Significance
Escherichia coli secrete polysaccharides, protein adhesins, and fibers to promote adhesion and biofilm formation. Curli are functional amyloid fibers produced by many human and environmental E. coli isolates. Furthermore, we recently discovered that E. coli produces a chemically modified form of cellulose, phosphoethanolamine cellulose. Here, we use a custom-built live-cell monolayer rheometer, housed within a microscope, to examine curli-based adhesion and reveal the molecular role of the modified cellulose in influencing adhesion of uropathogenic E. coli to bladder epithelial cells. In a high-shear environment, phosphoethanolamine cellulose confers a mortar-like function in maintaining cellular association of curli. Curli are readily dissociated from the cell in the absence of cellulose. This approach is applicable to other cellular and host-pathogen interfaces.
Keywords: E. coli, bacterial adhesion, phosphoethanolamine cellulose, curli, rheology
Abstract
Uropathogenic Escherichia coli (UPEC) are the major causative agents of urinary tract infections, employing numerous molecular strategies to contribute to adhesion, colonization, and persistence in the bladder niche. Identifying strategies to prevent adhesion and colonization is a promising approach to inhibit bacterial pathogenesis and to help preserve the efficacy of available antibiotics. This approach requires an improved understanding of the molecular determinants of adhesion to the bladder urothelium. We designed experiments using a custom-built live cell monolayer rheometer (LCMR) to quantitatively measure individual and combined contributions of bacterial cell surface structures [type 1 pili, curli, and phosphoethanolamine (pEtN) cellulose] to bladder cell adhesion. Using the UPEC strain UTI89, isogenic mutants, and controlled conditions for the differential production of cell surface structures, we discovered that curli can promote stronger adhesive interactions with bladder cells than type 1 pili. Moreover, the coproduction of curli and pEtN cellulose enhanced adhesion. The LCMR enables the evaluation of adhesion under high-shear conditions to reveal this role for pEtN cellulose which escaped detection using conventional tissue culture adhesion assays. Together with complementary biochemical experiments, the results support a model wherein cellulose serves a mortar-like function to promote curli association with and around the bacterial cell surface, resulting in increased bacterial adhesion strength at the bladder cell surface.
Urinary tract infection (UTI) is one of the most common infectious diseases, affecting 150 million people worldwide annually (1) and with estimated healthcare costs exceeding $3 billion in the United States alone (2). Almost half of all women will experience at least one UTI (3). Many cases are uncomplicated and do not result in long-term sequelae. However, some infections result in more serious medical consequences, including pyelonephritis, renal damage in pediatric patients, and premature delivery and fetal mortality in pregnant women (2, 3). Chronic and recurrent infections require long-term antibiotic therapy and can lead to antibiotic resistance and even sepsis (4–7). Strategies to prevent adhesion, a crucial step in the initial interactions and molecular crosstalk at the host−pathogen interface, are attractive for the development of new antiinfectives (2, 8, 9).
Uropathogenic Escherichia coli (UPEC) are the major causative agents of UTI (10). UPEC pathogenesis in the bladder is initiated by bacterial adhesion to the bladder epithelium (11). Adhesion can be followed by bacterial invasion into the superficial epithelial cells, which is uniquely dependent upon the production of adhesive fimbriae termed type 1 pili (12). Type 1 pili are polymeric fibers made up of repeating Ig-like subunits of FimA, presenting the adhesin FimH at the pilus tip to bind to mannosylated host cell receptors (11, 13). Inside a urothelial cell, UPEC can replicate to form intracellular bacterial communities (IBCs) that offer protection from antibiotic treatment and host defenses (14–16). Subsequently, bacteria can exit the host cell to initiate further rounds of invasion and IBC formation. UPEC can also form quiescent intracellular reservoirs in underlying bladder cells to promote long-term persistence, presenting a potential contribution to recurrent UTI (15, 17–19).
A major challenge in targeting UPEC adhesion is the diverse and seemingly redundant array of UPEC adhesins and fibers as well as polysaccharides that can promote adhesion and colonization (2). The type 1 pilus is perhaps the most well-studied virulence factor associated with UPEC infection. However, clinical isolates differ tremendously in their phenotypes in vitro and in vivo due to other molecular features that differentiate them and their interactions with the host (20). Indeed, evidence has been emerging demonstrating that curli amyloid fibers can contribute to UPEC pathogenesis. Curli are functional amyloid fibers that mediate bacterial adhesion and the formation of bacterial communities termed biofilms (21). Curli have also been considered for possible roles in UTI pathogenesis. Curli (i) confer a fitness advantage to curli-producing organisms in the mouse model of UTI (22), (ii) have been detected in human urine samples (23), and (iii) elicit modulation of immune responses (23–26). Curli production is also strongly correlated with progression of UTI to sepsis (27, 28). Outcomes of UTI pathogenesis experiments in mouse models can vary substantially depending on the UPEC clinical isolate being employed and the genetic and molecular features that are deployed by the bacterium during mouse bladder colonization (29). We hypothesize that the extent of curli production may be a factor in infection outcomes. Thus, efforts to inhibit curli production and biofilm formation are being actively pursued by the L.C. laboratory and others (22, 30). In this work, we sought to understand the relative contributions of curli versus type 1 pili and to examine the influence of the modified cellulose, phosphoethanolamine (pEtN) cellulose, produced by UPEC (31) in bladder cell adhesion.
