Significance
Conversion of ribonucleotides to the 2′-deoxyribonucleotides required for DNA biosynthesis is catalyzed by ribonucleotide reductases (RNRs) via a free-radical mechanism. Known types of RNRs all depend on redox-active transition metals—manganese, iron, or cobalt—for radical initiation. Pathogenic bacteria are challenged by transition metal sequestration and infliction of oxidative stress by their hosts, and the deployment of multiple RNRs with different metal requirements and radical-initiating oxidants is a known bacterial countermeasure. A class I RNR from two bacterial pathogens completely lacks transition metals in its active state and uses a tyrosine-derived dihydroxyphenylalanine radical as its initiator, embodying a novel tactic to combat transition metal- and oxidant-mediated innate immunity and reinforcing bacterial RNRs as potential antibiotic targets.
Keywords: DNA biosynthesis, semiquinone, DOPA
Abstract
All cells obtain 2′-deoxyribonucleotides for DNA synthesis through the activity of a ribonucleotide reductase (RNR). The class I RNRs found in humans and pathogenic bacteria differ in (i) use of Fe(II), Mn(II), or both for activation of the dinuclear-metallocofactor subunit, β; (ii) reaction of the reduced dimetal center with dioxygen or superoxide for this activation; (iii) requirement (or lack thereof) for a flavoprotein activase, NrdI, to provide the superoxide from O2; and (iv) use of either a stable tyrosyl radical or a high-valent dimetal cluster to initiate each turnover by oxidizing a cysteine residue in the α subunit to a radical (Cys•). The use of manganese by bacterial class I, subclass b-d RNRs, which contrasts with the exclusive use of iron by the eukaryotic Ia enzymes, appears to be a countermeasure of certain pathogens against iron deprivation imposed by their hosts. Here, we report a metal-free type of class I RNR (subclass e) from two human pathogens. The Cys• in its α subunit is generated by a stable, tyrosine-derived dihydroxyphenylalanine radical (DOPA•) in β. The three-electron oxidation producing DOPA• occurs in Escherichia coli only if the β is coexpressed with the NrdI activase encoded adjacently in the pathogen genome. The independence of this new RNR from transition metals, or the requirement for a single metal ion only transiently for activation, may afford the pathogens an even more potent countermeasure against transition metal-directed innate immunity.
Ribonucleotide reductases (RNRs) catalyze the only known biochemical reaction—replacement of the 2′ hydroxyl group of a ribonucleoside 5′-diphosphate or -triphosphate by hydrogen—that provides the substrates for DNA replication and repair (1). RNRs use a largely conserved pathway for C2′ reduction. A transient thiyl radical produced by one-electron oxidation of a cysteine residue (Cys•) removes a hydrogen atom (H•) from C3′ of the substrate (2). The 3′ radical activates C2′ for dehydration, and ensuing electron, proton, and/or hydrogen-atom transfers (from sulfur-containing amino acids and/or formate) then generate a 2′-deoxy 3′-radical, which retakes the H• from the cysteine to complete the reduction and regenerate the Cys• (3). Importantly, the Cys• is not present in any resting RNR but must be generated at the outset and quenched at the end of each turnover.
The imperative for Cys• production engendered at least three distinct classes (I–III) of RNRs and four known subclasses (a–d) within class I (1, 4, 5). Different transition metal-dependent strategies for Cys• generation define the (sub)classes. Class II (6) and class III (7) RNRs use cobalamin and iron-sulfur cofactors, respectively, to generate the Cys•. The class III enzymes undergo preactivation by a separate protein, a radical-SAM (S-adenosyl-l-methionine) activase (7). The activase uses its iron-sulfur cluster to reductively cleave SAM, and the resultant 5′-dA• abstracts H• from a glycine residue in the RNR. The stable glycyl radical generates the Cys• to initiate each turnover.
Class I RNRs, such as the enzymes found in all eukarya and many pathogenic bacteria, use a separate ferritin-like protein subunit, denoted β, as radical initiator. RNR β subunits contain four core helices that provide six metal ligands, two histidines and four carboxylates (Fig. 1 A and B), to a dinuclear, divalent metal cluster (M2II/II) (4). M2II/II-β reacts with a form of dioxygen to install a stable oxidant that generates the Cys• in the catalytic subunit, α, by a long-range intersubunit radical-translocation (RT) process mediated by a chain of conserved aromatic residues that spans the subunit interface and connects the active sites (8). RT is bidirectional, occurring in the first and last steps of each turnover to generate and quench the 3′-H–cleaving Cys•.
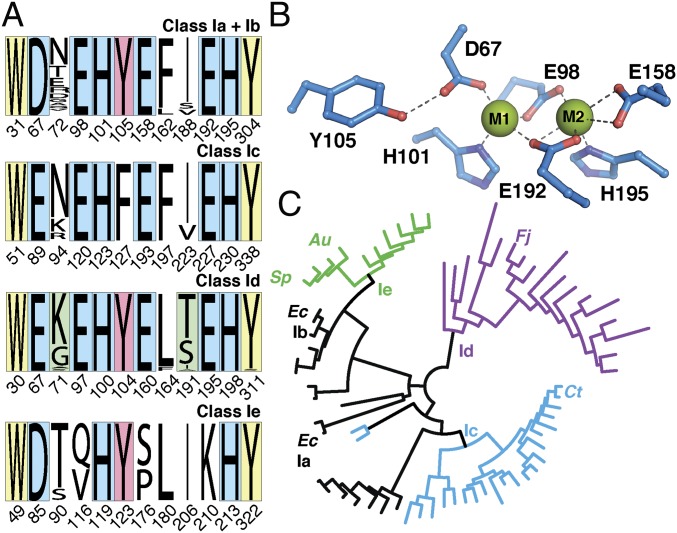
Fig. 1.
