley DG, Anderson WW, Schaffer HE. Gene frequency changes at the alpha-amylase locus in experimental populations of Drosophila pseudoobscura. Genetics. 1977;87:357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Jong G, Scharloo W. Environmental determination of selective significance or neutrality of amylase variants in Drosophila melanogaster. Genetics. 1976;84:77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doane WW. Selection for amylase allozymes in Drosophila melanogaster: some questions. Evolution. 1980;34:868–874. [DOI] [PubMed] [Google Scholar]
- 13. Hickey DA. Selection on amylase allozymes in Drosophila melanogaster: selection experiments using several independently derived pairs of chromosomes. Evolution. 1979;33:1128–1137. [DOI] [PubMed] [Google Scholar]
- 14. Powell JR, Amato GD. Population genetics of Drosophila amylase. V.Genetic background and selection on different carbohydrates. Genetics. 1984;106:625–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamazaki T, Matsuo Y. Genetic variability and selection for inducibility at the amylase locus in Drosophila melanogaster. Jpn J Genet. 1983;58:383–386. [Google Scholar]
- 16. Araki H, Yoshizumi S, Inomata N, Yamazaki T. Genetic coadaptation of the amylase gene system in Drosophila melanogaster: evidence for the selective advantage of the lowest AMY activity and of its epistatic genetic background. J Hered. 2005;96:388–395. [DOI] [PubMed] [Google Scholar]
- 17. Da Lage J-L, Klarenberg A, Cariou M-L. Variation in sex-, stage- and tissue-specific expression of the amylase genes in Drosophila ananassae. Heredity. 1996;76:9–18. [DOI] [PubMed] [Google Scholar]
- 18. Eguchi Y, Matsuo Y. Divergence of the regulation of alpha-amylase activity in Drosophila melanogaster, Drosophila funebris, and Drosophila saltans. Biochem Genet. 1999;37:41–52. [DOI] [PubMed] [Google Scholar]
- 19. Fujimoto J, Kanou C, Eguchi Y, Matsuo Y. Adaptation to a starch environment and regulation of alpha-amylase in Drosophila. Biochem Genet. 1999;37:53-62. [DOI] [PubMed] [Google Scholar]
- 20. Hickey DA. Regulation of amylase activity in Drosophila melanogaster: variation in the number of enzyme molecules produced by different amylase genotypes. Biochem Genet. 1981;19:783–796. [DOI] [PubMed] [Google Scholar]
- 21. Hoorn AJW, Scharloo W. The functional significance of amylase polymorphism in Drosophila melanogaster. V. The effect of food components on amylase and alpha-glucosidase activity. Genetica. 1978;49:181–187. [Google Scholar]
- 22. Kotkar HM, Bhide AJ, Gupta VS, Giri AP. Amylase gene expression patterns in Helicoverpa armigera upon feeding on a range of host plants. Gene. 2012;501:1–7. [DOI] [PubMed] [Google Scholar]
- 23. Matsuo Y, Yamazaki T. Genetic analysis of natural populations of Drosophila melanogaster in Japan. VI. Differential regulation of duplicated amylase loci and degree of dominance of amylase activity in different environments. Jpn J Genet. 1986;61:543–558. [Google Scholar]
- 24. Prigent S, Renard E, Cariou M-L. Electrophoretic mobility of amylase in Drosophilids indicates adaptation to ecological diversity. Genetica. 2003;119:133–145. [DOI] [PubMed] [Google Scholar]
- 25. Commin C, Aumont-Nicaise M, Claisse G, Feller G, Da Lage J-L. Enzymatic characterization of recombinant α-amylase in the Drosophila melanogaster species subgroup: is there an effect of specialization on digestive enzyme? Genes Genet Syst. 2013;88:251–259. [DOI] [PubMed] [Google Scholar]
- 26. Shibata H, Yamazaki T. A comparative study of the enzymological features of alpha-amylase in the Drosophila melanogaster species subgroup. Jpn J Genet. 1994;69:251–258. [DOI] [PubMed] [Google Scholar]
