Significance
Many species are evolving in response to human-induced global change, often at a pace that exceeds natural rates of trait change. This difference could be due to generally stronger phenotypic selection in human-impacted environments, to which populations might be relatively maladapted. We conducted a large-scale test of this hypothesis by synthesizing 1,366 selection estimates from populations exposed to a variety of disturbances. Surprisingly, this meta-analysis revealed that human disturbances might slightly decrease selection by increasing mean fitness, except for some fish stocks showing very strong harvest selection. This synthesis provides new insights into the evolutionary response of populations to global change, and suggests that only some human disturbances might have large immediate evolutionary impacts in nature.
Keywords: contemporary evolution, global change, anthropogenic stressor, fitness, harvest selection
Abstract
Human activities are driving rapid phenotypic change in many species, with harvesting considered to be a particularly potent evolutionary force. We hypothesized that faster evolutionary change in human-disturbed populations could be caused by a strengthening of phenotypic selection, for example, if human disturbances trigger maladaptation and/or increase the opportunity for selection. We tested this hypothesis by synthesizing 1,366 phenotypic selection coefficients from 37 species exposed to various anthropogenic disturbances, including harvest. We used a paired design that only included studies measuring selection on the same traits in both human-disturbed and control (not obviously human-disturbed “natural”) populations. Surprisingly, this meta-analysis did not reveal stronger selection in human-disturbed environments; in fact, we even found some evidence that human disturbances might slightly reduce selection strength. The only clear exceptions were two fisheries showing very strong harvest selection. On closer inspection, we discovered that many disturbances weakened selection by increasing absolute fitness and by decreasing the opportunity for selection—thus explaining what initially seemed a counterintuitive result. We discuss how human disturbances can sometimes weaken rather than strengthen selection, and why measuring the total effect of disturbances on selection is exceedingly difficult. Despite these challenges, documenting human influences on selection can reveal disturbances with particularly strong effects (e.g., fishing), and thus better inform the management of populations exposed to these disturbances.
The anthropogenic alteration of ecosystems is a well-recognized evolutionary force (1, 2). Disturbances such as climate change, habitat degradation, species introduction, and the harvest of wild animals and plants reconfigure adaptive landscapes, altering selective pressures acting on populations and shifting intraspecific trait values toward new trait optima (3–5). Ample data are now available on phenotypic change associated with such human pressures, and quantitative syntheses of these data have revealed that trait change (in part genetically based) is often faster and of greater magnitude in human-impacted populations than in more “natural” populations (5–7). Most dramatically, populations experiencing direct harvest (e.g., hunting and fishing) are often typified by particularly high rates of phenotypic change (8, 9), leading some authors to conclude that human predation might be one of the most potent selective agents in nature (10). Such large-scale trait change caused by human activities has important consequences for the dynamics of populations and communities, the functioning of ecosystems, and the services that we derive from them (11–14).
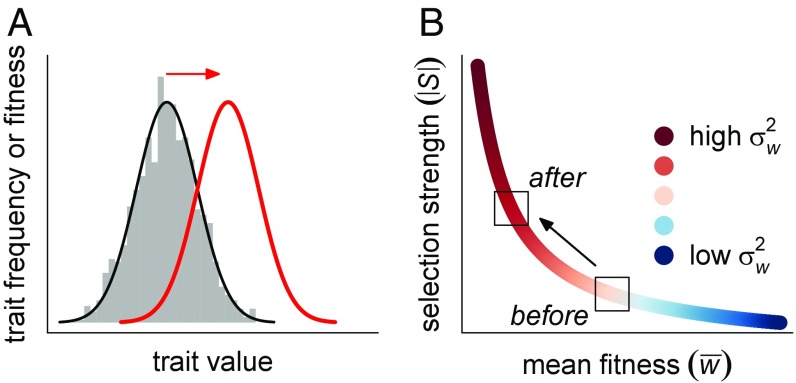
If phenotypic change is faster in disturbed environments, human disturbances need to affect at least one of the two drivers of evolutionary rates, namely the amount of heritable variation in, and the strength of selection acting on, traits. We suggest two main mechanisms through which disturbances could affect the latter by generally strengthening selection, thus accelerating trait change (Fig. 1). The first mechanism is a modification of the adaptive landscape (15) leading to maladaptation, whereby the most common trait values in a population no longer yield the highest fitness in the new, human-modified environment (4). Specifically, if we assume that most populations are reasonably well adapted to their current environments (16), most changes in the environment should increase the mismatch between a population’s current trait distribution and the fitness function for that trait in the new environment (Fig. 1A). Such fluctuations in adaptive landscapes and resulting levels of maladaptation are not limited to human-disturbed environments—they are also well documented in populations experiencing natural disturbances (16, 17). However, given the pronounced and well-documented impacts that humans have had on many abiotic and biotic variables (18), we should expect rates of environmental change (and thus the mean level of maladaptation) to be greater in the case of human disturbances, thus potentially strengthening selection.
Fig. 1.
Anthropogenic disturbances could generally strengthen phenotypic selection by increasing maladaptation and/or the opportunity for selection. (A) The disturbance causes maladaptation by shifting fitness peaks away from current trait distributions. The gray histogram illustrates the trait distribution of a fictive population which is well adapted to predisturbance conditions, and the black and red curves show the hypothetical pre- and postdisturbance fitness functions of the same trait, respectively. (B) The disturbance diminishes mean absolute fitness (e.g., number of offspring), thus increasing the variance in relative fitness and the opportunity for selection.