Bioinformatics analysis of class I RNR β subunits and features of the new Ie subclass. (A) Conservation of residues with roles in radical translocation/electron transfer (yellow), metal coordination (blue), radical formation (red), and control of activation (green) in subclass a–e βs. The residue numbering shown in the four rows of A is based on the sequences of the Ec Ib (Ia/b), Chlamydia trachomatis (Ic), F. johnsoniae (Id), and Au (Ie) β subunits. (B) Generalized structural depiction of the dimetal site (Ec Ib numbering). (C) Cladogram of representative class I RNR β sequences based on overall sequence identity. Clade coloring: black, class Ia/b with Asp at the first coordination site and Tyr at the radical site; blue, class Ic with Glu and Phe at these sites; purple, class Id with Glu and Tyr; green, class Ie lacking three coordinating Glu residues.
The identities of the transition metals (manganese or iron), activating oxidants (dioxygen or superoxide), and Cys• generators (high-valent dimetal center or tyrosyl radical) are the basis for the division of class I enzymes into subclasses a–d (4, 5). The Ia and Ib enzymes use their dimetal clusters (Fe2 and Mn2, respectively) to oxidize a nearby tyrosine residue to a stable radical (Tyr•), which serves as the catalytic initiator. The Ic and Id enzymes use a stable, oxidized form of the dimetal cluster itself (Mn/Fe and Mn2, respectively) directly for Cys• generation. The presence of FeII in the Ia and Ic proteins allows them to undergo activation by O2; this reaction converts the Fe2II/II-β and MnII/FeII-β complexes to their Fe2III/III-Tyr• and MnIV/FeIII active states. The Mn2II/II complexes of the Ib and Id βs are unreactive toward O2 and must instead be oxidized by superoxide. For the Ib βs, the superoxide is provided by a flavoprotein activase, NrdI, that binds to β and forms a hydrophilic channel connecting their active sites. The NrdI flavin reduces O2, and the channel shepherds the product to the Mn2II/II cluster in β. The ensuing reaction generates the Mn2III/III-Tyr• active state. The β subunit of the class Id RNR from Flavobacterium johnsoniae (Fj) can scavenge superoxide directly from solution to oxidize the Mn2II/II-β complex to the active Mn2III/IV state (5). Accordingly, the genomes of Fj and many other organisms with Id RNRs do not encode NrdI homologs.
The RNR(s) of an organism equip it for its environmental niche (9, 10). For example, both the glycyl radical of a class III RNR and the iron-sulfur cluster of its activase are unstable in the presence of O2, limiting the enzymes to anaerobes, whereas class I RNRs require O2 for activation and are functional only in aerobes, including all eukaryotes. Escherichia coli (Ec) has both types, expressing its class III enzyme during anaerobic growth and its class Ia enzyme during aerobic growth. Analogously, whereas eukarya possess exclusively iron-dependent class Ia enzymes, the use of manganese-dependent class I (b–d) RNRs appears to equip bacteria to cope with stresses encountered in their more dynamic niches. For example, the class Ib Ec enzyme sustains replication only under conditions of iron limitation or oxidative stress (11, 12). Thus, Ec mutants with a nonfunctional class Ia RNR subunit are not viable in the presence of O2 under normal growth conditions, and an Ec strain with a mutation that renders its class Ia α subunit inactive at elevated temperatures (42 °C) has been used as a functional selection for heterologous RNRs (13, 14). The facts that (i) the immune systems of warm-blooded hosts inflict both iron limitation and oxidative stress on an invading bacterium (15) and (ii) class Ib enzymes are highly represented in pathogens (16) have led to the hypothesis that the use of manganese is a specific countermeasure against iron-directed innate immunity. Consistent with this notion, Streptococcus sanguinis, an opportunistic pathogen that is normally present in the human mouth but can be unleashed by dental procedures to cause subacute bacterial endocarditis, requires its class Ib RNR, but not its class III enzyme, for virulence in an animal model of the disease (17, 18). It has been suggested that the divergent metal requirements of the human and bacterial RNRs could be leveraged for antibiotic development (18).
In view of the likely relevance of the diverse radical-initiation chemistry of RNRs to bacterial pathogenesis, we continue to mine sequence databases for orthologs that deploy hitherto unknown strategies. We show here that the β subunits of a pair of class I RNRs from two human pathogens, Streptococcus pyogenes (Sp) (19) and Aerococcus urinae (Au) (20), lack any transition metal in their active states and use as their initiator a previously unknown stable dihydroxyphenyalanine radical (DOPA•) derived posttranslationally from tyrosine in an NrdI- and O2-dependent reaction. These proteins are representative of a larger sequence group of class I RNR βs (class Ie) that are expected to function in the same way and are prevalent in parasitic and commensal organisms (SI Appendix, Fig. S1). Although the Ie βs might still require a single, as-yet unidentified transition metal ion transiently for activation, in either case they would exemplify the most sparing metal use of any known RNR and thus could afford the most effective RNR-based countermeasure yet discovered against transition metal restriction by the host.
Results and Discussion
A Group of Active Class I RNRs Lacking Three Glu Metal Ligands in β.
To identify RNRs with unusual cofactors or activation mechanisms, we compiled β sequences (14,807 unique UniProt queries, July 2018) and generated a multiple sequence alignment to mine computationally for examples that fail to conserve residues with established functions (e.g., metal ligands, radical Tyr). Members of a group of 430 sequences all lack three otherwise-conserved metal-coordinating glutamates (Fig. 1 A and B, light blue), which are replaced by two hydrophobic residues and a cationic Lys. These β sequences are adjacent to the Ib subclass in molecular phylogeny (Fig. 1C), and, accordingly, almost all of them are encoded close to genes for apparent NrdI activases. Importantly, the associated operon (encoding β-NrdI-α) from Sp, the causative agent of strep throat and scarlet fever, can complement the aforementioned Ia RNR-α temperature-sensitive mutant at the restrictive temperature, as shown by Sjöberg et al. (14), thereby ruling out the possibility that they are nonfunctional, vestigial sequences. We verified that synthetic operons with all three components from either Sp or Au give robust complementation in this assay (SI Appendix, Table S1). We also found that the NrdI component from Au can functionally substitute for Sp NrdI in supporting the in vivo function of the Sp RNR.