Curli can mediate adhesion to biotic and abiotic surfaces and are able to bind to host proteins, including laminin (32), fibronectin (33), and major histocompatibility complex class I molecules (34). However, less is known regarding the potential involvement of cellulose in adhesion. Moreover, we recently discovered that E. coli produces a chemically modified cellulose: pEtN cellulose (31). This discovery was made possible using solid-state NMR analysis with intact material. When the material is digested with acid, as is common in conventional studies with E. coli cellulose, the pEtN modification is hydrolyzed and thus not detected as containing modified glucose using mass spectrometry, for example. We also identified the genetic basis for pEtN installation, requiring the bcsEFG operon, where BcsG appears to be the pEtN transferase which installs the modification to newly synthesized cellulose in the periplasm. Thus, we now appreciate that most studies that have examined cellulose production among curli-producing strains of E. coli likely involved pEtN cellulose. Previous studies have suggested that the coexpression of curli and cellulose can reduce adhesion to bladder, renal, and intestinal epithelial cells compared with adhesion when curli are expressed alone (23, 35). However, the coexpression of curli and cellulose was reported to synergistically increase adhesion, in a separate study involving only intestinal epithelial cells (36).
Here, we present measurements using a live-cell monolayer rheometer (LCMR) (37, 38), coupled with traditional bladder cell adhesion assays and a biochemical assay, to define contributions of type 1 pili, curli, and pEtN cellulose in UPEC adhesion to bladder cells. Our results indicate that curli can play a significant role in bladder cell adhesion, and that pEtN cellulose facilitates curli-mediated adhesion in high-shear conditions that are relevant in the bladder niche.
Results
Evaluation of UPEC Adhesion to Bladder Cells Through Traditional Adhesion Assays.
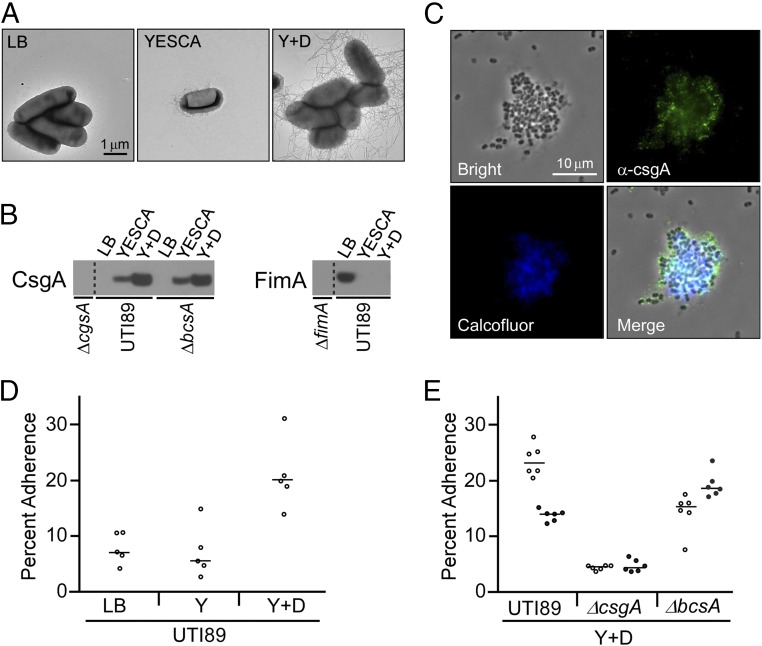
A hallmark UPEC strain, UTI89 (39), and its isogenic mutants were used to consider the individual and combined contributions of type 1 pili, curli, and pEtN cellulose to adhesion of 5637 bladder cells. Distinct growth conditions have been defined for promoting the production of type 1 pili (static LB broth) (40) and curli [YESCA (0.5 g/L yeast extract, 10 g/L casamino acids) nutrient broth] and increased curli production (YESCA supplemented with DMSO, abbreviated Y + D in figures) (41). Type 1 pili and curli protein production was characterized by electron microscopy and Western blot analysis for the exact bacterial strains and conditions used in this work and confirmed expectations for the selected growth conditions (Fig. 1 A and B). Fluorescence microscopy of UTI89 grown shaking in YESCA nutrient broth supplemented with DMSO at 26 °C confirmed the presence of both curli (CsgA immunofluorescence) and pEtN cellulose (Calcofluor fluorescence) as shown in Fig. 1C, with colocalization of the two signals. Growth in YESCA nutrient broth or agar generally results in curli and pEtN cellulose production due to gene regulation by the global regulator, CsgD, which activates curli and cellulose gene transcription in stationary phase (42).
Fig. 1.