The second mechanism is related to the opportunity for selection, that is, the variance in relative fitness in a population (19, 20). Because phenotypic selection is defined (21) as the covariance between trait values and relative fitness (the absolute fitness of an individual divided by the mean absolute fitness of the population), human disturbances that affect the mean fitness or the variance in fitness of a population could influence the strength of selection via an altered opportunity for selection (22). Anthropogenic stressors often negatively impact population sizes (23, 24); thus, it seems reasonable to hypothesize that many disturbances would also affect population mean fitness. When a disturbance reduces fitness in absolute terms (e.g., all individuals produce one fewer offspring), a decrease in mean absolute fitness could lead to an increase in the relative fitness of a better-than-average individual and a decrease in the relative fitness of a worse-than-average individual, thereby increasing the variance in relative fitness (SI Appendix, Fig. S1). Such an increase in the opportunity for selection would result in higher covariance between the trait and relative fitness and, consequently, stronger selection (Fig. 1B). In contrast, if fitness loss across individuals is purely proportional (e.g., all individuals lose exactly 10% of their absolute fitness), then the opportunity for selection remains unchanged (SI Appendix, Fig. S1). A recent meta-analysis of experimental studies of selection revealed that manipulations of the biotic or abiotic environment often affect the opportunity for selection (25). Hence, it seems likely that at least some forms of human-induced environmental change would, too, influence the opportunity for selection, and thus that anthropogenic stressors could impact selection strength not only via maladaptation (as above) but also by changing the variance in relative fitness.
Several case studies have indeed documented an increase in selection strength due to various disturbances such as anthropogenic fire and invasive species (26, 27). Other studies, however, have found weaker (relaxed) selection in disturbed than undisturbed populations (28–30), such that it remains unclear whether an overall trend exists of generally stronger selection in human-altered environments. To assess whether such a trend exists, we synthesized the recent literature that reported estimates of human impacts on selection strength. Since Lande and Arnold’s (21) paper on how to measure phenotypic selection, myriad studies have quantified selection across a variety of taxa, and several recent quantitative syntheses and meta-analyses have reviewed this literature to describe the strength, direction, and dynamics of natural selection in the wild (25, 31–39). Our analysis differs from those conducted previously in that (i) we specifically focus on populations disturbed by human activities, and (ii) we include only data from disturbed populations for which selection was also measured in at least one control (not obviously human-disturbed; henceforth natural) population (Methods). We thus assembled a synthetic dataset of selection coefficients from recent studies of anthropogenic selection and analyzed it to test two simple predictions: (i) On average, when considering data from many species and human disturbances, phenotypic selection is stronger in human-disturbed populations than it is in natural populations; and (ii) harvest selection causes even stronger selection than other forms of anthropogenic selection, in line with trends reported for phenotypic change (10).
Of the 4,115 publications that we reviewed, 40 satisfied our inclusion criteria (Methods). We extracted linear selection differentials (S) and gradients (β) measured in both natural and disturbed conditions from these publications. These selection coefficients correspond to the slopes of linear regressions between relative fitness and standardized trait values, and they provide a standardized measure of phenotypic selection that can be compared across studies (21). The absolute value of these slopes indicates the strength of selection irrespective of its sign, which was the focus of our analysis. The assembled database comprises 1,366 linear selection coefficients (710 S and 656 β) measured on 102 traits from 37 different species (SI Appendix, Table S1). Coefficients were grouped into 37 “systems,” defined as specific taxon–disturbance combinations (e.g., pipefish or morning glory exposed to xenobiotics, lemon sharks experiencing habitat degradation, etc.). Harvested and nonharvested systems were also distinguished. We were able to obtain SEs for 67% of S and 82% of β-estimates; we used this subset of the data to fit a Bayesian formal meta-analytical model testing if selection strength varied with disturbance conditions (natural vs. “disturbed”), disturbance type (“harvest” vs. “other”), and their interaction. We also repeated a similar analysis using the full dataset instead but ignoring uncertainty over individual selection estimates, that is, using all estimates without considering SEs (below; the “informal model”). We also replicated all analyses using a frequentist statistical framework, and also modeled maximum (as opposed to mean) selection strength using quantile regression (Methods and SI Appendix). Finally, we were able to collect fitness estimates from 62% of systems to assess how disturbances influenced mean absolute fitness and the opportunity for selection.