To understand how these β proteins can be active despite lacking three metal ligands present in all other orthologs, we expressed (in Ec) N-terminally affinity-tagged Sp and Au β subunits with and without their NrdI activases and purified them chromatographically (SI Appendix, Fig. S2). We also expressed and purified affinity-tagged versions of the cognate α subunits for RNR activity measurements. The β subunits purified following coexpression with an NrdI, including the Sp β coexpressed with the Au activase, were capable of supporting reduction of CDP to dCDP in the presence of the cognate α subunit and DTT (SI Appendix, Figs. S3 and S4). Maximal turnover rates of 0.35 s−1•β−1 were observed (SI Appendix, Fig. S3) for the Sp β. In contrast, preparations of β proteins obtained following expression in Ec in the absence of a NrdI exhibited undetectable activity (<2 × 10−5 s−1•β−1). The Sp-β/Au-NrdI coexpression system more readily yielded β with minimal contaminating NrdI, with its intensely absorbing flavin (Fig. 2A and SI Appendix, Fig. S3); thus, the heterologous system was used to generate the active protein for the spectroscopic analysis presented below.
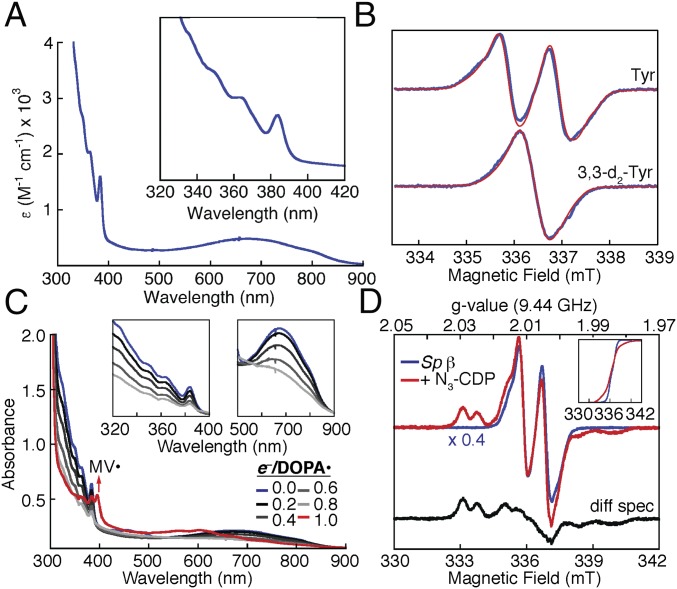
Fig. 2.
Chemical and spectroscopic analysis of the radical oxidant in the class Ie Sp RNR β. (A) UV-visible absorption spectrum. (B) EPR spectra of Sp β (blue) and global simulations thereof (red), with natural-abundance Tyr (Top) and β-[2H2]-Tyr (Bottom). (C) Spectrophotometrically monitored reductive titration by dithionite. (D) Loss of the radical oxidant and production of the nitrogen-centered radical (N•; black spectrum) on treatment of the holoenzyme with with N3-CDP. (Insets) The double integral of the −/+ N3-dCDP spectra, accounting for >98% of the total spin. Procedures and sample compositions are described in SI Appendix.
The Active Class Ie βs Lack Metal and Contain a Dihydroxyphenylalanine Radical (DOPA•).
Preparations of activated (NrdI-coexpressed) Sp and Au βs exhibited a broad absorption feature at 690 nm and a complex of four sharper peaks at approximately 383, 364, 349, and 337 nm (Fig. 2A and SI Appendix, Fig. S4), which were absent from the preparations of unactivated (expressed alone) proteins. The sharpest peak of the complex (383 nm) resembles the hallmark feature of a Tyr•, but its wavelength is less than that for any Tyr• in a class Ia/b RNR (405–417 nm) (SI Appendix, Fig. S5). X-band EPR spectra of active preparations also revealed a g = 2.0 (free-radical) EPR signal accounting for 0.4–0.7 spin/β monomer (Fig. 2B and SI Appendix, Fig. S4), comparable to the levels of Tyr• reported in class Ia/b enzymes (4). Use of protein containing β-[2H2]-Tyr caused a narrowing characteristic of the replacement of a strong doublet splitting from a 1H nucleus with a smaller (unresolved) splitting from 2H (21). Reductive titration of the active Sp β protein with dithionite (Fig. 2C) or treatment with the radical-quenching class I RNR inhibitor, hydroxyurea (SI Appendix, Fig. S6) (22), caused loss of these spectral signatures, implying that they all arise from the same Tyr-derived radical species, which the difference in absorption energies suggested could not be a simple Tyr•. The importance of this radical to catalysis was ascertained by use of the 2′-azido-2′-deoxynucleoside 5′-diphosphate (N3-NDP) substrate analog (Fig. 2D) (23, 24). N3-NDPs (N3-CDP, N3-UDP) irreversibly reduce the initiator in β (23, 24) and produce a metastable nitrogen-centered radical (N•) (25) in α when processed by a class I RNR (5, 24, 26). Analogously, treatment of the active Sp enzyme with N3-CDP resulted in loss of approximately 60% of the original EPR signal and development of equivalent integrated intensity of the N• signal, thus establishing the Tyr-derived radical as the initiator for the new Ie subclass. Analysis of the active preparations for relevant metals revealed none present at >0.15 equiv per radical (SI Appendix, Table S2). Furthermore, the total transition metal content was less than the quantity of radical. The results indicate that class Ie βs are metal-independent after activation in vivo.