- 27. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. [DOI] [PubMed] [Google Scholar]
- 28. Bahn E. Crossing over in the chromosomal region determining amylase isozymes in Drosophila melanogaster. Hereditas. 1967;58:1–12. [DOI] [PubMed] [Google Scholar]
- 29. Inomata N, Yamazaki T. Evolution of nucleotide substitutions and gene regulation in the amylase multigenes in Drosophila kikkawai and its sibling species. Mol Biol Evol. 2000;17:601–615. [DOI] [PubMed] [Google Scholar]
- 30. Da Lage J-L, Maczkowiak F, Cariou M-L. Molecular characterization and evolution of the amylase multigene family of Drosophila ananassae. J Mol Evol. 2000;51:391–403. [DOI] [PubMed] [Google Scholar]
- 31. Brown CJ, Aquadro CF, Anderson WW. DNA sequence evolution of the amylase multigene family in Drosophila pseudoobscura. Genetics. 1990;126:131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steinemann S, Steinemann M. The amylase gene cluster on the evolving sex chromosomes of Drosophila miranda. Genetics. 1999;151:151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCommas S, Shornick LP. The effect of carbohydrate sources on the level of amylase activity in Musca domestica. Biochem Genet. 1990;28:585–589. [DOI] [PubMed] [Google Scholar]
- 34. Kikkawa H. Biochemical genetics of Bombyx mori (Silkworm). Adv Genet. 1953;5:107–140. [DOI] [PubMed] [Google Scholar]
- 35. Pytelková J, Hubert J, Lepsík M, et al. Digestive alpha-amylases of the flour moth Ephestia kuehniella – adaptation to alkaline environment and plant inhibitors. FEBS J. 2009;276:3531–3546. [DOI] [PubMed] [Google Scholar]
- 36. Bhide A, Channale SM, Patil SS, Gupta VS, Ramasamy S, Giri AP. Biochemical, structural and functional diversity between two digestive α-amylases from Helicoverpa armigera. Bioch Biophy Acta. 2015;1850:1719–1728. [DOI] [PubMed] [Google Scholar]
- 37. Yezdani E, Sendi JJ, Zibaee A, Ghadamyari M. Enzymatic properties of alpha-amylase in the midgut and the salivary glands of mulberry moth, Glyphodes pyloalis Walker (Lepidoptera: Pyralidae). C R Biol. 2010;333:17–22. [DOI] [PubMed] [Google Scholar]
- 38. Chen M-S, Feng G, Zen KC, et al. α-Amylases from three species of stored grain coleoptera and their inhibition by wheat and corn proteinaceous inhibitors. Insect Biochem Mol Biol. 1992;22:261–268. [Google Scholar]
- 39. Valencia A, Bustillo AE, Ossa GE, Chrispeels MJ. Alpha-amylases of the coffee berry borer (Hypothenemus hampei) and their inhibition by two plant amylase inhibitors. Insect Biochem Mol Biol. 2000;30:207–213. [DOI] [PubMed] [Google Scholar]
- 40. Baker JE, Halliday WR, Lum PTM. Genetics of alpha amylase in Sitophilus oryzae (L.) (Coleoptera: Curculionidae). J Stored Prod Res. 1990;26:7–10. [Google Scholar]
- 41. Titarenko E, Chrispeels MJ. cDNA cloning, biochemical characterization and inhibition by plant inhibitors of the alpha-amylases of the Western corn rootworm, Diabrotica virgifera virgifera. Insect Biochem Mol Biol. 2000;30:979–990. [DOI] [PubMed] [Google Scholar]
- 42. Oliveira-Neto OB, Batista JAN, Rigden DJ, et al. Molecular cloning of α-amylases from cotton boll weevil, Anthonomus grandis and structural relations to plant inhibitors: an approach to insect resistance. J Protein Chem. 2003;22:77–87. [DOI] [PubMed] [Google Scholar]
- 43. Viktorinova I, Kucerova L, Bohmova M, et al. Characterization of two closely related α-amylase paralogs in the bark beetle, Ips typographus (L.). Arch Insect Biochem Physiol. 2011;77:179–198. [DOI] [PubMed] [Google Scholar]
- 44. Franco OL, Melo FR, Mendes PA, et al. Characterization of two Acanthoscelides obtectus α-amylases and their inactivation by wheat inhibitors. J Agric Food Chem. 2005;53:1585–1590. [DOI] [PubMed] [Google Scholar]
