Significance
Neural development is essential for the formation of neuronal networks and brain function. cTAGE5/MEA6 plays a critical role in the secretion of proteins, including very low density lipoprotein and insulin; however, its role in the transport of cellular (nonsecretory) components and in brain development has not been previously explored. Here, we show that cTAGE5/MEA6 is essential for neural development, and knockout of cTAGE5/MEA6 in the mouse brain leads to severe neural developmental defects. The underlying mechanisms for the role of cTAGE5/MEA6 in brain development have been explored in this study. Interestingly, mutations in cTAGE5/MEA6 have been found in patients with Fahr’s disease. Thus, our study also provides insight into the possible pathogenesis mechanisms of this neurological disorder.
Keywords: cTAGE5, MEA6, brain development, COPII, vesicle trafficking
Abstract
Normal neural development is essential for the formation of neuronal networks and brain function. Cutaneous T cell lymphoma-associated antigen 5 (cTAGE5)/meningioma expressed antigen 6 (MEA6) plays a critical role in the secretion of proteins. However, its roles in the transport of nonsecretory cellular components and in brain development remain unknown. Here, we show that cTAGE5/MEA6 is important for brain development and function. Conditional knockout of cTAGE5/MEA6 in the brain leads to severe defects in neural development, including deficits in dendrite outgrowth and branching, spine formation and maintenance, astrocyte activation, and abnormal behaviors. We reveal that loss of cTAGE5/MEA6 affects the interaction between the coat protein complex II (COPII) components, SAR1 and SEC23, leading to persistent activation of SAR1 and defects in COPII vesicle formation and transport from the endoplasmic reticulum to the Golgi, as well as disturbed trafficking of membrane components in neurons. These defects affect not only the transport of materials required for the development of dendrites and spines but also the signaling pathways required for neuronal development. Because mutations in cTAGE5/MEA6 have been found in patients with Fahr’s disease, our study potentially also provides insight into the pathogenesis of this disorder.
During neural development, along with the outgrowth of dendrites and axons, neurons form synaptic connections with other neurons to construct neural circuits and networks (1–4). During these processes, neuronal morphology increases in complexity, with cell membrane areas expanding thousands of folds, requiring the synthesis and trafficking of large amounts of proteins and lipids (4–7).
In general, following protein synthesis, trafficking of membrane proteins and secretory proteins from the endoplasmic reticulum (ER) to the Golgi apparatus is mediated by the coat protein complex II (COPII) vesicles (8–10). SAR1, a small GTPase belonging to the Ras family, is the key factor mediating the formation of COPII vesicles. Specifically, the exchange of SAR1 from its GDP-bound form to its GTP-bound form, is mediated by guanine nucleotide exchange factor (GEF)–SEC12, leading to binding of SAR1-GTP to the ER membrane. SAR1-GTP then recruits the COPII inner membrane protein, SEC23-SEC24, to form SAR1-GTP/SEC23/SEC24 prebudding vesicles (11–14). SEC23 is a GTPase activating protein (GAP), which activates the GTPase activity of SAR1 to promote the exchange of SAR1-GTP to SAR1-GDP, while SEC24 functions as the cargo receptor for the recognition of different substances in transit from the ER to the Golgi apparatus (15, 16). Subsequently, the COPII outermembrane protein, SEC31/SEC13 dimer, is recruited to form functioning COPII vesicles. These COPII vesicles then detach from ER at the ER exit site (ERES) and translocate to transport compartments. The levels of SAR1-GTP and SAR1-GDP are strictly controlled in cells, as overexpression of either dominant-negative form or constitutively active form would affect ER export and Golgi apparatus function (17, 18).