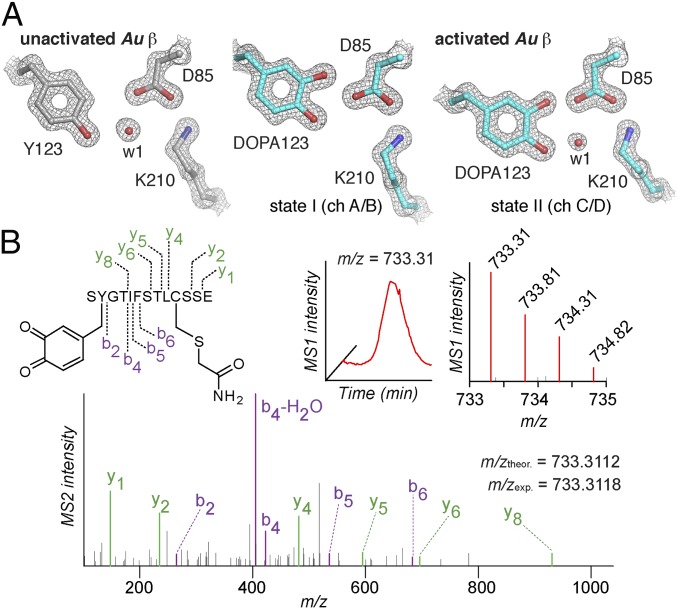
We verified the absence of any transition metal in the active state and garnered insight into the structure of the Tyr-derived radical by X-ray crystallographic analysis (SI Appendix, Table S3) of activated and unactivated forms of Au β, which formed high-quality crystals more readily than the Sp protein. Structures solved at 1.6-Å resolution showed no density attributable to a metal ion in either form (Fig. 3A and SI Appendix, Fig. S7) and confirmed that the substitution of the three Glu ligands is not compensated for by a change in protein fold that introduces other ligands to preserve metal site 2. Moreover, the structures revealed that the ammonium group of the Lys that replaces a Glu ligand comes to occupy the usual location of the site 1 metal ion (SI Appendix, Fig. S8) (27–29), thus rationalizing the complete absence of metal. Exclusively in the structure of the activated form, clear Fo-Fc difference density could be observed near one of the carbons (C3) ortho to the hydroxyl group of Tyr123 (SI Appendix, Fig. S7B), revealing hydroxylation of Tyr123 to dihydroxyphenylalanine (DOPA) in the activation process. Evidence of the modification was observed in all four β subunits of the asymmetric unit (SI Appendix, Figs. S7C and S9A), but differences in the electron density maps suggested that one or more of the monomers could have been in an intermediate state of the activation process or have lost its radical.
Fig. 3.
Hydroxylation of Tyr123 associated with activation of class Ie RNR β. (A) Comparison of the structure in the vicinity of Tyr123 in unactivated (PDB ID code 6EBO) (Left) and activated (PDB ID code 6EBP) (Right) forms of the Au β protein reveals the dihydroxyphenylalanine (DOPA) modification in two different configurations. A 2Fo-Fc electron density map contoured to 2.0σ is shown in gray mesh. Selected side chains and water molecules are shown as ball-and-stick models and spheres, respectively, and colored by atom type. (B) Identification of the cofactor-containing peptide in activated Sp β. The anticipated structure of the active site peptide with DOPA-quinone modification at Tyr123 is shown at Top Left. The extracted parent ion chromatogram and corresponding isotopic envelope for the doubly charged peptide are shown at Top Center and Top Right, respectively. As described in detail in the SI Appendix, protein from a preparation of activated Sp β was denatured in air, reduced with TCEP, alkylated with iodoacetamide, and digested sequentially with trypsin and GluC. The DOPA-quinone modification at Tyr123, sequence position 2 of the 13-mer trypsin-GluC peptide, was identified by detection of a parent peptide with mass within 0.87 ppm of the theoretical m/z of that predicted for the indicated, DOPA-quinone–containing peptide. The high-resolution MS2 spectrum consistent with DOPA-quinone is shown at Bottom.
We next used mass spectrometry to characterize the Tyr123 modification. Initial whole-protein analysis of Au and Sp β suggested gains in mass of 15.7 ± 0.7 Da and 15.5 ± 0.6 Da, respectively, on activation (SI Appendix, Table S4), consistent with the +16-Da change expected for hydroxylation. Proteolysis of activated and unactivated forms of both Sp and Au β (sequentially treated with trypsin and endopeptidase GluC), chromatographic separation of peptides, and sequencing by high-resolution mass-spectrometric fragmentation (LC-MS/MS) provided data in support of the site of modification and the chemical identity deduced by crystallography (Fig. 3B). An unbiased search (30) of all MS2 spectra for four or more of the y-ions expected to result from the predicted 13-mer peptide containing Tyr123 allowed its identification in both activated and unactivated preparations. The masses of the dominant ions for the peptides from the activated Sp and Au β preparations were within 1.0 and 1.7 ppm of those predicted for the Cys-carbamidomethylated, Tyr123-hydroxylated 13-mer in its quinone form ([M+H]+ = 1,465.6164 Da for Sp and 1479.6333 Da for Au) (Fig. 3B and SI Appendix, Fig. S10 and Table S5). Observation of the quinone, rather than DOPA itself, is expected, given that the analysis was performed in air. For the Sp protein, an additional modification, consistent with iodination of the DOPA123 quinone during Cys-alkylation by excess iodoacetamide, was also observed (SI Appendix, Fig. S10). In neither system could a Tyr123-modified peptide be detected in the unactivated β sample expressed without NrdI.
Without tuning by its active-site environment (e.g., by control of protonation state), the DOPA radical (DOPA•) implicated by the crystallographic and LC-MS/MS data would not be expected to have a reduction potential sufficient for its generation of Cys• (31, 32). Thus, it was important to verify the structure of the modified Tyr123 species in its functional, radical state. We performed a series of Q-band 1H ENDOR measurements on activated Sp β prepared with various Tyr isotopologs (Fig. 4). We detected six distinct 1H hyperfine interactions and, for all but one, determined the associated coupling constants. Representative spectra measured at the field position corresponding to the maximum EPR absorption of the radical are shown in Fig. 4; the complete set of orientation-selected ENDOR and high-field EPR measurements, along with associated analysis, are also provided (SI Appendix, Figs. S11–S14).
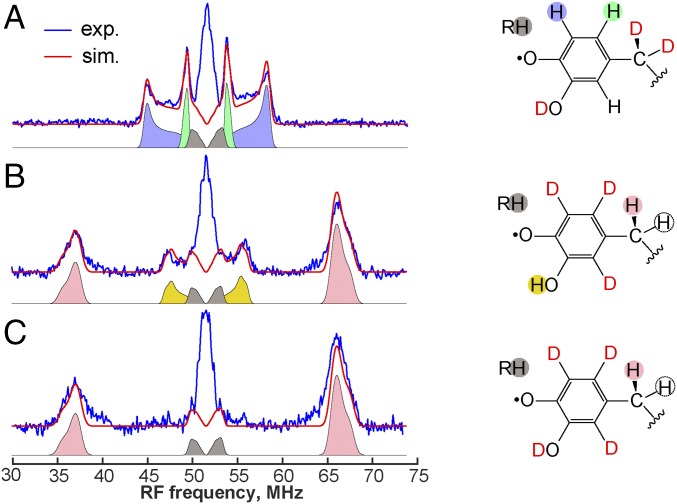
Fig. 4.