- 45. Lagarda-Diaz I, Geiser D, Guzman-Partida AM, Winzerling J, Vazquez-Moreno L. Recognition and binding of the PF2 lectin to α-amylase from Zabrotes subfasciatus (Coleoptera:Bruchidae) larval midgut. J Insect Sci. 2014;14:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohashi K, Natori S, Kubo T. Expression of amylase and glucose oxidase in the hypopharyngeal gland with an age-dependent role change of the worker honeybee (Apis mellifera L.). Eur J Biochem. 1999;265:127–133. [DOI] [PubMed] [Google Scholar]
- 47. Tamaki FK, Pimentel AC, Dias AC, et al. Physiology of digestion and the molecular characterization of the major digestive enzymes from Periplaneta americana. J Insect Physiol. 2014;70:22–35. [DOI] [PubMed] [Google Scholar]
- 48. Ghamari M, Hosseininaveh V, Darvishzadeh A, Chougule NP. Carbohydrases in the digestive system of the spined soldier bug, Podisus maculiventris (Say) (Hemiptera: Pentatomidae). Arch Insect Biochem Physiol. 2014;85:195–215. [DOI] [PubMed] [Google Scholar]
- 49. Zhu Y-C, Yao J, Luttrell R. Identification of genes potentially responsible for extra-oral digestion and overcoming plant defense from salivary glands of the Tarnished plant bug (Hemiptera: Miridae) using cDNA sequencing. J Insect Sci. 2016;16:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Daïnou O, Cariou ML, David JR, Hickey D. Amylase gene duplication: an ancestral trait in the Drosophila melanogaster species subgroup. Heredity. 1987;59:245–251. [DOI] [PubMed] [Google Scholar]
- 51. Doane WW, Thompson DB, Norman RA, Hawley SA. Molecular genetics of a three-gene cluster in the Amy region of Drosophila. In: Ogita ZI, Markert CL. (eds) Isozymes: Structure, Function and Use in Biology and Medicine. New York, NY: Wiley-Liss; 1990:19–48. [PubMed] [Google Scholar]
- 52. Popadic A, Norman RA, Doane WW, Anderson WW. The evolutionary history of the amylase multigene family in Drosophila pseudoobscura. Mol Biol Evol. 1996;13:883–888. [DOI] [PubMed] [Google Scholar]
- 53. Magoulas C, Loverre-Chyurlia A, Abukashawa S, Bally-Cuif L, Hickey DA. Functional conservation of a glucose-repressible amylase gene promoter from Drosophila virilis in Drosophila melanogaster. J Mol Evol. 1993;36:234–242. [DOI] [PubMed] [Google Scholar]
- 54. Da Lage J-L, Renard E, Chartois F, Lemeunier F, Cariou ML. Amyrel, a paralogous gene of the amylase gene family in Drosophila melanogaster and the Sophophora subgenus. Proc Natl Acad Sci U S A. 1998;95:6848–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maczkowiak F, Da Lage J-L. Origin and evolution of the Amyrel gene in the alpha-amylase multigene family of Diptera. Genetica. 2006;128:145–158. [DOI] [PubMed] [Google Scholar]
- 56. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Perry GH, Dominy NJ, Claw CG, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Axelsson E, Ratnakumar A, Arendt ML, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. [DOI] [PubMed] [Google Scholar]
- 61. Bally-Cuif L, Payant V, Abukashawa S, Benkel BF, Hickey DA. Molecular cloning and partial sequence characterization of the duplicated amylase genes from Drosophila erecta. Genet Sel Evol. 1990;22:57–64. [Google Scholar]
- 62. Hickey DA, Bally-Cuif L, Abukashawa S, Payant V, Benkel BF. Concerted evolution of duplicated protein-coding genes in Drosophila. Proc Natl Acad Sci U S A. 1991;88:1611–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Darvishzadeh A, Bandani AR, Mousavi SQ. Biochemical characterisation of α-amylase in two aphid species, Aphis fabae Scopoli (Hemiptera: Aphididae) and A. gossypii Glover (Hemiptera: Aphididae). Plant Protect Sci. 2014;50:84–89. [Google Scholar]