Cutaneous T cell lymphoma-associated antigen 5 (cTAGE5), also known as meningioma-expressed antigen 6 (MEA6), was found to be up-regulated in different tumor tissues and cell lines (19–21). cTAGE5/MEA6 has several different splicing isoforms, and its main structural domains contain a single transmembrane domain, two coiled-coil domains, and a proline-rich domain (22). cTAGE5/MEA6 is expressed at much higher levels in organs with secretory functions, including the liver and pancreas, and at more moderate levels in the brain, spleen, lung, intestine, and muscle (22, 23). cTAGE5/MEA6 has been shown to localize in the ER exit site, and interact with TANGO1 and SEC12 to take part in the regulation of collagen secretion (24, 25). Moreover, cTAGE5/MEA6 was also found to regulate the assembly of the COPII complex and to regulate the transport and secretion of very low density lipoprotein (VLDL) in the liver, as well as to interact and regulate the localization of SEC22, thus ultimately playing an important role in insulin secretion from the pancreas (22, 23). Notably, SEC22b has been shown to form a trans-SNARE complex with syntaxin1 and has a conserved function in plasma membrane expansion (26). Although the function of cTAGE5/MEA6 in protein secretion is known, its possible involvement in the transport of cellular (nonsecretory) components of neurons and in brain development, as well as the underlying molecular mechanisms involved in these processes have not been explored.
Here, we generated a mouse model with conditional knockout of cTAGE5/MEA6 in the brain to study the specific roles of cTAGE5/MEA6 in brain development. We found that knockout of cTAGE5/MEA6 in the brain leads to severe developmental defects including movement disorders. Results indicate that loss of cTAGE5/MEA6 leads to defects in COPII vesicle formation and in the transport of proteins and lipids from the ER to the Golgi apparatus in neurons. These defects not only affect the development and maintenance of dendrites and spines, but also affect the signaling pathways essential for neuronal development.
Results
Conditional Knockout of cTAGE5/MEA6 in the Brain Leads to Severe Developmental Defects.
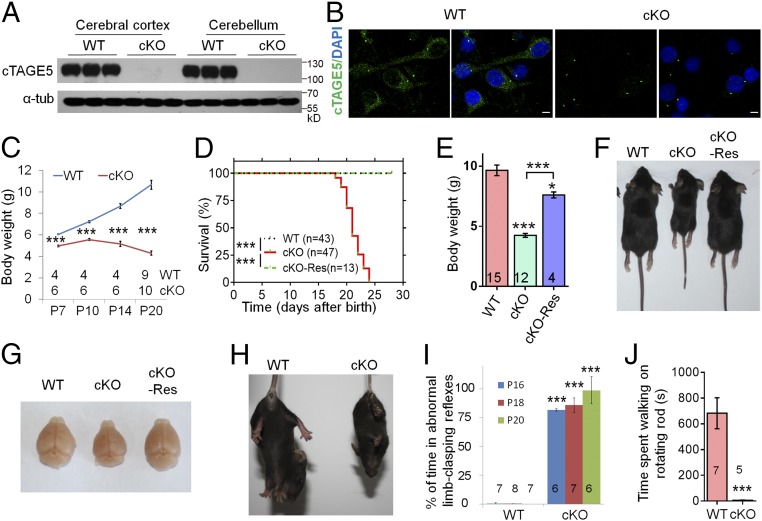
To study the specific function of cTAGE5/MEA6 in the brain, we generated conditional knockout (cKO) of cTAGE5/MEA6 in the mouse brain by mating cTAGE5F/F mice with Nestin-Cre transgenic mice. Because cTAGE5/MEA6 is located on chromosome 12 and was cross-linked with Nestin-Cre, cTAGE5F, Nestin-Cre//cTAGE5+ mice were mated with cTAGE5F/F mice to generate cTAGE5F/+ mice (WT mice) and cTAGE5F, Nestin-Cre//cTAGE5F mice (cTAGE5/MEA6 cKO mice). Both Western blot and real-time PCR analyses indicated that cTAGE5/MEA6 was efficiently knocked out in postnatal day 20 (P20) cKO mouse brains at both the protein and mRNA level, respectively (Fig. 1A and SI Appendix, Fig. S1A), but not in other tissues, including the heart, liver, and kidney, despite a subtle decrease in cTAGE5/MEA6 expression in the heart (SI Appendix, Fig. S1B). Moreover, immunostaining analysis revealed the near complete loss of cTAGE5/MEA6 in cKO neurons, although the signal at the centrosome remained, and was possibly due to the nonspecific signaling of the cTAGE5/MEA6 antibody (Fig. 1B and SI Appendix, Fig. S1C).