Selected Q-band 1H ENDOR measurements (blue) of the DOPA• in activated Sp β. Protein was prepared with β-[2H2]-Tyr in 2H2O (A), phenyl-[2H4]-Tyr in H2O (B), and phenyl-[2H4]-Tyr in 2H2O (C). Simulations are shown as red lines (the 1H hyperfine coupling constants are listed in SI Appendix, Table S5). Structures of DOPA• on the right indicate the proposed assignment of ENDOR signals. In total, five 1H hyperfine coupling constants were extracted, including four corresponding to 1H nuclei on DOPA• and one corresponding to a hydrogen on an unresolved amino acid residue (designated RH). Experimental conditions: Davies ENDOR pulse sequence; tinv = 60 ns; temperature, 80 K; magnetic field, 1211.8 mT; MW frequency, 34.006 GHz. The large signal at 51.6 MHz arises from weak unresolved hyperfine interactions from 1H nuclei of the protein.
The most prominent pair of peaks in the 1H ENDOR spectra corresponds to a coupling of approximately 30 MHz (SI Appendix, Table S6). This interaction is responsible for the doublet splitting that dominates the continuous-wave EPR spectrum. A sample prepared with β-[2H2]-Tyr did not exhibit the signal for this most strongly coupled 1H, confirming that this pair of ENDOR peaks arises from one of the two 1H atoms on Cβ. We did not observe a second strong coupling from the other Cβ hydrogen; we estimate that it must be weaker than 2 MHz to have eluded detection. The strength of coupling for the Cβ hydrogens is proportional to the sine of the dihedral angle between the plane of the phenyl ring and the corresponding Cβ-H bond (33). The results thus imply that one of the two Cβ-H bonds lies nearly parallel to the phenyl plane, consistent with the orientation of the DOPA seen in the Au β structure ([phenyl]-[Cβ-Cα] angle of ∼60°–70°).
Spectra of a sample made with β-[2H2]-Tyr and in 2H2O lack one pair of peaks present in the equivalent sample in 1H2O, indicating that these peaks arise from an exchangeable hydrogen on the radical (see below). Two remaining signals from a pair of anisotropically coupled 1H nuclei are absent from spectra of a sample prepared with phenyl-[2H4]-Tyr (in either 2H2O or 1H2O), associating these signals with two ring hydrogens (Fig. 4 B and C). The weak-coupling region of the spectra reveals at least one more 1H coupling from the aromatic side chain; however, we could not confidently determine its hyperfine constant, owing to interference from weakly coupled, nonexchangeable 1H atoms of the protein matrix. A pair of ENDOR signals identified in all studied samples was unaffected by the isotopolog of Tyr or solvent and accounted for in simulations (see below) by a dipolar coupling of approximately 1.0–1.5 MHz. This observation implies the presence of at least one hydrogen nucleus at 3.7–4.2 Å from the center of spin density of the radical (according to the point-dipole approximation), likely a –CHx unit from a neighboring hydrophobic amino acid.
The hyperfine interaction from the exchangeable 1H, most obvious in the spectra of the phenyl-[2H4]-Tyr samples (Fig. 4 B and C), has a substantial isotropic component (∼5 MHz; SI Appendix, Table S6), indicating that it is bonded to an atom harboring significant spin density. This signal, absent from corresponding spectra of known Tyr•s, can be assigned to the 1H on the C3–OH of the neutral DOPA•. The retention of this 1H can be rationalized based on the active-site H-bonding interactions seen in the crystal structure (see below) and is almost certainly essential for the tuning of the DOPA• for α-Cys oxidation.
The parameters used in a global simulation of all the ENDOR and continuous-wave EPR spectra (SI Appendix, Table S6) are consistent with the assignment of the radical as neutral DOPA•. This conclusion is further supported by the consistency of the couplings with those from density functional theory calculations of a model of the radical (SI Appendix, Tables S6 and S7). The combined structural data thus imply that class Ie RNRs initiate catalysis with a DOPA•, which is generated by a three-electron oxidation of a Tyr residue mediated by the flavoprotein NrdI.
Deep Mutational Scanning of Au β Reveals Sites Essential for DOPA• Installation and Function.
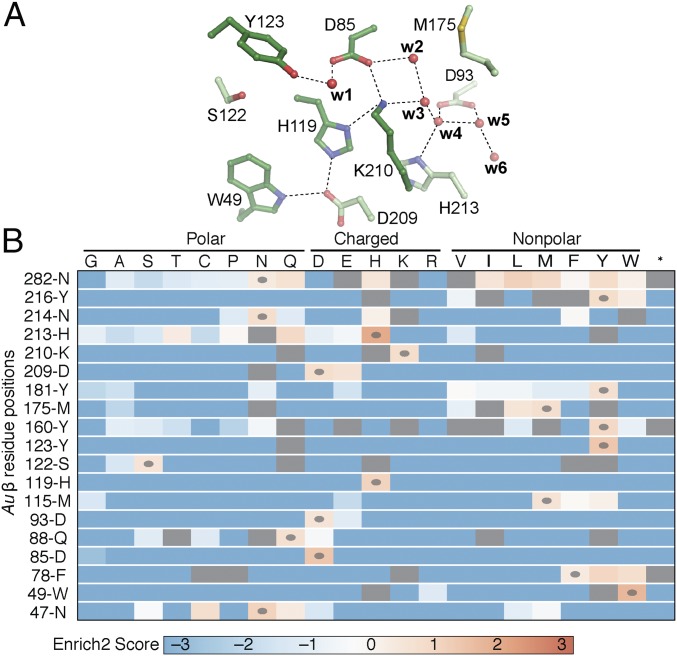
The distinctive modus operandi of the new Ie subclass upends expectations of the roles of conserved (and subclass e-specific) residues in activation and radical initiation. To assess the functional importance of selected residues, we leveraged the aforementioned Ec strain with the temperature-sensitive class Ia RNR α (13) as a selection for a deep mutational scanning (DMS) experiment (SI Appendix, Fig. S15) (34). We selected 19 residues in Au β (SI Appendix, Fig. S16), constructed a library of 32 genetic variants (encoding all 20 amino acids plus a stop codon) for each site, and assembled them into the complementation plasmid. After transformation of the selection strain, KK444, amplicon libraries from preselection (28 °C) and postselection (42 °C) populations were pooled and sequenced on the Illumina HiSeq platform. No functionally competent substitution for Tyr123, the site of DOPA• installation, was identified (Fig. 5B) in this experiment.