- 64. Mury FB, da Silva JR, Ferreira LS, et al. Alpha-glucosidase promotes hemozoin formation in a blood-sucking bug: an evolutionary history. PLoS ONE. 2009;4:e6966. doi:6910.1371/journal.pone.0006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. de Figueroa JM, Trenzado CE, Lopez-Rodriguez MJ, Sanz A. Digestive enzyme activity of two stonefly species (Insecta, Plecoptera) and their feeding habits. Comp Biochem Physiol A Mol Integr Physiol. 2011;160:426–430. [DOI] [PubMed] [Google Scholar]
- 66. Boer PH, Hickey DA. The alpha-amylase gene in Drosophila melanogaster: nucleotide sequence, gene structure and expression motifs. Nucleic Acids Res. 1986;14:8399–8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bandani AR, Kazzazi M, Mehrabadi M. Purification and characterization of midgut α-amylases of Eurygaster integriceps. Entomol Sci. 2009;12:25–32. [Google Scholar]
- 68. Dojnov B, Božic N, Nenadović V, Ivanović J, Vujčić Z. Purification and properties of midgut alpha-amylase isolated from Morimus funereus (Coleoptera: Cerambycidae) larvae. Comp Biochem Physiol B Biochem Mol Biol. 2008;149:153–160. [DOI] [PubMed] [Google Scholar]
- 69. Rafiei B, Ghadamyari M, Imani S, Hosseininaveh V, Ahadiyat A. Purification and characterization of α-amylase in Moroccan locust, Dociostaurus maroccanus Thunberg (Orthoptera: Acrididae) and its inhibition by inhibitors from Phaseolus vulgaris L. Toxin Rev. 2016;35:90–97. [Google Scholar]
- 70. Sharifloo A, Zibaee A, Sendi JJ, Talebi Jaroumi K. Characterization of a digestive α-amylase in the midgut of Pieris brassicae L. (Lepidoptera: Pieridae). Front Physiol. 2016;7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Grossman GL, Campos Y, Severson DW, James AA. Evidence for two distinct members of the amylase gene family in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 1997;27:769–781. [DOI] [PubMed] [Google Scholar]
- 72. Terra WR, Ferreira C. Insect digestive enzymes: properties, compartmentalization and function. Comp Biochem Physiol B Biochem Mol Biol. 1994;109:1–62. [Google Scholar]
- 73. Da Lage J-L, Maczowiak F, Cariou M-L. Phylogenetic distribution of intron positions in alpha-amylase genes of bilateria suggests numerous gains and losses. PLoS ONE. 2011;6:e19673. doi:19610.11371/journal.pone.0019673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stam MR, Danchin EGJ, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Prot Eng Des Sel. 2006;19:555–562. [DOI] [PubMed] [Google Scholar]
- 75. Strobl S, Maskos K, Betz M, et al. Crystal structure of the yellow meal worm alpha-amylase at 1.64Å resolution. J Mol Biol. 1998;278:617–628. [DOI] [PubMed] [Google Scholar]
- 76. Claisse G, Feller GMB, Da Lage J-L. A single amino-acid substitution toggles chloride dependence of the alpha-amylase paralog amyrel in Drosophila melanogaster and Drosophila virilis species. Insect Biochem Mol Biol. 2016;75:70–77. [DOI] [PubMed] [Google Scholar]
- 77. Abraham I, Doane WW. Genetic regulation of tissue-specific expression of amylase structural genes in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1978;75:4446–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mehrabadi M, Franco OL, Bandani AR. Plant proteinaceous alpha-amylase and proteinase inhibitors and their use in insect pest control. In: Bandani AR. (ed.) New Perspect Plant Protec. Rijeka: InTech; 2012:229–246. [Google Scholar]
- 79. Rocha AA, Pinto CJC, Samuels RI, Alexandre D, Silva CP. Digestion in adult females of the leaf-footed bug Leptoglossus zonatus (Hemiptera: Coreidae) with emphasis on the glycoside hydrolases α-amylase, α-galactosidase, and α-glucosidase. Arch Insect Biochem Physiol. 2014;85:152–163. [DOI] [PubMed] [Google Scholar]
- 80. Kluh I, Horn M, Hyblova J, et al. Inhibitory specificity and insecticidal selectivity of alpha-amylase inhibitor from Phaseolus vulgaris. Phytochemistry. 2005;66:31–39. [DOI] [PubMed] [Google Scholar]