Growth conditions modulate type 1 pili, curli, and pEtN cellulose production and influence adhesion to bladder cells in traditional adhesion assays. (A) Transmission electron microscopy (TEM) images of UTI89 grown to promote differential production of extracellular fibers. (B) Whole cell levels of CsgA and FimA determined by Western blot analysis. (C) Fluorescence images of UTI89 grown in YESCA broth supplemented with 4% DMSO and examined with the cellulose-binding dye Calcofluor White and immunofluorescent staining for the major curli subunit CsgA. (D) Bacterial adherence to bladder cells in the traditional tissue culture adhesion assay with UTI89 grown under different conditions. (E) Bacterial adherence comparing UTI89, UTI89∆csgA, and UTI89∆bcsA. Open and solid circles represent data from two separate experiments. Percent adherence was calculated as the number of adherent bacteria in one test well divided by the average number of bacteria from control wells used for total bacterial counts. Bars represent the median of each data set. Growth conditions were as follows: LB: LB broth, 37 °C, static culture; YESCA: YESCA broth, 26 °C, shaking culture; Y + D: YESCA broth supplemented with 4% DMSO, 26 °C, shaking culture.
Traditional tissue culture adhesion assays were employed to compare adhesion by UTI89 grown under the distinct type 1 pili and curli-promoting conditions. When grown under type 1 pili conditions (LB broth), UTI89 enumeration after a 10-min incubation revealed that ∼7% of bacteria remained adherent to bladder cells (Fig. 1D). Bacterial adhesion to bladder cells has been previously demonstrated to depend highly on type 1 pilus expression and the adhesin FimH (12, 43–45), which was observed in our experiments as well (SI Appendix, Fig. S1). Assays with moderately curliated cells (YESCA nutrient broth) yielded a similar percent of adherent bacteria as the type 1 pili producing UTI89 (Fig. 1D). UTI89 with increased curliation (YESCA broth supplemented with DMSO) yielded an approximately fourfold increase in the percent of adherent bacteria over UTI89 grown in LB and YESCA (Fig. 1D). Adhesion of the double mutant UTI89ΔfimHΔcsgA was similar to UTI89ΔfimH and UTI89ΔcsgA in the LB and YESCA conditions, respectively, further supporting the specific roles of type 1 pili and curli for adhesion in those conditions and demonstrating that some minimal adhesion occurs even in the absence of type 1 pili and curli (SI Appendix, Fig. S1). Experiments were then performed to compare UTI89 with the curli mutant UTI89∆csgA (lacking the major curli subunit CsgA) and the cellulose mutant UTI89∆bcsA (lacking the major cellulose synthase protein BcsA) when grown in the increased curliation conditions of DMSO-supplemented YESCA broth. This comparison revealed that bacteria lacking curli were compromised in mediating adhesion to bladder cells. Loss of pEtN cellulose had a less dramatic effect, and UTI89∆bcsA was observed to exhibit a slight decrease or increase in different trials relative to UTI89, whereas adherence trends were always maintained for other strain and condition comparisons (Fig. 1E). Thus, these traditional tissue culture adhesion assays revealed that increased curli production by UTI89 yields an increased number of adherent bacteria associated with a bladder cell monolayer. However, by microscopy, clusters of bacteria were often observed associated with bladder cells (SI Appendix, Fig. S2), and adherence of UTI89∆bcsA was observed to be more variable than other strains (Fig. 1E). This presented the possibility that increased bacterial numbers in this adhesion assay could be attributed to aggregative interbacterial interactions rather than reflecting intrinsically more bacteria in specific contact with bladder cells. In addition, the traditional assay utilizes an uncontrolled, operator-dependent rinsing procedure to expel unattached bacteria from the bladder cells. This means that the procedure can provide a useful index of bacteria adherence but not a standardized quantification of adhesion. Thus, we sought to develop an approach for the quantitative evaluation of the strength of bacterial adhesion to mammalian cells.
The LCMR for Direct Measurement of Bacterial Adhesion to Mammalian Cells.
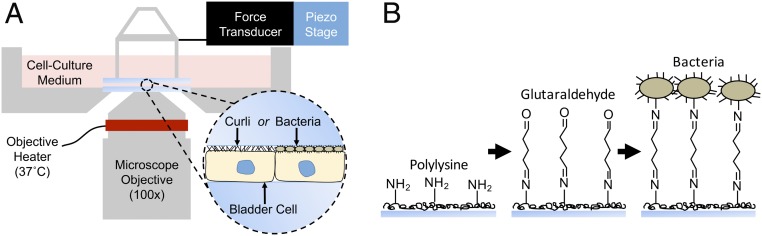
Several technologies enable measurement of bacterial adhesion to surfaces, including atomic force microscopy (AFM) and the quartz crystal microbalance (46). However, these approaches can require many trials to obtain statistically significant results and are not readily amenable to measuring bacterial adhesion to mammalian cells. In addition, techniques such as AFM only probe a small portion of a sample at a time. The LCMR (37, 38) was designed and built to enable quantitative measurements of adhesion involving mammalian cells. The LCMR enables the rapid determination of the average mechanical properties of an entire cell monolayer, while maintaining cell−cell contacts and the adherent geometry of living cells. We introduce the LCMR here as being uniquely suited to the examination of host−pathogen interfaces. As shown in Fig. 2A, the LCMR consists of a parallel glass plate assembly mounted over an inverted microscope that can be submerged in cell culture medium. In this work, bladder cells were cultured as a monolayer on the bottom plate. Either extracellular bacterial fibers or whole bacteria cells were attached to the top plate. The complete LCMR design and operation is provided in SI Appendix.