Results and Discussion
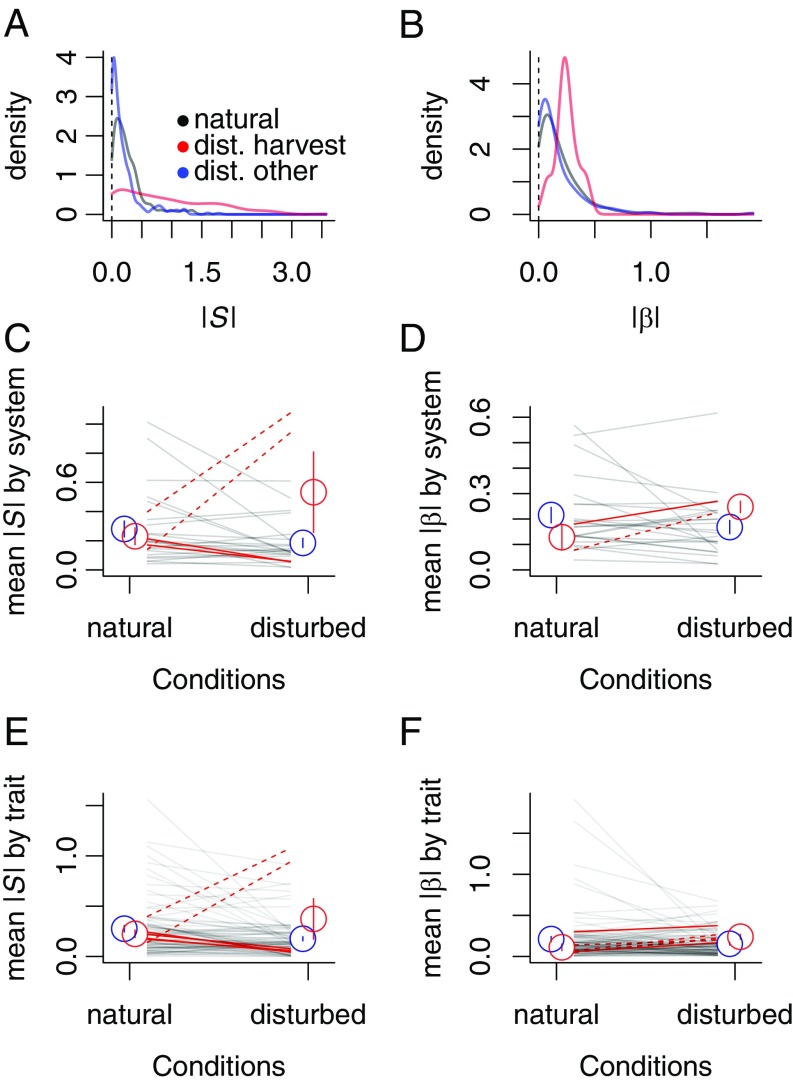
The distribution of estimates of |S| and |β| (selection strength) indicated slightly weaker selection (smaller values) in disturbed than natural conditions, and much stronger selection in disturbed conditions for harvested populations (Fig. 2 A and B). Median selection strength was 0.19 (|S|) or 0.13 (|β|) in natural conditions, 0.11 (|S|) or 0.09 (|β|) in disturbed conditions in nonharvested systems, and 0.69 (|S|) or 0.24 (|β|) in harvested populations. As a comparison, previous data compilations have reported median selection strength (|β|) ranging from 0.08 to 0.26 depending on fitness components (36, 39). The formal meta-analytical Bayesian model (but not the informal model) as well as all frequentist models indicated that mean selection strength was significantly weaker in disturbed than natural conditions (Fig. 2 C–F, Table 1, and SI Appendix, Table S2). However, disturbance type (harvest vs. other) and disturbance conditions (natural vs. disturbed) had a significant interactive effect on mean selection strength measured as |S|, with harvested systems showing stronger selection in disturbed than natural conditions (Fig. 2 C and E, Table 1, and SI Appendix, Table S2). This effect was entirely due to two fisheries (cod and pike), as other harvested systems (sheep and ginseng) had weaker mean phenotypic selection in disturbed than natural conditions. Disturbance conditions did not have a statistically significant effect on selection strength measured as |β| in the Bayesian models but did in the frequentist models, again suggesting slightly weaker selection in disturbed conditions (Fig. 2 D and F, Table 1, and SI Appendix, Table S2). However, only two harvested systems had β-coefficients such that the effect of disturbance type could not be tested.
Fig. 2.
Effects of human disturbances on the strength of phenotypic selection. (A and B) Probability density functions of absolute values of linear selection differentials (|S|) and gradients (|β|). Coefficients are pooled across systems and color-coded by disturbance conditions and disturbance type (harvest vs. other). (C–F) Mean selection strength by system or by trait, across disturbance conditions. Lines correspond to individual systems or traits, color-coded by disturbance type (red, harvest; gray, other). Dotted lines correspond to the subset of harvested systems that are fisheries (cod and pike). Circles indicate mean selection strength in natural vs. disturbed conditions for harvested systems (red) and other systems (blue). Error bars represent SEM.
Table 1.
Model predictions for mean selection strength in natural vs. disturbed conditions, for harvested vs. other systems
| Formal meta-analysis | Full dataset | ||||
| Estimate | Disturbance | Natural | Disturbed | Natural | Disturbed |
| |S| | Other | 0.21 [0.16 to 0.28] | 0.09 [0.06 to 0.16] | 0.21 [0.18 to 0.25] | 0.19 [0.17 to 0.22] |
| Harvest | 0.2 [0.13 to 0.41] | 0.85 [0.68 to 1.08] | 0.28 [0.21 to 0.47] | 0.95 [0.78 to 1.19] | |
| |β| H+ | 0.15 [0.13 to 0.18] | 0.13 [0.11 to 0.16] | 0.18 [0.16 to 0.21] | 0.19 [0.17 to 0.22] | |
| |β| H− | 0.16 [0.13 to 0.19] | 0.14 [0.11 to 0.17] | 0.19 [0.16 to 0.22] | 0.2 [0.17 to 0.23] | |
Results indicate the mode and 95% credible intervals of posterior distributions of mean |S| or |β|. Separate analyses were conducted with the subset of data with SEs (formal meta-analysis) and with the full dataset (ignoring measurement error). Effects of disturbance type on |β| could not be assessed due to data limitations (only two harvested systems); we thus conducted separate analyses including (H+) or excluding (H−) harvested systems.
The results for maximum selection strength modeled with quantile regression followed the same general pattern (i.e., disturbance lowered selection strength; SI Appendix, Fig. S2 and Table S3). When distinguishing systems or traits using a variety of grouping factors (focal taxa, study designs, types of disturbance, fitness components, and major trait types), we also observed that disturbance had small (but nonsignificant) negative effects on selection strength across most system/trait categories, with the only noticeable exception again being the two fisheries noted above (SI Appendix, Figs. S3 and S4). Therefore, the dominant trend across most analyses was slightly weaker selection in disturbed conditions. However, this effect is both small and uncertain (e.g., not significant for |β|), and could vanish as the database grows to include additional records. Thus, the main conclusion that we draw is that, contrary to our initial prediction, phenotypic selection is not stronger in human-disturbed populations, with the potential exception of fisheries. Expanding the dataset is unlikely to modify this basic conclusion, as this would require a substantial reversal of the trends reported here.