- 81. Ravan S, Mehrabadi M, Bandani AR. Biochemical characterization of digestive amylase of wheat bug, Eurygaster maura (Hemiptera: Scutelleridae). Afric J Biotech. 2009;8:3640–3648. [Google Scholar]
- 82. Gutierrez C, Sanchez-Monge R, Gomez L, Ruiz-Tapiador M, Castañera P, Salcedo G. α-Amylase activities of agricultural insect pests are specifically affected by different inhibitor preparations from wheat and barley endosperms. Plant Sci. 1990;72:37–44. [Google Scholar]
- 83. Grossi de, Sa MF, Chrispeels MJ. Molecular cloning of Bruchid (Zabrotes subfasciatus) α-amylase cDNA and interactions of the expressed enzyme with bean amylase inhibitors. Insect Biochem Mol Biol. 1997;27:271–281. [DOI] [PubMed] [Google Scholar]
- 84. Ishimoto M, Chrispeels MJ. Protective mechanism of the Mexican bean weevil against high levels of alpha-amylase inhibitor in the common bean. Plant Physiol. 1996;111:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Franco OL, Rigden DJ, Melo FR, Bloch C, Jr, Silva CP, Grossi de, Sa MF. Activity of wheat α-amylase inhibitors towards bruchid α-amylases and structural explanation of observed specificities. Eur J Biochem. 2000;267:2166–2173. [DOI] [PubMed] [Google Scholar]
- 86. Mattar da, Silva MC, Perseghini del Sarto R, Lucena WA, et al. Employing in vitro directed molecular evolution for the selection of α-amylase variant inhibitors with activity toward cotton boll weevil enzyme. J Biotech. 2013;167:377–385. [DOI] [PubMed] [Google Scholar]
- 87. Mendiola-Olaya E, Valencia-Jiménez A, Valdés-Rodriguez S, Délano-Frier J, Blanco-Labra A. Digestive amylase from the larger grain borer, Prostephanus truncatus Horn. Comp Biochem Physiol B Biochem Mol Biol. 2000;126:425–433. [DOI] [PubMed] [Google Scholar]
- 88. Mehrabadi M, Bandani AR, Saadati F, Mahmudvand M. α-Amylase activity of stored products insects and its inhibition by medicinal plant extracts. J Agr Sci Tech. 2011;13:1173–1182. [Google Scholar]
- 89. Lopes KVG, Silva LB, Reis AP, Oliveira MGA, Guedes RNC. Modified α-amylase activity among insecticide-resistant and -susceptible strains of the maize weevil, Sitophilus zeamais. J Insect Physiol. 2010;56:1050–1057. [DOI] [PubMed] [Google Scholar]
- 90. Applebaum SW, Jancovic M, Birk Y. Studies on the midgut amylase activity of Tenebrio molitor L. larvae. J Insect Physiol. 1961;7:100–108. [Google Scholar]
- 91. Pereira PJ, Lozanov V, Patthy A, et al. Specific inhibition of insect alpha-amylases: yellow meal worm alpha-amylase in complex with the amaranth alpha-amylase inhibitor at 2.0 Å resolution. Structure. 1999;7:1079–1088. [DOI] [PubMed] [Google Scholar]
- 92. Bhide AJ, Channale SM, Yadav Y, et al. Genomic and functional characterization of coleopteran insect-specific α-amylase inhibitor gene from Amaranthus species. Plant Mol Biol. 2017;94:319–332. [DOI] [PubMed] [Google Scholar]
- 93. Kasar SS, Marathe KR, Bhide AJ, et al. A glycoprotein α-amylase inhibitor from Withania somnifera differentially inhibits various α-amylases and affects the growth and development of Tribolium castaneum. Pest Manag Sci. 2017;73:1382–1390. [DOI] [PubMed] [Google Scholar]
- 94. Payan F. Structural basis for the inhibition of mammalian and insect α-amylases by plant protein inhibitors. Bioch Biophy Acta. 2004;1696:171–180. [DOI] [PubMed] [Google Scholar]
- 95. Morton RL, Schroeder HE, Bateman KS, Chrispeels MJ, Armstrong E, Higgins TJV. Bean α-amylase inhibitor 1 in transgenic peas (Pisum sativum) provides complete protection from pea weevil (Bruchus pisorum) under field conditions. Proc Natl Acad Sci U S A. 2000;97:3820–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Baker JE. Purification and partial characterization of α-amylase allozymes from the lesser grain borer, Rhyzopertha dominica. Insect Biochem. 1991;21:303–311. [Google Scholar]