Fig. 2.
LCMR instrument and adhesion measurements. (A) Diagram of the LCMR, illustrating its utility in measuring the adhesion of either isolated curli fibers or bacterial whole cells to a bladder cell monolayer. (B) Schematic representation of the covalent linking strategy implemented to attach whole bacterial cells to the top plate of the LCMR.
During experiments, the top plate was translated horizontally by movement of a piezoelectric stage connected to a force transducer, effectively shearing the plate across the bladder cells. This shearing motion, or step strain, will impart a greater stress in the bladder cell layer if there is adhesion between the top plate and bladder cells. This was quantified by recording the force on the top plate and distance of displacement, which was used to calculate a relaxation modulus and strain, providing a quantitative measure of adhesion. Following the step strain, stress in the bladder cell layer can be relieved via both internal structural rearrangements of the bladder cells and progressive detachment of the bladder cells from the top plate. As the bladder cell layer was of the same cell type and grown to confluency for every experiment, mechanics of the cell layer were assumed to be consistent from trial to trial. Differences in relaxation moduli were therefore due directly to differences in adhesion, with the initial magnitude and decay providing information on both the strength of adhesion and detachment dynamics.
Purified Curli Enhance Bladder Cell Adhesion.
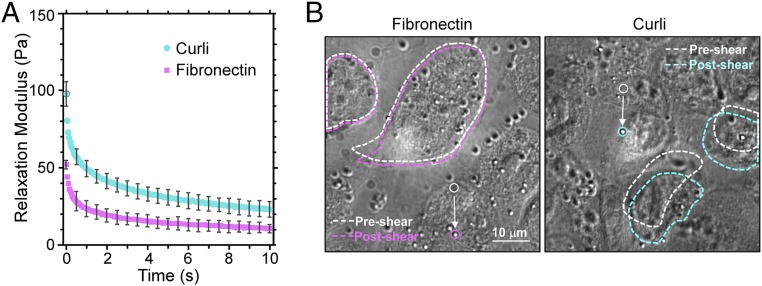
To investigate the role of curli in bladder cell adhesion, we first performed experiments with isolated curli (47) attached to the top plate of the LCMR. We utilized the known fibronectin-binding property of curli to associate curli with a fibronectin-coated top plate. As shown in Fig. 3A, the presence of curli resulted in a significant increase of the relaxation modulus over the entire measurement time compared with control experiments with the top plate coated in fibronectin alone. It is therefore evident that curli can mediate physical adhesion to bladder cells. In addition, this phenomenon could be visualized through the inverted microscope as shown in Fig. 3B. In these images, the top plate is in focus and the upper surfaces of the bladder cells before and after shearing are outlined. In the absence of curli (fibronectin coating only), the upper cell surface is not significantly displaced by motion of the top plate. The top plate could essentially glide and slip across the cell layer, indicating low adhesion. On the other hand, with curli present on the top plate, the upper cell surface was significantly deformed and observed to translate in the direction of shear, revealing obvious and strong adhesive interactions (Movies S1 and S2).
Fig. 3.
Interactions of isolated curli and bladder cells in the LCMR. (A) Relaxation moduli from LCMR experiments reveal the adhesiveness of curli to bladder cells compared with the fibronectin alone (control). Error bars represent the SD of three trials. The applied strain was 1.3 ± 0.3. (B) Images show the bladder cell deformation. White dashed lines outline bladder cells before top plate movement; colored lines outline cells directly after top plate motion. Solid circles identify the starting and final positions of a 1-μm tracking bead attached to the top plate.
The absolute magnitude of the moduli measured in these experiments, with maxima ranging from ∼50 Pa to ∼100 Pa, also indicates that the bladder cell layer is a relatively soft material. While cell moduli values generally can range from tens of pascals to tens of kilopascals depending on the cell type and measurement technique, the order of magnitude of our values agrees with previous studies on both bladder cells (48, 49) and other epithelial cell lines (50–53).
Experiments with Intact Bacteria Reveal Different Roles for Cell Surface Structures.