Fig. 1.
cTAGE5 cKO in the brain leads to severe defects in development, which can be rescued by transgenic expression of human FLAG-cTAGE5. (A) Western blotting results showing the knockout of cTAGE5 in the cerebral cortex and cerebellum of Nestin-cKO mice at P20. Three independent replicates were performed. α-Tub was used as loading control. (B) Primary neurons at 14 div dissected from around E15.5 WT and cKO brains were stained with cTAGE5 antibody. (Scale bars, 5 μm.) Nucleus was stained with DAPI. (C) Body weight of WT and cKO mice from P7 to P20. t test; ***P < 0.001. (D) Postnatal survival curve of WT, cKO, and cKO-Res mice. One-way ANOVA; ***P < 0.001. (E) Measurement of body weight of WT, cKO, and cKO-Res mice at P20. One-way ANOVA; **P < 0.01; ***P < 0.001. (F and G) Images of the bodies (F) and brains (G) of the WT, cKO and cKO-Res mice at P20. (H and I) cKO mice showing abnormal limb clasping reflexes in tail suspension test. t test; ***P < 0.001. (J) The time of WT and cKO mice at P16 spent on the rotating rod (10 rotations per min) during Rota-rod test. t test; ***P < 0.001. Numbers of mice tested are labeled in C–E, I, and J.
Compared with WT mice, cKO mice showed severe growth defects. The body weight of cKO mice decreased gradually after P10, with all cKO mice dying before P25 (Fig. 1 C and D). Indeed, cKO mice at P20 had significantly smaller brain and body sizes than WT mice, suggesting an essential role of cTAGE5/MEA6 in development (SI Appendix, Fig. S2 A and B). Although cTAGE5/MEA6 was reported to form a hybrid gene (Tali) with its upstream gene (Mia2), we confirmed with RT-PCR analysis that Tali is not expressed in the brain (SI Appendix, Fig. S2C) (27, 28). To further demonstrate that the severe growth defects in cKO mice were due to cTAGE5/MEA6 deletion exclusively in the brain, we constructed a human FLAG-cTAGE5 conditional transgenic mouse. We crossed FLAG-hcTAGE5 transgenic mice with cTAGE5F, Nestin-Cre//cTAGE5+ mice to generate cTAGE5F, Nestin-Cre//cTAGE5F, FLAG-hcTAGE5 mice (cKO-Res mouse). The expression of full-length human FLAG-cTAGE5 (809 aa), which has a larger molecular weight than endogenous mouse brain cTAGE5/MEA6 (772 aa for the full-length protein), was detected in the brains of FLAG-hcTAGE5 transgenic mice and cKO-Res mice (SI Appendix, Fig. S2 D and E). Surprisingly, the expression of FLAG-hcTAGE5 suppressed the expression of endogenous cTAGE5/MEA6 in the brain, implying that cTAGE5/MEA6 level is strictly controlled in the brain (SI Appendix, Fig. S2D). More importantly, the postnatal lethality and developmental defects, including reduced brain and body sizes of cKO mice, could be rescued by transgenic expression of FLAG-hcTAGE5 (Fig. 1 D–G).