- 97. Channale SM, Bhide AJ, Yadav Y, et al. Characterization of two coleopteran α-amylases and molecular insights into their differential inhibition by synthetic α-amylase inhibitor, acarbose. Insect Biochem Mol Biol. 2016;74:1–11. [DOI] [PubMed] [Google Scholar]
- 98. Pelegrini PB, Lay FT, Murad AM, Anderson MA, Franco OL. Novel insights on the mechanism of action of α-amylase inhibitors from the plant defensin family. Proteins. 2008;73:719–729. [DOI] [PubMed] [Google Scholar]
- 99. Hivrale VK, Chougule NP, Giri AP, Chhabda PJ, Kachole MS. Biochemical characterisation of α-amylase inhibitors from Achyranthes aspera and their interactions with digestive amylases of coleopteran and lepidopteran insects. J Sci Food Agric. 2011;91:1773–1780. [DOI] [PubMed] [Google Scholar]
- 100. Pelegrini PB, Murad AM, Grossi-de-Sá MF, et al. Structure and enzyme properties of Zabrotes subfasciatus alpha-amylase. Arch Insect Biochem Physiol. 2006;61:77–86. [DOI] [PubMed] [Google Scholar]
- 101. Franco OL, Rigden DJ, Melo FR, Grossi-de-Sá MF. Plant alpha-amylase inhibitors and their interaction with insect alpha-amylases; structure, function and potential for crop protection. Eur J Biochem. 2002;269:397–412. [DOI] [PubMed] [Google Scholar]
- 102. Valencia-Jimenez A, Arboleda Valencia JW, Grossi de, Sa MF. Activity of α-amylase inhibitors from Phaseolus coccineus on digestive α-amylases of the coffee berry borer. J Agric Food Chem. 2008;56:2315–2320. [DOI] [PubMed] [Google Scholar]
- 103. Albuquerque EVS, Bezerra CA, Romero JV, et al. Seed-specific stable expression of the α-AI1 inhibitor in coffee grains and the in vivo implications for the development of the coffee berry borer. Trop Plant Biol. 2015;8:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shabarari M, Naseri B, Zibaee A, Hajizadeh J. Characterization of digestive α-amylase in the midgut of willow leaf beetle Plagiodera versicolora (Coleoptera: Chrysomelidae). J Crop Protec. 2014;3:245–254. [Google Scholar]
- 105. Cruz WO, Sinhori GGC, de Lima CAR, Pontes EG. Biochemical properties of α-amylase from midgut of Alphitobius diaperinus (Panzer) (Coleoptera: Tenebrionidae) larvae. Neotrop Entomol. 2018;47:698–708. [DOI] [PubMed] [Google Scholar]
- 106. Darvishzadeh A, Hosseininaveh V, Ghamari M. Identification and biochemical characterisation of α-amylase in the alimentary tract of Mediterranean fruit fly, Ceratitis capitata (Wiedemann) (Diptera: Tephritidae). Arch Phytopath Plant Protec. 2013;46:1061–1069. [Google Scholar]
- 107. Vale VF, Moreira BH, Moraes CS, Pereira MH, Genta FA, Gontijo NF. Carbohydrate digestion in Lutzomyia longipalpis’ larvae (Diptera – Psychodidae). J Insect Physiol. 2012;58:1314–1324. [DOI] [PubMed] [Google Scholar]
- 108. Abraham EG, Nagaraju J, Datta RK. Biochemical studies of amylases in the silkworm, Bombyx mori L.: comparative analysis in diapausing and nondiapausing strains. Insect Biochem Mol Biol. 1992;22:867–873. [Google Scholar]
- 109. Zibaee A, Bandani AR, Kafil M, Ramzi S. Characterization of α-amylase in the midgut and the salivary glands of rice striped stem borer, Chilo suppressalis Walker (Lepidoptera: Pyralidae). J Asia-Pacif Entomol. 2008;11:201–205. [Google Scholar]
- 110. Dow JA. Extremely high pH in biological systems: a model for carbonate transport. Am J Physiol. 1984;246:R633–R636. [DOI] [PubMed] [Google Scholar]
- 111. Nagaraju J, Abraham EG. Purification and characterization of digestive amylase from the tasar silkworm, Antheraea mylitta (Lepidoptera: Saturniidae). Comp Biochem Physiol B Biochem Mol Biol. 1995;110:201–209. [Google Scholar]