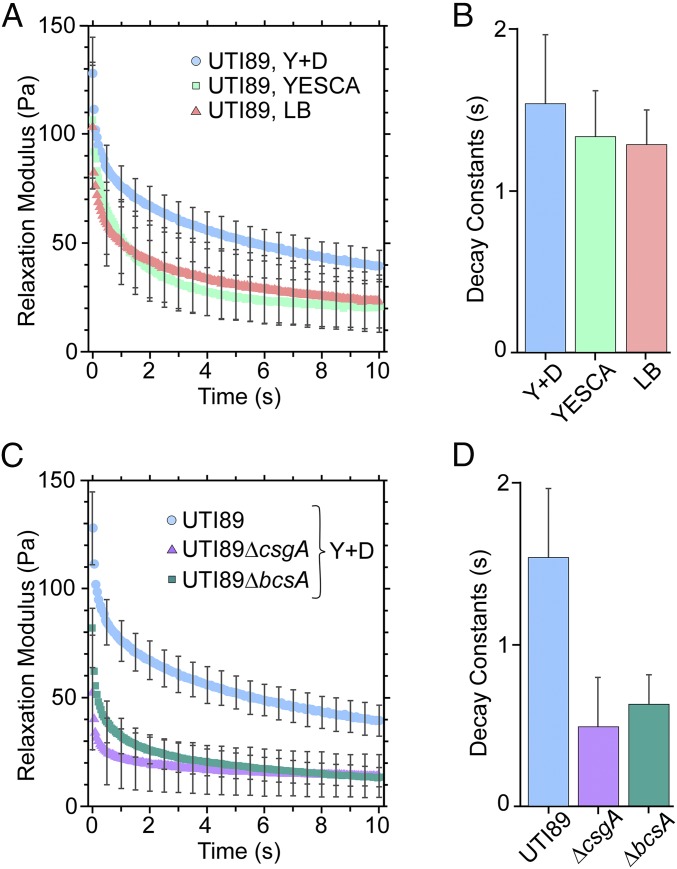
To utilize the LCMR instrument with intact bacteria, we developed a covalent attachment strategy to assemble a single compact layer of bacteria on the top plate (Fig. 2B). In addition, the amount of strain to be employed in each LCMR measurement was optimized and corresponded to strains of 1 to 1.5, i.e., a 5- to 7-µm movement of the top plate. This strain regime is larger than the diameter of individual bacteria, but not larger than the bladder cell diameter. We first used the LCMR to quantify adhesion of UTI89 to bladder cells when grown under type 1 pili (LB broth) and curli-promoting (YESCA and YESCA supplemented with DMSO) conditions, as in the traditional tissue culture assays presented in Fig. 1. As observed in Fig. 4A, adhesion of type 1 piliated and moderately curliated bacteria resulted in similar relaxation moduli, while bacteria with increased curliation had a higher relaxation modulus. We additionally demonstrated that reduced adhesion in the LCMR measurement was observed for UTI89ΔfimH grown in LB broth compared with UTI89, consistent with the contribution of type 1 pili to adhesion and without curli production in this growth condition (SI Appendix, Fig. S3). These measurements independently support the conclusion from the traditional adhesion assay: Strongly curliated bacteria exhibit enhanced bladder cell adhesion capacity compared with type 1 piliated bacteria.
Fig. 4.
Interactions of whole bacteria and bladder epithelial cells in the LCMR. (A) Relaxation moduli comparing the adhesiveness of UTI89 producing type 1 pili (LB), curli (YESCA), and an increased level of curli (Y + D) to bladder cells. The applied strain in these experiments was 1.4 ± 0.5. (B) Decay constants were obtained from fitting data of the individual trials in A. The data were fit with a double exponential decay, and the decay constants shown are for the faster relaxation process. (C) Relaxation moduli comparing the adhesiveness of UTI89, UTI89∆bcsA, and UTI89∆csgA to bladder cells. The applied strain was 1.4 ± 0.5. (D) Decay constants were obtained and fit as in B. All error bars represent the SD of three trials.
We next sought to investigate the potential influence of pEtN cellulose in bladder cell adhesion using the LCMR platform. UTI89, UTI89∆csgA, and UTI89∆bcsA were each grown in YESCA supplemented with DMSO and prepared on top plates for the LCMR measurements. As shown in Fig. 4C, UTI89, expressing both curli and pEtN cellulose, exhibited the highest relaxation modulus. UTI89∆csgA, lacking curli, had a much lower relaxation modulus than UTI89 over the entire measurement time, indicating that loss of curli resulted in a reduction in adhesion. This contribution of curli to bladder cell adhesion was consistent with results from the traditional adhesion assay. For experiments with UTI89∆bcsA, lacking pEtN cellulose, the maximum relaxation modulus at time zero was intermediate between that of UTI89 and UTI89∆csgA. At later times (6 s to 10 s), the relaxation modulus of UTI89∆bcsA equaled that of UTI89∆csgA. This indicated that cells lacking pEtN cellulose exhibited some initial adhesion capacity (higher initial adhesion than UTI89∆csgA) but failed to maintain strong bladder cell adhesion after several seconds. These observations differed from the traditional adhesion assay, which was unable to differentiate bladder cell adhesion between UTI89 and UTI89∆bcsA.
Thus, we considered whether the LCMR data could provide further insight through examination of time-dependent processes occurring during the experiment. We discovered that the relaxation moduli for all experiments fit well to a double exponential decay model of the form y = Ae−t/α+ Be−t/β + C. The slower decay time, α, was similar for every bacterial strain and growth condition (∼15 s). In contrast, the faster decay time, β, was dependent on the bacterial strain. For experiments comparing type 1 piliated to curliated bacteria, values of β were 1.3 s to 1.5 s and did not vary significantly between conditions (Fig. 4B). However, for experiments examining the roles of curli and pEtN cellulose, the values of β were 0.5 s to 0.6 s for the two mutants (UTI89∆csgA and UTI89∆bcsA) and 1.5 s for UTI89 (Fig. 4D). We attribute the similar slow decay time, α, to the internal bladder cell mechanical response, which should be consistent between experiments. We attribute the different values for the faster decay time, β, to differences in detachment dynamics. This indicates that, although UTI89∆bcsA may have higher initial adhesion to the bladder cells than UTI89∆csgA, both detach more rapidly from the bladder cells than UTI89, which expresses both curli and pEtN cellulose.
pEtN Cellulose Enhances Bacterial Surface Association of Curli.