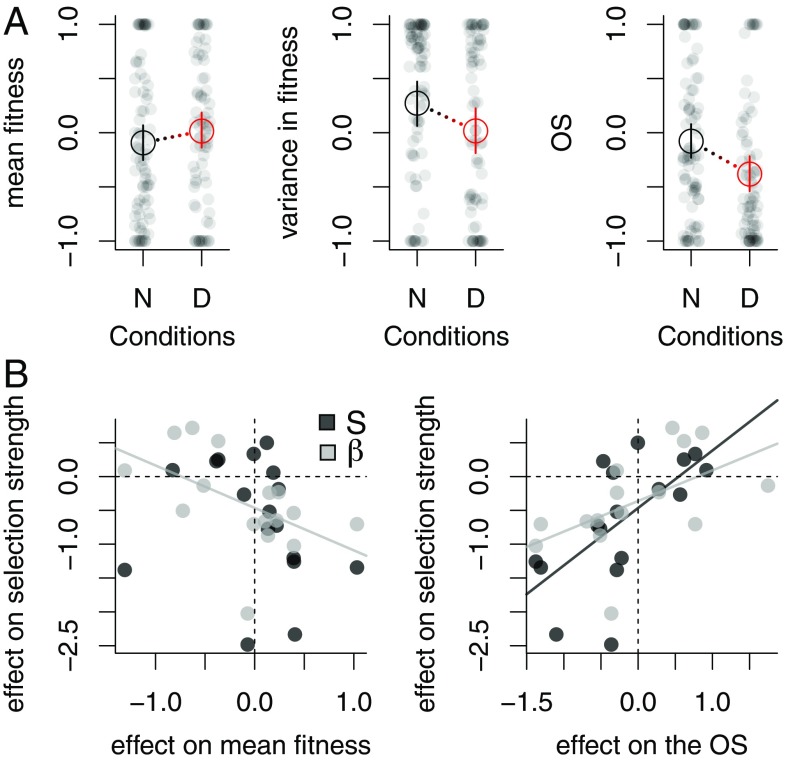
These results challenged our initial intuitions regarding human impacts on selection strength (Fig. 1). Perhaps human disturbances do not strengthen selection (on average) because they result in neither trait-level maladaptation (Fig. 1A) nor an increase in the opportunity for selection (Fig. 1B). The latter is possible if many disturbances included in our analysis increased absolute fitness and improved growth rate and population size (e.g., greater resource availability from eutrophication or elevated CO2, or warming up to a certain level), in which case we would not expect strengthened phenotypic selection (22, 25). By collecting estimates of mean absolute fitness, variance in absolute fitness, and opportunity for selection from studies in our database, we found that, on average, disturbance generally increased mean absolute fitness, decreased the variance in absolute fitness, and thus decreased the opportunity for selection (Fig. 3A; these trends were statistically significant for the variance in fitness and the opportunity for selection). Further, when the effect of disturbance on fitness or on the opportunity for selection is related to the effect of the same disturbance on selection strength, we find that disturbances that had a positive effect on fitness and a negative effect on the opportunity for selection significantly weakened selection (Fig. 3B and SI Appendix, Table S4; see also ref. 25). Therefore, most studies that we collated focused on disturbances that increased mean absolute fitness and decreased the opportunity for selection, thus leading to slightly weaker selection in disturbed conditions.
Fig. 3.
Human impacts on fitness and on the opportunity for selection influence selection strength. (A) Distribution of mean absolute fitness, the variance in absolute fitness, and the opportunity for selection in natural (N) and disturbed (D) conditions across the 22 systems that reported fitness values. Gray circles are individual estimates, scaled within systems. Connected black and red symbols indicate the results of Bayesian mixed models (mode and 95% credible intervals of posterior distributions) estimating mean values in either condition. OS, opportunity for selection. (B) Effect size (log response ratio) of disturbance on selection strength as a function of the effect size of the same disturbance on either mean absolute fitness (Left) or on the opportunity for selection (Right). Symbols are individual systems. Positive values indicate systems for which disturbance increased selection strength, mean/variance in fitness, or the opportunity for selection. Solid lines indicate results of linear regressions with statistically significant slopes at P < 0.05 (see SI Appendix, Table S4 for test statistics).
These findings are somewhat surprising in that we might expect investigators to choose disturbances with strong (potentially negative) effects on their study organisms. We are thus left with a puzzling question: How can human disturbances have, on average, no net effect (or a positive effect) on mean absolute fitness? Given widespread human impacts on the biosphere and the current sixth mass extinction (23, 24), it hardly seems possible that a majority of species would not be negatively affected by human-induced environmental change. Perhaps, then, the surprising trend in our dataset reflects a taxonomic bias affecting most studies of human-induced evolution; that is, we can only measure selection and adaptation in species that persist along anthropogenic gradients, at least long enough to be the focus of mark–recapture studies, rearing experiments, and so forth. None of the study populations included in this database went locally extinct following disturbance, yet human-induced extirpation is widespread (23). Thus, if we can only study species that fare relatively well in human-altered environments, accurately estimating the mean effect of humans on selection across taxa and types of disturbance will be difficult.
Quantifying the contribution of trait-level maladaptation to changes in selection (Fig. 1A) is also problematic. Most studies estimate selection based on a component of fitness (e.g., survival or mating success) or based on a proxy for fitness [which has limitations (40)], rather than total fitness, and all studies measure selection on one or a few traits only. Thus, when human disturbances weaken selection on one trait based on one fitness component, the same disturbance could in theory strengthen selection on the same trait via a different fitness component, and/or strengthen selection on another trait (SI Appendix, Fig. S5). Quantifying overall maladaptation in a human-altered environment (i.e., mapping the location of a population in a hyperdimensional adaptive landscape with a known topography) would require measuring total selection on all traits, which is impossible both logistically and conceptually (given the infinite number of traits that can be defined). The measured effect of humans on phenotypic selection coefficients is thus entirely dependent upon the traits and fitness components on which investigators choose to focus. For example, some studies are interested in how disturbances weaken selection (41), such that they specifically focus on traits hypothesized to be under stronger selection in natural conditions. Two such examples from the database include a eutrophication-induced breakdown of sexual selection on nuptial coloration in sticklebacks (28) and a loss of pollinator-mediated selection on floral traits due to forest degradation (29). In those examples, disturbance could relax selection on the traits under study but increase selection on other, unmeasured traits.
Finally, another complication is that human disturbances might influence hard and soft selection differently. Hard selection is density- and frequency-independent selection that can have a direct effect on population size, while soft selection is density- and frequency-dependent selection with generally limited consequences for population size (42, 43). Some disturbances could increase soft selection without influencing hard selection, and vice versa. For example, in a hypothetical harvested population in which a constant fraction of individuals with specific trait values are removed (e.g., the largest, 10%) but where harvest regulations maintain a stable population size and an invariant mean absolute fitness, one would expect strong (and persistent) soft selection and perhaps rapid phenotypic change without any change in hard selection. In contrast, all cases of population extirpation and biodiversity loss in human-altered environments are possible examples of strengthened (but unmeasured) hard selection, where individuals have low fitness regardless of population density or composition. This perhaps common scenario is not well represented in the database analyzed herein, as discussed above. Moreover, depending on whether a disturbance is predicted to impact hard or soft selection, different trait and fitness standardization approaches should be employed to adequately measure variation in selection strength (44). This is rarely done, which could mask human impacts on selection in some studies.