- 112. Ishaaya I, Moore I, Joseph D. Protease and amylase activity in larvae of the Egyptian cotton worm, Spodoptera littoralis. J Insect Physiol. 1971;17:945–953. [Google Scholar]
- 113. Kusano T, Tanabe S. Enzymatic properties of the midgut amylase activity and its changes in the development in the cabbage armyworm Mamestra brassicae. Koontyu. 1986;54:12–24. [Google Scholar]
- 114. Santos CD, Terra WR. Distribution and characterization of oligomeric digestive enzymes from Erinnyis ello larvae and inferences concerning secretory mechanisms and the permeability of the peritrophic membrane. Insect Biochem. 1986;16:691–700. [Google Scholar]
- 115. Valencia-Jiménez A, Arboleda JW, Lopez Avila A, Grossi-de-Sà MF. Digestive alpha-amylases from Tecia solanivora larvae (Lepidoptera: Gelechiidae): response to pH, temperature and plant amylase inhibitors. Bull Entomol Res. 2008;98:575–579. [DOI] [PubMed] [Google Scholar]
- 116. Bandani AR, Maleki F, Rahmani S, Fazeli-Dinan M. Characterization of α-amylase in the alimentary canal of Naranga aenescens Moore (Lepidoptera: Noctuidae), the rice green caterpillar. Mun Ent Zool. 2010;5:716–725. [Google Scholar]
- 117. Alfonso J, Ortego F, Sanchez-Monge R, et al. Wheat and barley inhibitors active towards α-amylase and trypsin-like activities from Spodoptera frugiperda. J Chem Ecol. 1997;23:1729–1741. [Google Scholar]
- 118. Esmaeily M, Bandani AR. Interaction between larval α-amylase of the tomato leaf miner, Tuta absoluta Meyrick (Lepidoptera: Gelechiidae) and proteinaceous extracts from plant seeds. J Plant Protec Res. 2015;55:278–286. [Google Scholar]
- 119. Dow JAT. Insect midgut function. Adv Insect Physiol. 1986;19:187–328. [Google Scholar]
- 120. Cipolla A, Delbrassine F, Da Lage J-L, Feller G. Temperature adaptations in psychrophilic, mesophilic and thermophilic chloride-dependent alpha-amylases. Biochimie. 2012;94:1943–1950. [DOI] [PubMed] [Google Scholar]
- 121. Terra WR, Ferreira C, Bastos F. Phylogenetic considerations of insect digestion: Disaccharidases and the spatial organization of digestion in the Tenebrio molitor larvae. Insect Biochem. 1985;15:443–449. [Google Scholar]
- 122. Klarenberg AJ, Vermeulen JWC, Jacobs PJM, Scharloo W. Genetic and dietary regulation of tissue-specific expression patterns of α-amylase in larvae of Drosophila melanogaster. Comp Biochem Physiol B Biochem Mol Biol. 1988;89:143–146. [Google Scholar]
- 123. Powell JR, Lichtenfels JM. Population genetics of Drosophila amylase. I. Genetic control of tissue-specific expression in D. pseudoobscura. Genetics. 1979;92:603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Haj-Ahmad Y, Hickey DA. A molecular explanation of frequency-dependent selection in Drosophila. Nature. 1982;299:350–352. [DOI] [PubMed] [Google Scholar]
- 125. Sakaguchi H, Suzuki MG. Drosophila melanogaster larvae control amylase secretion according to the hardness of food. Front Physiol. 2013;4:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Gregg TG, McCrate A, Reveal G, Hall S, Rypstra AL. Insectivory and social digestion in Drosophila. Biochem Genet. 1990;28:197–207. [DOI] [PubMed] [Google Scholar]
- 127. Chng WA, Sleiman MSB, Schüpfer F, Lemaitre B. Transforming growth factor β/activin signaling functions as a sugar-sensing feedback loop to regulate digestive enzyme expression. Cell Rep. 2014;9:336–348. [DOI] [PubMed] [Google Scholar]
- 128. Woodring J. The flow and fate of digestive enzymes in the field cricket, Gryllus bimaculatus. Arch Insect Biochem Physiol. 2017;95. DOI:10.1002/arch.21398 [DOI] [PubMed] [Google Scholar]