Based on the above results, we hypothesized that the role of pEtN cellulose in enhancing bladder cell adhesion could be to improve the association of curli with the bacterial cell surface. That is, a weaker association between curli and the bacterial cell surface could result in reduced curli-mediated adhesion to a bladder cell. This hypothesis was supported by the LCMR data: UTI89 and UTI89∆bcsA produce the same amount of curli per cell (Fig. 1B), yet UTI89∆bcsA exhibited rapid detachment after shearing.
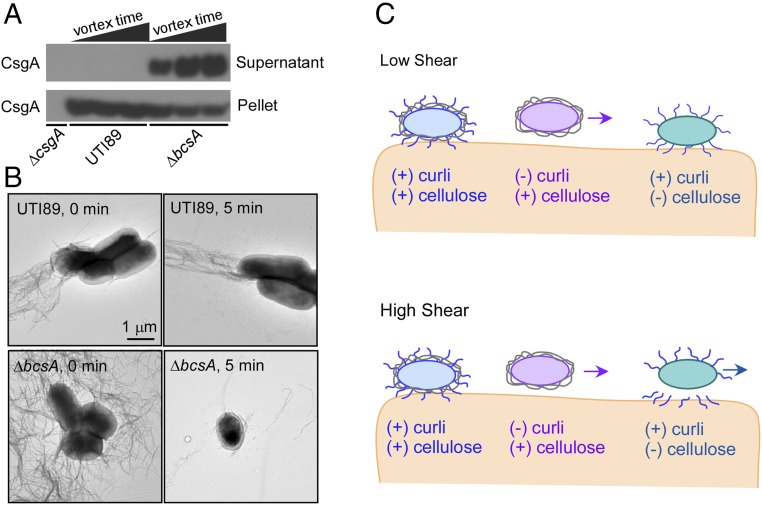
To test this hypothesis, we designed an experiment to determine the extent to which curli are released from the bacterial cell surface when subjected to shearing forces in suspension. We mechanically perturbed bacterial cells through vortexing and, after pelleting the cells by centrifugation, performed a Western blot to detect curli CsgA protein in both the bacterial cell pellets and supernatants. Curli remained associated with UTI89 even after vortexing and were only detected in the cell pellet (Fig. 5A). In contrast, fewer curli were found in the UTI89∆bcsA cell pellet and more were detected in the supernatant as a function of increasing vortex time. Thus, curli remain cell-associated for pEtN cellulose-producing bacteria upon mechanical perturbation, but are less tightly cell-associated in the absence of cellulose. Electron microscopy confirmed the dramatic loss of curli from the bacterial cell surface in the absence of cellulose (Fig. 5B).
Fig. 5.
Effect of pEtN cellulose production on curli association with bacterial cells. (A) Western blot analysis of curli abundance in bacterial cell pellets versus supernatant as evaluated by detection of the major curli subunit CsgA. Cellular suspensions of UTI89 and UTI89∆bcsA were vortexed for increasing times of 10 s, 1 min, or 5 min, and then separated into cell pellet and supernatant samples by centrifugation. Curli remained cell-associated in UTI89, in which pEtN cellulose is coproduced with curli. Curli were released from cells in the absence of pEtN cellulose production (UTI89∆bcsA). (B) TEM images of bacterial cells vortexed for either 0 min or 5 min. (C) Schematic summary of the potential roles of curli and pEtN cellulose in bladder cell adhesion under conditions of low and high shear.
Discussion
Recent and emerging discoveries with UPEC and UTI underscore the necessity to consider the role of curli in host pathogenesis. Specific molecular studies have identified a fitness advantage for curliated bacteria and indicate that curli are immunogenic and elicit immune activation in the host. Moreover, transcriptomics data indicate that curli genes are expressed in vivo (54). We developed the LCMR as a unique measurement tool to define the adhesion strength between bladder cell monolayers and isolated bacterial fibers or intact bacteria. Our results demonstrate the significant contribution that curli can make in mediating bladder cell adhesion and reveal a specific functional role for pEtN cellulose in promoting curli-mediated adhesion, particularly under shear forces, which are relevant in the bladder niche. Our experiments were performed with a commonly employed immortalized cell line, and future work could employ primary bladder facet cells integrated with animal models of UTI pathogenesis to consider possible differences in the presentation of mannosylated cell surface receptors that could influence type 1 pilus mediated adhesion, for example.