Despite the challenges of quantifying human impacts on overall selection, studies of anthropogenic selection are still useful for revealing effects of specific disturbances that are large and consistent enough to be detected amid all of the noise—effects that might also be the most relevant from a conservation perspective. For example, fishery-induced selection on body size is clearly a strong effect that deserves the attention of managers. Indeed, the two fisheries in our database showed markedly stronger anthropogenic than natural selection (45–47). Other studies of fishery selection that could not be included here due to a lack of control (unfished) selection coefficients also report some very high selection estimates (e.g., ref. 48), as do recent results from experimental fisheries (49). The distinct nature of fishing is also supported by phenotypic change data. For example, the well-known result of Darimont et al. (10) that trait change is fastest in harvested populations is based on the large number of fisheries in their analysis (30 out of 40 systems); excluding fisheries leads to similar rates of trait change in natural and harvested populations (a mean Darwin numerator of ∼0.1 in both natural and other harvested populations, as opposed to >0.2 for fisheries). Sharpe and Hendry (50) also found high rates of phenotypic change of life-history traits in the most heavily exploited fish stocks in their dataset. We speculate that fishing induces particularly strong selection because a large fraction of the population is often harvested (i.e., mortality is greatly increased), because fishing gear is designed specifically to be size-selective, and because selection keeps removing the largest individuals of the population despite adaptive trait change in response to prior harvest (50–52). We also suggest that additional studies of fisheries are strongly needed to confirm the pattern that we report here, and that such studies will be most informative if they also include matching data from control (unharvested) populations of the same species.
Conclusion
Overall, our analysis does not support the notion that most human disturbances strengthen phenotypic selection. However, we interpret this finding with caution, because the large number of selection coefficients that we synthesized originated from a limited number of study systems. Studies of human impacts on phenotypic selection have only recently become available in the literature (mean publication year of studies included in the database, 2010; range, 1999 to 2018). We predict that many more such studies will be published in upcoming years, and we will keep collating selection coefficients from human-disturbed populations as they arise. We suggest that it is paramount to gather more estimates of selection in populations exposed to a variety of disturbances if we are to uncover those human activities that consistently induce strong selection (e.g., fishing). This information is especially relevant when anthropogenic selection acts on traits that have implications for demography or community/ecosystem-level processes, for example, body size (9). As authors of previous meta-analyses have argued (36, 39), we too urge authors to report SEs of selection coefficients in future studies of phenotypic selection; many studies included in this database failed to do so (even recent ones), and most authors that we contacted could not retrieve SEs. Reporting mean and SE of absolute and relative/standardized fitness and trait values alongside selection estimates should also be routine, and would help document human impacts on the constituents of selection estimates and the relationships among them.
Finally, solving the paradox of faster evolutionary rates in human-impacted populations despite no apparent change in the strength of phenotypic selection will require more integrative studies measuring additive genetic variance, phenotypic selection, and rates of trait change along anthropogenic gradients. Some disturbances might have opposing effects on selection and genetic variation, leading to evolutionary stasis, whereas other disturbances could have synergistic effects on both variables, leading to positive feedback loops and even faster phenotypic change (53). To alleviate the taxonomic bias discussed above, future studies could also include experimental manipulations and/or monitoring of populations expected to undergo a disturbance, tracking their evolutionary response even if they eventually become extirpated. Lastly, incorporating demographic data in studies of phenotypic selection would be key in determining whether fluctuations in population abundance associated with human disturbances can influence the strength of selection, for example, via a change in population mean fitness and in the opportunity for selection (e.g., due to density-dependent processes). Understanding how human disturbances alter fundamental evolutionary processes such as selection is essential if we are to predict and mitigate population extirpation in the face of global change.
Methods
Database.
We found studies documenting selection in human-disturbed environments by reviewing all articles citing Lande and Arnold (21). This paper described a method to quantify linear selection differentials (S: the total directional selection acting on a trait) and linear selection gradients (β: the directional selection acting on a trait after controlling for selection on correlated traits) using simple and multiple linear regression, respectively. These regressions use standardized trait values as the predictor(s) and relative fitness as the response (the latter is most often a fitness component such as survival or mating success rather than total lifetime fitness). The absolute value of S or β indicates “selection strength,” the magnitude of directional selection irrespective of its sign. Most (if not all) studies measuring phenotypic selection in this way reference Lande and Arnold (21), such that reviewing its citations provides a convenient and unbiased method for finding a large number of S and β (henceforth “selection coefficients”).
Our literature survey found 4,115 articles published between 1983 and 2018. We screened these publications and retained those reporting selection coefficients for at least one population experiencing some form of human disturbance. We defined “human disturbance” as human activities noted by the authors that were likely to have modified the adaptive landscape experienced by the population in which selection was measured (disturbance types are described in SI Appendix). Moreover, we only included studies that also reported selection coefficients for the same traits in at least one control population found in natural conditions, namely in the absence of obvious human impacts noted by the authors (this could be selection measured in the same population before a disturbance, in a control group in an experiment, in a population in a natural habitat near a disturbed habitat, etc.). This paired design, meant to control for differences in selection strength across traits and species even in the absence of disturbance, led to the exclusion of many studies lacking selection estimates in natural conditions, such as some time series of harvest-induced selection in fisheries. We also excluded time series linking trends in selection with trends in climate, as those studies lack a clear delineation between natural and disturbed conditions, precluding a paired analysis. Therefore, climate change studies included herein were experiments that used warming chambers or greenhouses to contrast selection in control vs. treatment conditions. Finally, we also included only studies that standardized trait values based on the SD of the trait (21), as this is the most common form of standardization and is thus most appropriate for meta-analyses.