Fig. 5.
Results of DMS of Au β at 19 selected sites. (A) Sites near the DOPA• that are intolerant to substitution. Selected side chains and water molecules are shown as ball-and-stick models and spheres, respectively, and color-coded by atom type and degree of substitution tolerance (dark green, intolerant to substitution; light green, moderately tolerant to conservative substitutions). (B) Amino acid enrichment/depletion map of inferred mutational tolerance of targeted residues. Red boxes (positive scores) indicate mutations highly enriched by selective growth at 42 °C; blue boxes (negative scores), mutations highly depleted by selective growth; gray boxes, mutations not tested due to incomplete coverage during library construction. Ovals indicate the wild-type residues at each site. One-letter amino acid codes are used. The stop codon TAG is indicated by an asterisk (*).
To assess the basis for the redox tuning of the DOPA• expected to be required for it to generate the Cys• in α, we included the adjacent residues, Asp85 and Lys210, in the DMS assay. Neither residue can be substituted with any other natural amino acid present in the initial library without loss of in vivo function, as judged by the ability to complement the temperature-sensitive mutant strain at the restrictive temperature. Inspection of the X-ray structure suggests that one of the Asp85 side-chain carboxylate O-atoms makes a short H-bond with the newly installed O-atom of DOPA123 (SI Appendix, Fig. S17). Comparison of the positioning of DOPA relative to Asp85 in each of the four chains of the asymmetric unit in the activated Au β crystals (Fig. 3A and SI Appendix, Fig. S7) reveals two distinct states, I and II, of the DOPA/Asp85 dyad. One of these states could represent an intermediate, as in a form that undergoes hydroxylation but not further oxidation to the radical, in a two-step activation process. Alternatively, failed activation events or adventitious reduction to the inactive DOPA state in a fraction of the monomers could account for the differences. In all structures, the other Asp85 side-chain oxygen H-bonds to the side chain of Lys210. These interactions would maintain both the DOPA• and the Lys210 nitrogen in their protonated forms (SI Appendix, Fig. S17). In general, a proximal positive charge should elevate the reduction potential of the DOPA•. Residues His119 and Trp49, also intolerant to every substitution sampled in the DMS experiment (Fig. 5 and SI Appendix, Fig. S16), form a water-mediated H-bonding network between Lys210 and the protein surface (SI Appendix, Fig. S17), and they likely play roles in controlling proton transfer(s) coupled to the α-Cys-to-DOPA• electron transfer that initiates turnover (8). The radical-translocation process in class Ia RNRs involves the transfer of a proton from a water molecule coordinated to the Fe(III) ion in site 1 to the oxygen of the Tyr• (35). The lack of a transition metal in the active state of the class Ie RNR necessitates a different proton donor and pathway if, as expected, reduction of the DOPA• is coupled to proton transfer to the C4 O-atom.
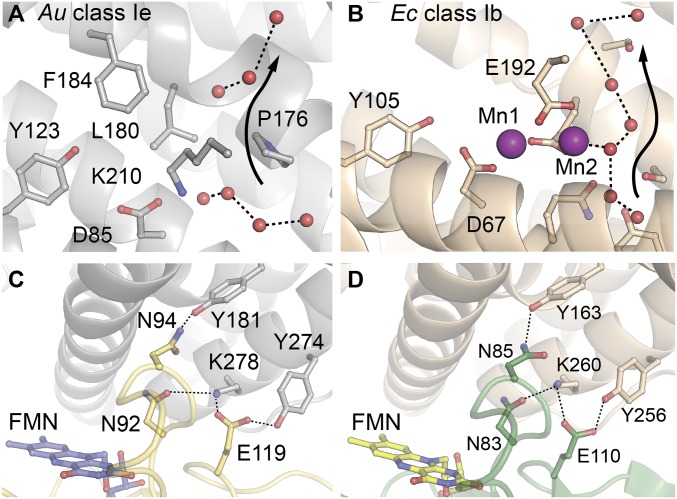
The inferred transformation of the class Ie β protein by its NrdI activase—formally, hydroxylation and removal of an additional electron from a Tyr residue—is quite different from that effected in the class Ib system (Mn2II/II oxidation on the path to Tyr•) (4, 36), raising the possibility that the two NrdIs might have different modes of action. However, the structure of the Au NrdIox (1.95-Å resolution) (SI Appendix, Fig. S18) shares with previously characterized Ib NrdI proteins (especially the Ec ortholog) (28) several functionally important features (SI Appendix, Figs. S18–S20). For example, a Gly-rich FMN-binding loop (50s loop) in Au NrdI donates a backbone N-H to H-bond with N5 of the flavin, an interaction that significantly occludes the O2-reactive C4a position (SI Appendix, Fig. S18). In Ec NrdI, the 50s loop opens on FMN reduction, and the C4a position becomes more exposed. The redox-dependent change controls reaction of the flavin with oxygen/superoxide (28, 36). Conservation of the loop in Au NrdI weighs in favor of O2 reduction to superoxide or peroxide by the class Ie activase. Moreover, a model for the Au β•NrdI complex, which we generated using the structure of the corresponding Ib Ec complex [Protein Data Bank (PDB) ID code 3N3A] (28) as a template, shows that the Ie β protein shares two defining structural adaptations of the Ib βs that enable (i) their complexation with NrdI and (ii) guidance of the reactive, hydrophilic oxidant (superoxide) to the buried cofactor site. The model is consistent with use of identical protein–protein interfaces in the Ib and Ie β•activase complexes; residues predicted to interact at the interface are highly conserved (Fig. 6 A and B). The second feature, a chain of hydrogen-bonded water molecules in Ie β lining the proposed pathway for superoxide migration, is also retained, interrupted only by the side chain of the Pro that replaces a Glu metal ligand in Ie proteins (Fig. 6 C and D). In addition, a strictly conserved surface Lys residue in Ib β (K260) (Fig. 6D), a key component of the NrdI interface proposed to guide the anionic superoxide to the Mn2II/II cluster, is also present in the Ie β. These observations are all consistent with similar roles for the Ie and Ib NrdI proteins.