As illustrated in Fig. 5C, our collective LCMR and independent biochemical data support a fundamental model in which curli can serve as adhesive fibers and mediate adhesion to bladder cells, but also require pEtN cellulose for tight association with the bacterial cell surface to maintain this adhesion. Conventional tissue culture adhesion assays are characterized by the undefined and low-shear forces of gentle washes and did not reveal a clear role for pEtN cellulose. However, experiments with our customized LCMR allow for quantitative strength of adhesion measurements by applying systematically controlled strains onto the adhesive layer. This revealed a clear role for pEtN cellulose production in mediating bladder cell adhesion by UPEC. Our model is supported by the time-dependent data obtained from the LCMR measurements in which the pEtN cellulose mutant, UTI89∆bcsA, exhibited initial adhesion but detached rapidly over time, yielding a final relaxation modulus and reduction in adhesion that was comparable to the curli mutant, UTI89∆csgA. The model is further supported by an independent biochemical experiment that revealed more facile detachment and dissociation of curli from the UTI89∆bcsA cell surface than from pEtN cellulose-expressing UTI89.
Collectively, pEtN cellulose acts as a “glue” or mortar-like scaffold to maintain curli association with the bacterial cell surface. This molecular function of the combined contributions of pEtN cellulose and curli identifies a potentially important role for pEtN cellulose in UPEC adhesion and UTI pathogenesis. Furthermore, the LCMR approach introduced here can be implemented broadly to examine interactions at the host−pathogen interface.
Materials and Methods
LCMR design and implementation, together with cell growth conditions, adhesion assays, and other protocols, are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. Michael Hsieh for providing bladder epithelial cells; Prof. Matthew Chapman for the UTI89ΔbcsA strain; Prof. David Hunstad for the UTI89ΔfimH strain; the Physics Machine Shop and Crystal Shop at Stanford for helpful discussion and fabrication; and the Cell Sciences Imaging Facility for TEM assistance. E.C.H. and A.A. are recipients of fellowships from the Center for Molecular Analysis and Design at Stanford. E.C.H. is the recipient of a fellowship from the Achievement Rewards for College Scientists Foundation. L.C. acknowledges support by the National Science Foundation Career Award 1453247.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801564115/-/DCSupplemental.
References
- 1.Stamm WE, Norrby SR. Urinary tract infections: Disease panorama and challenges. J Infect Dis. 2001;183(Suppl 1):S1–S4. doi: 10.1086/318850. [DOI] [PubMed] [Google Scholar]
- 2.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foxman B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Am J Med. 2002;113(Suppl 1A):5S–13S. doi: 10.1016/s0002-9343(02)01054-9. [DOI] [PubMed] [Google Scholar]
- 4.Kurtaran B, et al. Antibiotic resistance in community-acquired urinary tract infections: Prevalence and risk factors. Med Sci Monit. 2010;16:CR246–CR251. [PubMed] [Google Scholar]
- 5.Pallett A, Hand K. Complicated urinary tract infections: Practical solutions for the treatment of multiresistant Gram-negative bacteria. J Antimicrob Chemother. 2010;65(Suppl 3):iii25–iii33. doi: 10.1093/jac/dkq298. [DOI] [PubMed] [Google Scholar]
- 6.Bryce A, et al. Global prevalence of antibiotic resistance in paediatric urinary tract infections caused by Escherichia coli and association with routine use of antibiotics in primary care: Systematic review and meta-analysis. BMJ. 2016;352:i939. doi: 10.1136/bmj.i939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta K, Hooton TM, Stamm WE. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann Intern Med. 2001;135:41–50. doi: 10.7326/0003-4819-135-1-200107030-00012. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien VP, Hannan TJ, Nielsen HV, Hultgren SJ. Drug and vaccine development for the treatment and prevention of urinary tract infections. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.UTI-0013-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. 2010;7:653–660. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- 11.Mulvey MA, et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 12.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X-R, Sun T-T, Medina JJ. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: Relation to urinary tract infections. Proc Natl Acad Sci USA. 1996;93:9630–9635. doi: 10.1073/pnas.93.18.9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson GG, et al. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 15.Justice SS, et al. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci USA. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- 17.Blango MG, Ott EM, Erman A, Veranic P, Mulvey MA. Forced resurgence and targeting of intracellular uropathogenic Escherichia coli reservoirs. PLoS One. 2014;9:e93327–e93329. doi: 10.1371/journal.pone.0093327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mysorekar IU, Hultgren SJ. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc Natl Acad Sci USA. 2006;103:14170–14175. doi: 10.1073/pnas.0602136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eto DS, Sundsbak JL, Mulvey MA. Actin-gated intracellular growth and resurgence of uropathogenic Escherichia coli. Cell Microbiol. 2006;8:704–717. doi: 10.1111/j.1462-5822.2006.00691.x. [DOI] [PubMed] [Google Scholar]