Forty publications satisfied our search criteria, from which we obtained S and β values and, when reported, their SEs. We also compiled nonlinear selection coefficients (21), but those were too few for reliable analysis here. The assembled database comprises 1,366 linear selection coefficients (710 S and 656 β) measured on 102 traits from 37 different species (SI Appendix, Table S1). Coefficients were grouped into 37 systems, defined as taxon–disturbance combinations. We also classified selection coefficients based on major trait categories and systems based on focal taxa, types of disturbance, experimental designs, and fitness component(s), to explore whether these moderator variables could influence human impacts on selection strength (see SI Appendix for a description of grouping classes). SEs were reported for 43% of S and 52% of β in the original publications from which the coefficients were obtained. To increase these proportions, we contacted authors and, when possible, calculated SEs based on reported P values or confidence intervals. After these efforts to collect additional information, 67% of S and 82% of β in the final database had SEs (SI Appendix, Table S1).
Analyses.
We analyzed the database using both formal and informal meta-analytical approaches, using R version 3.4.1 (54) for all analyses. To first visualize the distribution of selection coefficients (|S| or |β|), we plotted probability density functions for both natural and disturbed coefficients, ignoring the paired structure in the dataset, pooling coefficients from all systems, and distinguishing harvest vs. other disturbances for disturbed coefficients. To quantitatively compare selection strength in natural vs. disturbed environments, we used the subset of data with SEs to fit a formal meta-analytical (mixed effects) model. The fact that the variable of interest is a transformed (absolute) value of reported coefficients introduces some complexity, as SEs of estimated S and β do not directly correspond to measurement errors for |S| and |β|. To circumvent this problem, a recently developed approach consists of modeling the raw (not absolute) coefficients taking into account measurement error, and then using the estimated effects and variances to parameterize folded normal distributions that predict the distribution of absolute coefficients (55). We used this approach and first modeled S (not |S|) with a mixed model including disturbance conditions (natural vs. disturbed), disturbance type (other vs. harvest), and their interaction as fixed effects, and the system and trait on which selection was measured (nested within system) as random effects. The model was ŷijk ∼ µ + β1*X1ijk + β2*X2ijk + β3*X1ijk*X2ijk + systemi + traitij + mijk + eijk, where ŷijk is the k-th selection coefficient measured on the j-th trait in the i-th system, µ is mean selection in natural conditions in nonharvested systems, the three βs are estimates of the fixed effects, X1 and X2 are dummy (binary) variables coding for disturbed conditions and harvest, and mijk and eijk are observation-level sampling errors and residuals.
We fitted this model in a Bayesian framework, estimating parameters with Markov chain Monte Carlo implemented in the R package MCMCglmm (56). We ran the model over 103,000 iterations, discarded the first 3,000 iterations as burn-in, and used a thinning interval of 10. We used noninformative priors for fixed effects and weakly informative priors with a low degree of belief for random effects. Fixed effects are estimated by the model. Random effects and residuals are drawn from normal distributions with a mean of zero and a variance estimated by the model, with residual variances allowed to differ among levels of fixed effects. Sampling errors are drawn from normal distributions with a mean of zero and a known variance given by the squared SE of the selection estimate. We generated posterior predictive distributions of S in all four contexts (natural vs. disturbed, harvest vs. other) and then transformed these distributions using folded normal distributions, to predict distributions of |S| in each context. We report the mode and 95% credible intervals of these predicted distributions.
To use all data available, we also fitted a model using the full dataset instead of only the subset with SEs. This model is like the one described above but omitted the term for sampling error (mijk), thus ignoring uncertainty associated with individual selection estimates. We compared the output of this model with the formal meta-analytical model. We then repeated this analysis with β, again fitting a formal meta-analytical model with the subset of data including SEs and a (informal) model with the full dataset, and then using the posterior distributions to model the expected value of |β| by sampling folded normal distributions. However, due to data limitations (only two harvested systems), models for β had a single fixed effect (disturbance conditions). We fitted models by either including or excluding the two harvested systems, to determine the mean impact of disturbances other than harvest. We then repeated this set of analyses using maximum likelihood-based parameter estimation instead. We fitted weighted and unweighted, general and generalized (gamma) linear mixed models on either untransformed or log-transformed absolute selection estimates using the R package lme4 (57). These alternative approaches are described in SI Appendix, Table S2. We also modeled maximum selection strength using quantile regression (SI Appendix, Fig. S2 and Table S3), and tested whether effects of disturbance on selection strength varied across taxa, study designs, types of disturbance, fitness components, and trait types (SI Appendix, Figs. S3 and S4).
Finally, we also assessed the impact of human disturbances on mean fitness by extracting fitness estimates from studies included in the database. Out of 37 systems, 23 provided at least one mean absolute fitness estimate per disturbance condition (total, 159 estimates; reported in figures, tables, or text). We also noted the variance in absolute fitness when available, to calculate the opportunity for selection (the variance in relative fitness, given by the variance in absolute fitness divided by mean absolute fitness squared). In some cases, the opportunity for selection was directly reported by the authors. We rescaled estimates from −1 to 1 within each system because values vary widely in scale depending on the choice of fitness measure. We then analyzed these data with Bayesian mixed models of the form ŷijk ∼ conditionsi + systemj + eijk, where ŷijk is the k-th estimate (mean absolute fitness, variance in absolute fitness, or the opportunity for selection) from the j-th system (a random effect) when under the i-th condition (natural or disturbed; a fixed effect). Fitting parameters (number of iterations, burn-in, etc.) were identical to other models described above. The one harvested system that reported fitness estimates was excluded from the analysis.
To link impacts on fitness and impacts on selection strength, we also calculated for each system an effect size of human disturbance on mean absolute fitness, on the opportunity for selection, and on selection strength. These effect sizes were log response ratios computed as the natural logarithm of the mean value in disturbed conditions over the mean value in natural conditions. We averaged all selection and fitness estimates within system and disturbance conditions rather than using a hierarchical model, because the number of fitness and selection estimates generally differed within studies (e.g., a system could have many selection estimates based on many traits but a single reported fitness measurement in natural and in disturbed conditions). After averaging, each system contributed a single data point (an effect size of disturbance) for each variable. We tested for an association between the effect size of disturbance on fitness or the opportunity for selection and the effect size of disturbance on selection strength using linear regressions, fitting separate models for |S| and for |β| (SI Appendix, Table S4).