- 129. Zhang J, Zhang Y, Li J, Liu M, Liu Z. Midgut transcriptome of the cockroach Periplaneta americana and its microbiota: digestion, detoxification and oxidative stress response. PLoS ONE. 2016;11:e0155254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Li W, Zhao X, Yuan W, Wu K. Activities of digestive enzymes in the omnivorous pest Apolygus lucorum (Hemiptera: Miridae). J Econ Entomol. 2017;110:101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Erthal M, Jr, Silva CP, Samuels RI. Digestive enzymes in larvae of the leaf cutting ant, Acromyrmex subterraneus (Hymenoptera: Formicidae: Attini). J Insect Physiol. 2007;53:1101–1111. [DOI] [PubMed] [Google Scholar]
- 132. Saltzmann KD, Saltzmann KA, Neal JJ, Scharf ME, Bennett GW. Characterization of BGTG-1, a tergal gland-secreted alpha-amylase, from the German cockroach, Blattella germanica. Insect Mol Biol. 2006;15:425–433. [DOI] [PubMed] [Google Scholar]
- 133. Myers AJ, Gondhalekar AD, Fardisi M, Saltzmann KD, Bennett GW, Scharf ME. RNA interference and functional characterization of a tergal gland alpha amylase in the German cockroach, Blattella germanica L. Insect Mol Biol. 2018;27:143–153. [DOI] [PubMed] [Google Scholar]
- 134. Hickey DA, Benkel BF. Regulation of amylase activity in Drosophila melanogaster: effects of dietary carbohydrate. Biochem Genet. 1982;20:1117–1129. [DOI] [PubMed] [Google Scholar]
- 135. Benkel BF, Hickey DA. Glucose repression of amylase gene expression in Drosophila melanogaster. Genetics. 1986;114:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Inomata N, Kanda K, Cariou M-L, Tachida H, Yamazaki T. Evolution of the response patterns to dietary carbohydrates and the developmental differentiation of gene expression of alpha-amylase in Drosophila. J Mol Evol. 1995;41:1076-1084. [DOI] [PubMed] [Google Scholar]
- 137. Tejima T, Ohba S. Genetic regulation of amylase activity in Drosophila virilis. I. Activity variation among laboratory strains. Jpn J Genet. 1981;56:457–468. [Google Scholar]
- 138. Sivakumar S, Mohan M, Franco OL, Thayumanavan B. Inhibition of insect pest α-amylases by little and finger millet inhibitors. Pest Biochem Physiol. 2006;85:155–160. [Google Scholar]
- 139. Prescott VE, Campbell PM, Moore A, et al. Transgenic expression of bean alpha-amylase inhibitor in peas results in altered structure and immunogenicity. J Agric Food Chem. 2005;53:9023–9030. [DOI] [PubMed] [Google Scholar]
- 140. Solleti SK, Bakshi S, Purkayastha J, Panda SK, Sahoo L. Transgenic cowpea (Vigna unguiculata) seeds expressing a bean α-amylase inhibitor 1 confer resistance to storage pests, bruchid beetles. Plant Cell Rep. 2008;27:1841–1850. [DOI] [PubMed] [Google Scholar]
- 141. Barbosa AEAD, Albuquerque EVS, Silva MCM, et al. α-Amylase inhibitor-1 gene from Phaseolus vulgaris expressed in Coffea arabica plants inhibits α-amylases from the coffee berry borer pest. BMC Biotech. 2010;10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Strobl S, Maskos K, Wiegand G, Huber R, Gomis-Rüth F-X, Glockshuber R. A novel strategy for inhibition of α-amylases: yellow meal worm α-amylase in complex with the Ragi bifunctional inhibitor at 2.5 Å resolution. Structure. 1998;6:911–921. [DOI] [PubMed] [Google Scholar]
- 143. Da Silva MCM, Grossi de, Sa MF, Chrispeels MJ, Togawa RC, Neshich G. Analysis of structural and physico-chemical parameters involved in the specificity of binding between α-amylases and their inhibitors. Protein Eng. 2000;13:167–177. [DOI] [PubMed] [Google Scholar]
- 144. Grossi de Sa MF, Mirkov TE, Ishimoto M, Colucci G, Bateman KS, Chrispeels MJ. Molecular characterization of a bean α-amylase inhibitor that inhibits the α-amylase of the Mexican bean weevil Zabrotes subfasciatus. Planta. 1997;203:295–303. [DOI] [PubMed] [Google Scholar]