- 20.Lim JY, Pinkner JS, Cegelski L. Community behavior and amyloid-associated phenotypes among a panel of uropathogenic E. coli. Biochem Biophys Res Commun. 2014;443:345–350. doi: 10.1016/j.bbrc.2013.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cegelski L, et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kai-Larsen Y, et al. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010;6:e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tükel C, et al. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol. 2005;58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- 25.Tükel C, et al. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol. 2010;12:1495–1505. doi: 10.1111/j.1462-5822.2010.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter AM, Povolotsky TL, Wieler LH, Hengge R. Cyclic-di-GMP signalling and biofilm-related properties of the Shiga toxin-producing 2011 German outbreak Escherichia coli O104:H4. EMBO Mol Med. 2014;6:1622–1637. doi: 10.15252/emmm.201404309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 28.Hung C, Marschall J, Burnham C-AD, Byun AS, Henderson JP. The bacterial amyloid curli is associated with urinary source bloodstream infection. PLoS One. 2014;9:e86009. doi: 10.1371/journal.pone.0086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garofalo CK, et al. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect Immun. 2007;75:52–60. doi: 10.1128/IAI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maher MC, Lim JY, Gunawan C, Cegelski L. Cell-based high-throughput screening identifies rifapentine as an inhibitor of amyloid and biofilm formation in Escherichia coli. ACS Infect Dis. 2015;1:460–468. doi: 10.1021/acsinfecdis.5b00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thongsomboon W, et al. Phosphoethanolamine cellulose: A naturally produced chemically modified cellulose. Science. 2018;359:334–338. doi: 10.1126/science.aao4096. [DOI] [PubMed] [Google Scholar]
- 32.Olsén A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 33.Olsén A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 34.Olsén A, Wick MJ, Mörgelin M, Björck L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect Immun. 1998;66:944–949. doi: 10.1128/iai.66.3.944-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, et al. Impact of biofilm matrix components on interaction of commensal Escherichia coli with the gastrointestinal cell line HT-29. Cell Mol Life Sci. 2006;63:2352–2363. doi: 10.1007/s00018-006-6222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saldaña Z, et al. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ Microbiol. 2009;11:992–1006. doi: 10.1111/j.1462-2920.2008.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elkins CM, Qi QM, Fuller GG. Corneal cell adhesion to contact lens hydrogel materials enhanced via tear film protein deposition. PLoS One. 2014;9:e105512. doi: 10.1371/journal.pone.0105512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elkins CM, Shen W-J, Khor VK, Kraemer FB, Fuller GG. Quantification of stromal vascular cell mechanics with a linear cell monolayer rheometer. J Rheol. 2015;59:33–50. [Google Scholar]
- 39.Chen SL, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc Natl Acad Sci USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hultgren SJ, Schwan WR, Schaeffer AJ, Duncan JL. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect Immun. 1986;54:613–620. doi: 10.1128/iai.54.3.613-620.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim JY, May JM, Cegelski L. Dimethyl sulfoxide and ethanol elicit increased amyloid biogenesis and amyloid-integrated biofilm formation in Escherichia coli. Appl Environ Microbiol. 2012;78:3369–3378. doi: 10.1128/AEM.07743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mika F, Hengge R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 2014;11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wellens A, et al. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS One. 2008;3:e2040. doi: 10.1371/journal.pone.0002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo AWH, et al. Suppression of type 1 pilus assembly in uropathogenic Escherichia coli by chemical inhibition of subunit polymerization. J Antimicrob Chemother. 2014;69:1017–1026. doi: 10.1093/jac/dkt467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stærk K, Khandige S, Kolmos HJ, Møller-Jensen J, Andersen TE. Uropathogenic Escherichia coli express type 1 fimbriae only in surface adherent populations under physiological growth conditions. J Infect Dis. 2016;213:386–394. doi: 10.1093/infdis/jiv422. [DOI] [PubMed] [Google Scholar]
- 46.Camesano TA, Liu Y, Datta M. Measuring bacterial adhesion at environmental interfaces with single-cell and single-molecule techniques. Adv Water Resour. 2007;30:1470–1491. [Google Scholar]
- 47.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lekka M, et al. Elasticity of normal and cancerous human bladder cells studied by scanning force microscopy. Eur Biophys J. 1999;28:312–316. doi: 10.1007/s002490050213. [DOI] [PubMed] [Google Scholar]
- 49.Ramos JR, Pabijan J, Garcia R, Lekka M. The softening of human bladder cancer cells happens at an early stage of the malignancy process. Beilstein J Nanotechnol. 2014;5:447–457. doi: 10.3762/bjnano.5.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada S, Wirtz D, Kuo SC. Mechanics of living cells measured by laser tracking microrheology. Biophys J. 2000;78:1736–1747. doi: 10.1016/S0006-3495(00)76725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alcaraz J, et al. Microrheology of human lung epithelial cells measured by atomic force microscopy. Biophys J. 2003;84:2071–2079. doi: 10.1016/S0006-3495(03)75014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laurent VM, et al. Partitioning of cortical and deep cytoskeleton responses from transient magnetic bead twisting. Ann Biomed Eng. 2003;31:1263–1278. doi: 10.1114/1.1616932. [DOI] [PubMed] [Google Scholar]
- 53.Hoffman BD, Massiera G, Van Citters KM, Crocker JC. The consensus mechanics of cultured mammalian cells. Proc Natl Acad Sci USA. 2006;103:10259–10264. doi: 10.1073/pnas.0510348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subashchandrabose S, et al. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc Natl Acad Sci USA. 2014;111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.