Fig. 6.
(A and B) Solvent access channels in β and β-NrdI interfaces in the class Ie Au and class Ib Ec RNR systems. Channels in Au β (A) and Ec class Ib β (PDB ID code 3N37) (B). (C and D) NrdI-β interfaces proposed for the Au complex from the structural model constructed in this study (C) and observed in the structure of Ec Ib β•NrdI (PDB ID code 3N3A) (D).
Important aspects of the mechanism of activation of the new class Ie β remain to be resolved. For example, it is not clear whether the three-electron oxidation of Tyr123 to DOPA• requires a transition metal—as in the posttranslational installation of most known quinocofactors (37)—that subsequently dissociates or requires no metal assistance at all. In the former case, the transient metal requirement would be reminiscent of the activation mechanism of a class III RNR [iron-dependent installation of a catalytic Gly• in a metal-free RNR (7)] and would render the class Ie enzymes the aerobic counterpart of this catalytic metal usage. Similarly, the identity and stoichiometry of the (presumably) O2-derived oxidant(s) are not known, although the structural analogy to the Ib systems and the fact that DOPA• production is a three-electron oxidation make the use of a single equivalent of superoxide the simplest possibility. Interestingly, a structure of Au β solved with thiocyanate as additive reveals the ion bound noncovalently in a pocket immediately adjacent to the Lys210-Asp85-Tyr123 triad (SI Appendix, Fig. S21). The similarity of thiocyanate to superoxide in size, structure, and charge raises the possibility that this pocket binds the oxidant before its reaction with Tyr123 or a transiently bound metal ion.
Methods
Detailed information on metal analysis, spectroscopy, X-ray crystallography, mass spectrometry, and mutational scanning is provided in SI Appendix. A summary is provided here.
Purification of Active Ie β Subunits.
N-terminally His6-tagged Sp and Au β proteins were coexpressed with Au NrdI as described in SI Appendix. Supplementation of rich or minimal growth medium with transition metals was not required to obtain activated protein samples. Proteins were purified by NiII- or CoII-nitrilotriacetate-agarose affinity chromatography and used for subsequent activity, spectroscopic, or MS analysis in this form. Au β was further purified for crystallography as described in SI Appendix.
Activity Assays.
Ribonucleotide reduction reactions were performed at room temperature in 50 mM Tris⋅HCl pH 7.6 and 5% glycerol with DTT (10 mM). Mixtures contained 250 μM α subunit, 25 mM activated β subunit, 12.6 mM MgSO4, 2 mM CDP (substrate), and 1 mM ATP (allosteric effector). Reactions were initiated by addition of the limiting β subunit, present at a 1:10 molar ratio with its α counterpart. Product formation was detected by a previously validated LC-MS/MS assay (5), as described in SI Appendix.
Supplementary Material
Acknowledgments
We thank JoAnne Stubbe for donation of N3-CDP and Benjamin Garcia and his group for instrument time and helpful discussions. Support was provided by grants from the Searle Scholars Program (to A.K.B.) and National Institutes of Health (NIH) Grant GM119707 (to A.K.B.). E.J.B. was supported by NIH Ruth L. Kirschstein Postdoctoral National Research Service Award GM116353. Portions of this work were performed at the Penn State Proteomics and Mass Spectrometry Core Facility and at the Advanced Photon Source (APS), the latter a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. GM/CA at APS has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Eiger 16M detector was funded by NIH Office of Research Infrastructure Programs High-End Instrumentation Grant 1S10OD012289-01A1. The use of LS-CAT Sector 21 was supported by Michigan Economic Development Corporation and Michigan Technology Tri-Corridor Grant 085P1000817.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6EBO, 6EBP, 6EBQ, and 6EBZ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811993115/-/DCSupplemental.