Supplementary Material
Acknowledgments
We are grateful to Gabriel Pigeon, Marco Festa-Bianchet, and Stephanie Carlson for sharing data. Michael Morrissey and anonymous reviewers provided constructive comments that improved the manuscript. V.F. was supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada (NSERC). A.P.H. was supported by an NSERC discovery grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The database reported in this paper has been deposited in FigShare (https://doi.org/10.6084/m9.figshare.7019822.v1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806013115/-/DCSupplemental.
References
- 1.Palumbi SR. Humans as the world’s greatest evolutionary force. Science. 2001;293:1786–1790. doi: 10.1126/science.293.5536.1786. [DOI] [PubMed] [Google Scholar]
- 2.Hendry AP, Gotanda KM, Svensson EI. Human influences on evolution, and the ecological and societal consequences. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160028. doi: 10.1098/rstb.2016.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allendorf FW, Hard JJ. Human-induced evolution caused by unnatural selection through harvest of wild animals. Proc Natl Acad Sci USA. 2009;106:9987–9994. doi: 10.1073/pnas.0901069106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendry AP, Millien V, Gonzalez A, Larsson HC. How humans influence evolution on adaptive landscapes. In: Svensson E, Calsbeek R, editors. The Adaptive Landscape in Evolutionary Biology. Oxford Univ Press; Oxford: 2012. pp. 180–202. [Google Scholar]
- 5.Pelletier F, Coltman DW. Will human influences on evolutionary dynamics in the wild pervade the Anthropocene? BMC Biol. 2018;16:7. doi: 10.1186/s12915-017-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendry AP, Farrugia TJ, Kinnison MT. Human influences on rates of phenotypic change in wild animal populations. Mol Ecol. 2008;17:20–29. doi: 10.1111/j.1365-294X.2007.03428.x. [DOI] [PubMed] [Google Scholar]
- 7.Alberti M, et al. Global urban signatures of phenotypic change in animal and plant populations. Proc Natl Acad Sci USA. 2017;114:8951–8956. doi: 10.1073/pnas.1606034114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuparinen A, Festa-Bianchet M. Harvest-induced evolution: Insights from aquatic and terrestrial systems. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160036. doi: 10.1098/rstb.2016.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palkovacs EP, Moritsch MM, Contolini GM, Pelletier F. Ecology of harvest-driven trait changes and implications for ecosystem management. Front Ecol Environ. 2018;16:20–28. [Google Scholar]
- 10.Darimont CT, et al. Human predators outpace other agents of trait change in the wild. Proc Natl Acad Sci USA. 2009;106:952–954. doi: 10.1073/pnas.0809235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinnison MT, Hairston NG., Jr Eco-evolutionary conservation biology: Contemporary evolution and the dynamics of persistence. Funct Ecol. 2007;21:444–454. [Google Scholar]
- 12.Palkovacs EP, Kinnison MT, Correa C, Dalton CM, Hendry AP. Fates beyond traits: Ecological consequences of human-induced trait change. Evol Appl. 2012;5:183–191. doi: 10.1111/j.1752-4571.2011.00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudman SM, Kreitzman M, Chan KMA, Schluter D. Evosystem services: Rapid evolution and the provision of ecosystem services. Trends Ecol Evol. 2017;32:403–415. doi: 10.1016/j.tree.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Des Roches S, et al. The ecological importance of intraspecific variation. Nat Ecol Evol. 2018;2:57–64. doi: 10.1038/s41559-017-0402-5. [DOI] [PubMed] [Google Scholar]
- 15.Arnold SJ, Pfrender ME, Jones AG. The adaptive landscape as a conceptual bridge between micro- and macroevolution. Genetica. 2001;112–113:9–32. [PubMed] [Google Scholar]
- 16.Hendry AP. Eco-Evolutionary Dynamics. Princeton Univ Press; Princeton: 2016. [Google Scholar]
- 17.Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin’s finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- 18.Lewis SL, Maslin MA. Defining the Anthropocene. Nature. 2015;519:171–180. doi: 10.1038/nature14258. [DOI] [PubMed] [Google Scholar]
- 19.Crow JF. Some possibilities for measuring selection intensities in man. Hum Biol. 1958;30:1–13. [PubMed] [Google Scholar]
- 20.Arnold SJ, Wade MJ. On the measurement of natural and sexual selection: Applications. Evolution. 1984;38:720–734. doi: 10.1111/j.1558-5646.1984.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 21.Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 22.Rundle HD, Vamosi SM. Selection may be strongest when resources are scarce: A comment on Wilson. Evol Ecol. 1996;10:559–563. [Google Scholar]
- 23.Ceballos G, Ehrlich PR, Dirzo R. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc Natl Acad Sci USA. 2017;114:E6089–E6096. doi: 10.1073/pnas.1704949114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas JA, et al. Comparative losses of British butterflies, birds, and plants and the global extinction crisis. Science. 2004;303:1879–1881. doi: 10.1126/science.1095046. [DOI] [PubMed] [Google Scholar]
- 25.Caruso CM, et al. What are the environmental determinants of phenotypic selection? A meta-analysis of experimental studies. Am Nat. 2017;190:363–376. doi: 10.1086/692760. [DOI] [PubMed] [Google Scholar]
- 26.Gómez-González S, Torres-Díaz C, Bustos-Schindler C, Gianoli E. Anthropogenic fire drives the evolution of seed traits. Proc Natl Acad Sci USA. 2011;108:18743–18747. doi: 10.1073/pnas.1108863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright JT, Gribben PE, Byers JE, Monro K. Invasive ecosystem engineer selects for different phenotypes of an associated native species. Ecology. 2012;93:1262–1268. doi: 10.1890/11-1740.1. [DOI] [PubMed] [Google Scholar]
- 28.Candolin U, Salesto T, Evers M. Changed environmental conditions weaken sexual selection in sticklebacks. J Evol Biol. 2007;20:233–239. doi: 10.1111/j.1420-9101.2006.01207.x. [DOI] [PubMed] [Google Scholar]
- 29.Murúa M, Espinoza C, Bustamante R, Marín VH, Medel R. Does human-induced habitat transformation modify pollinator-mediated selection? A case study in Viola portalesia (Violaceae) Oecologia. 2010;163:153–162. doi: 10.1007/s00442-010-1587-3. [DOI] [PubMed] [Google Scholar]
- 30.DiBattista JD, Feldheim KA, Garant D, Gruber SH, Hendry AP. Anthropogenic disturbance and evolutionary parameters: A lemon shark population experiencing habitat loss. Evol Appl. 2011;4:1–17. doi: 10.1111/j.1752-4571.2010.00125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoekstra HE, et al. Strength and tempo of directional selection in the wild. Proc Natl Acad Sci USA. 2001;98:9157–9160. doi: 10.1073/pnas.161281098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kingsolver JG, et al. The strength of phenotypic selection in natural populations. Am Nat. 2001;157:245–261. doi: 10.1086/319193. [DOI] [PubMed] [Google Scholar]
- 33.Kingsolver JG, Diamond SE, Siepielski AM, Carlson SM. Synthetic analyses of phenotypic selection in natural populations: Lessons, limitations and future directions. Evol Ecol. 2012;26:1101–1118. [Google Scholar]
- 34.Hereford J, Hansen TF, Houle D. Comparing strengths of directional selection: How strong is strong? Evolution. 2004;58:2133–2143. doi: 10.1111/j.0014-3820.2004.tb01592.x. [DOI] [PubMed] [Google Scholar]
- 35.Siepielski AM, DiBattista JD, Carlson SM. It’s about time: The temporal dynamics of phenotypic selection in the wild. Ecol Lett. 2009;12:1261–1276. doi: 10.1111/j.1461-0248.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- 36.Siepielski AM, DiBattista JD, Evans JA, Carlson SM. Differences in the temporal dynamics of phenotypic selection among fitness components in the wild. Proc Biol Sci. 2011;278:1572–1580. doi: 10.1098/rspb.2010.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siepielski AM, et al. The spatial patterns of directional phenotypic selection. Ecol Lett. 2013;16:1382–1392. doi: 10.1111/ele.12174. [DOI] [PubMed] [Google Scholar]
- 38.Siepielski AM, et al. Precipitation drives global variation in natural selection. Science. 2017;355:959–962. doi: 10.1126/science.aag2773. [DOI] [PubMed] [Google Scholar]
- 39.Kingsolver JG, Diamond SE. Phenotypic selection in natural populations: What limits directional selection? Am Nat. 2011;177:346–357. doi: 10.1086/658341. [DOI] [PubMed] [Google Scholar]
- 40.Franklin OD, Morrissey MB. Inference of selection gradients using performance measures as fitness proxies. Methods Ecol Evol. 2017;8:663–677. [Google Scholar]
- 41.Lahti DC, et al. Relaxed selection in the wild. Trends Ecol Evol. 2009;24:487–496. doi: 10.1016/j.tree.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 42.Wallace B. Hard and soft selection revisited. Evolution. 1975;29:465–473. doi: 10.1111/j.1558-5646.1975.tb00836.x. [DOI] [PubMed] [Google Scholar]
- 43.Saccheri I, Hanski I. Natural selection and population dynamics. Trends Ecol Evol. 2006;21:341–347. doi: 10.1016/j.tree.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 44.De Lisle SP, Svensson EI. On the standardization of fitness and traits in comparative studies of phenotypic selection. Evolution. 2017;71:2313–2326. doi: 10.1111/evo.13325. [DOI] [PubMed] [Google Scholar]
- 45.Carlson SM, et al. Four decades of opposing natural and human-induced artificial selection acting on Windermere pike (Esox lucius) Ecol Lett. 2007;10:512–521. doi: 10.1111/j.1461-0248.2007.01046.x. [DOI] [PubMed] [Google Scholar]
- 46.Olsen EM, Moland E. Fitness landscape of Atlantic cod shaped by harvest selection and natural selection. Evol Ecol. 2011;25:695–710. [Google Scholar]
- 47.Olsen EM, Heupel MR, Simpfendorfer CA, Moland E. Harvest selection on Atlantic cod behavioral traits: Implications for spatial management. Ecol Evol. 2012;2:1549–1562. doi: 10.1002/ece3.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kendall NW, Quinn TP. Quantifying and comparing size selectivity among Alaskan sockeye salmon fisheries. Ecol Appl. 2012;22:804–816. doi: 10.1890/11-1189.1. [DOI] [PubMed] [Google Scholar]
- 49.Nussle S, et al. 2017. Thirty-five experimental fisheries reveal the mechanisms of selection. bioRxiv doi:10.1101/141259. Preprint, posted May 26, 2017.
- 50.Sharpe DMT, Hendry AP. Life history change in commercially exploited fish stocks: An analysis of trends across studies. Evol Appl. 2009;2:260–275. doi: 10.1111/j.1752-4571.2009.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jørgensen C, et al. Ecology: Managing evolving fish stocks. Science. 2007;318:1247–1248. doi: 10.1126/science.1148089. [DOI] [PubMed] [Google Scholar]
- 52.Hutchings JA, Fraser DJ. The nature of fisheries- and farming-induced evolution. Mol Ecol. 2008;17:294–313. doi: 10.1111/j.1365-294X.2007.03485.x. [DOI] [PubMed] [Google Scholar]
- 53.Wood CW, Brodie ED., III Evolutionary response when selection and genetic variation covary across environments. Ecol Lett. 2016;19:1189–1200. doi: 10.1111/ele.12662. [DOI] [PubMed] [Google Scholar]
- 54.R Core Team 2017. R: A Language and Environment for Statistical Computing (R Found Stat Comput, Vienna), Version 3.4.1.
- 55.Morrissey MB. Meta-analysis of magnitudes, differences and variation in evolutionary parameters. J Evol Biol. 2016;29:1882–1904. doi: 10.1111/jeb.12950. [DOI] [PubMed] [Google Scholar]
- 56.Hadfield JD. MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R package. J Stat Softw. 2010;33:1–22. [Google Scholar]
- 57.Bates D, Maechler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.