References
- 1.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Licht S, Gerfen GJ, Stubbe J. Thiyl radicals in ribonucleotide reductases. Science. 1996;271:477–481. doi: 10.1126/science.271.5248.477. [DOI] [PubMed] [Google Scholar]
- 3.Mao SS, et al. A model for the role of multiple cysteine residues involved in ribonucleotide reduction: Amazing and still confusing. Biochemistry. 1992;31:9733–9743. doi: 10.1021/bi00155a029. [DOI] [PubMed] [Google Scholar]
- 4.Cotruvo JA, Jr, Stubbe J. Class I ribonucleotide reductases: Metallocofactor assembly and repair in vitro and in vivo. Annu Rev Biochem. 2011;80:733–767. doi: 10.1146/annurev-biochem-061408-095817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose HR, et al. Structural basis for superoxide activation of Flavobacterium johnsoniae class I ribonucleotide reductase and for radical initiation by its dimanganese cofactor. Biochemistry. 2018;57:2679–2693. doi: 10.1021/acs.biochem.8b00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence CC, Stubbe J. The function of adenosylcobalamin in the mechanism of ribonucleoside triphosphate reductase from Lactobacillus leichmannii. Curr Opin Chem Biol. 1998;2:650–655. doi: 10.1016/s1367-5931(98)80097-5. [DOI] [PubMed] [Google Scholar]
- 7.Gambarelli S, Luttringer F, Padovani D, Mulliez E, Fontecave M. Activation of the anaerobic ribonucleotide reductase by S-adenosylmethionine. ChemBioChem. 2005;6:1960–1962. doi: 10.1002/cbic.200500182. [DOI] [PubMed] [Google Scholar]
- 8.Minnihan EC, Nocera DG, Stubbe J. Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase. Acc Chem Res. 2013;46:2524–2535. doi: 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 10.Cotruvo JA, Jr, Stubbe J. Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: The class I ribonucleotide reductases as a case study. Metallomics. 2012;4:1020–1036. doi: 10.1039/c2mt20142a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin JE, Imlay JA. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol. 2011;80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotruvo JA, Jr, Stubbe J. Escherichia coli class Ib ribonucleotide reductase contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry. 2011;50:1672–1681. doi: 10.1021/bi101881d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platz A, Sjöberg B-M. Construction and characterization of hybrid plasmids containing the Escherichia coli nrd region. J Bacteriol. 1980;143:561–568. doi: 10.1128/jb.143.2.561-568.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roca I, Torrents E, Sahlin M, Gibert I, Sjöberg B-M. NrdI essentiality for class Ib ribonucleotide reduction in Streptococcus pyogenes. J Bacteriol. 2008;190:4849–4858. doi: 10.1128/JB.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson R, et al. High-resolution crystal structures of the flavoprotein NrdI in oxidized and reduced states: An unusual flavodoxin. FEBS J. 2010;277:4265–4277. doi: 10.1111/j.1742-4658.2010.07815.x. [DOI] [PubMed] [Google Scholar]
- 17.Makhlynets O, et al. Streptococcus sanguinis class Ib ribonucleotide reductase: High activity with both iron and manganese cofactors and structural insights. J Biol Chem. 2014;289:6259–6272. doi: 10.1074/jbc.M113.533554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes DV, et al. Genetic characterization and role in virulence of the ribonucleotide reductases of Streptococcus sanguinis. J Biol Chem. 2014;289:6273–6287. doi: 10.1074/jbc.M113.533620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens DL, et al. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N Engl J Med. 1989;321:1–7. doi: 10.1056/NEJM198907063210101. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen M. Aerococcus: An increasingly acknowledged human pathogen. Clin Microbiol Infect. 2016;22:22–27. doi: 10.1016/j.cmi.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Sjöberg B-M, Reichard P. Nature of the free radical in ribonucleotide reductase from Escherichia coli. J Biol Chem. 1977;252:536–541. [PubMed] [Google Scholar]
- 22.Karlsson M, Sahlin M, Sjöberg B-M. Escherichia coli ribonucleotide reductase. Radical susceptibility to hydroxyurea is dependent on the regulatory state of the enzyme. J Biol Chem. 1992;267:12622–12626. [PubMed] [Google Scholar]
- 23.Salowe SP, et al. Alternative model for mechanism-based inhibition of Escherichia coli ribonucleotide reductase by 2′-azido-2′-deoxyuridine 5′-diphosphate. Biochemistry. 1993;32:12749–12760. doi: 10.1021/bi00210a026. [DOI] [PubMed] [Google Scholar]
- 24.Skoog L, et al. 2′-deoxy-2′-azidocytidine, a new inhibitor of DNA replication in mammalian cells. Eur J Biochem. 1977;72:371–378. doi: 10.1111/j.1432-1033.1977.tb11261.x. [DOI] [PubMed] [Google Scholar]
- 25.Fritscher J, et al. Structure of the nitrogen-centered radical formed during inactivation of E. coli ribonucleotide reductase by 2′-azido-2′-deoxyuridine-5′-diphosphate: Trapping of the 3′-ketonucleotide. J Am Chem Soc. 2005;127:7729–7738. doi: 10.1021/ja043111x. [DOI] [PubMed] [Google Scholar]
- 26.Jiang W, et al. A manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Science. 2007;316:1188–1191. doi: 10.1126/science.1141179. [DOI] [PubMed] [Google Scholar]
- 27.Voegtli WC, et al. Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2. J Am Chem Soc. 2003;125:15822–15830. doi: 10.1021/ja0370387. [DOI] [PubMed] [Google Scholar]
- 28.Boal AK, Cotruvo JA, Jr, Stubbe J, Rosenzweig AC. Structural basis for activation of class Ib ribonucleotide reductase. Science. 2010;329:1526–1530. doi: 10.1126/science.1190187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordlund P, Sjöberg B-M, Eklund H. Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature. 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 30.Matthews ML, et al. Chemoproteomic profiling and discovery of protein electrophiles in human cells. Nat Chem. 2017;9:234–243. doi: 10.1038/nchem.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seyedsayamdost MR, Stubbe J. Site-specific replacement of Y356 with 3,4-dihydroxyphenylalanine in the beta2 subunit of E. coli ribonucleotide reductase. J Am Chem Soc. 2006;128:2522–2523. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 32.Lee W, et al. Properties of site-specifically incorporated 3-aminotyrosine in proteins to study redox-active tyrosines: Escherichia coli ribonucleotide reductase as a paradigm. Biochemistry. 2018;57:3402–3415. doi: 10.1021/acs.biochem.8b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolberg M, et al. A new tyrosyl radical on Phe208 as ligand to the diiron center in Escherichia coli ribonucleotide reductase, mutant R2-Y122H. Combined x-ray diffraction and EPR/ENDOR studies. J Biol Chem. 2005;280:11233–11246. doi: 10.1074/jbc.M414634200. [DOI] [PubMed] [Google Scholar]
- 34.Fowler DM, Fields S. Deep mutational scanning: A new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wörsdörfer B, et al. Function of the diiron cluster of Escherichia coli class Ia ribonucleotide reductase in proton-coupled electron transfer. J Am Chem Soc. 2013;135:8585–8593. doi: 10.1021/ja401342s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotruvo JA, Jr, Stich TA, Britt RD, Stubbe J. Mechanism of assembly of the dimanganese-tyrosyl radical cofactor of class Ib ribonucleotide reductase: Enzymatic generation of superoxide is required for tyrosine oxidation via a Mn(III)Mn(IV) intermediate. J Am Chem Soc. 2013;135:4027–4039. doi: 10.1021/ja312457t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klinman JP, Bonnot F. Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ. Chem Rev. 2014;114:4343–4365. doi: 10.1021/cr400475g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.