In addition, cKO mice also showed obvious abnormal limb-clasping reflexes in the tail suspension test and spent dramatically less time on the rotating rod in the rotarod test, which are typical behavioral phenotypes involving neurodegeneration (Fig. 1 H–J) (29–31). Interestingly, compared with WT mice, cKO mice had higher movement activity in the open-field test, with longer moving distances, faster speeds, longer movement durations, and shorter rest durations (SI Appendix, Fig. S3). These results demonstrate that cTAGE5/MEA6 plays an important role in brain development.
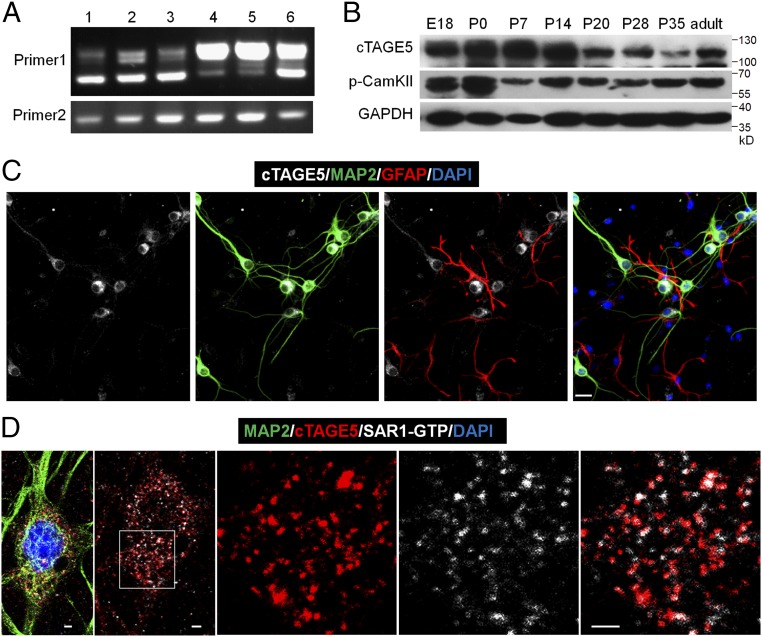
Because cTAGE5/MEA6 is important for brain development, we determined its expression in detail in the mouse brain. We also analyzed whether a cTAGE5/MEA6 isoform containing a ∼99-nt fragment alternatively spliced in the N terminus was present (SI Appendix, Fig. S4A). RT-PCR and Western blotting analyses showed that cTAGE5/MEA6 is primarily expressed as the shorter splicing isoform in the brain compared with other tissues such as the meninges, liver, and kidney, where it is primarily expressed as the longer splicing isoform (Fig. 2A and SI Appendix, Fig. S4B). This implies that cTAGE5/MEA6 may have a unique function in the brain. Moreover, cTAGE5/MEA6 is expressed in the cerebral cortex at different developmental stages, with relatively higher expression levels from around birth to P14 (Fig. 2B). We also inspected cellular subtypes expressing cTAGE5/MEA6 in the brain. Immunostaining results indicated that cTAGE5/MEA6 is highly expressed in neurons, but not in astrocytes (Fig. 2C). Consistent with previous observations (22, 24), cTAGE5/MEA6 is primarily located at the ERES where it colocalizes with SAR1-GTP (Fig. 2D); cTAGE5/MEA6 expression was also observed in the ERGIC of neurons, although to a lesser extent (SI Appendix, Fig. S4C).
Fig. 2.
cTAGE5 is expressed in neuron at the ER exit sites. (A) RT-PCR results showing different splicing isoforms in different tissues during the embryonic stages. The forward primer and reverse primer of “Primer 1” flank the spliced intron of cTAGE5, while “Primer 2” does not. 1: E14 brain; 2: E18 brain; 3: E14 primary neuron; 4: E18 kidney; 5: E18 liver; 6: E14 meninge. (B) Western blotting results showing the expression levels of cTAGE5 and p-CamKII (phospho T286) in the cerebral cortex at different developmental stages. GAPDH was used as loading control. (C and D) Primary neurons (14 div) dissected from E15.5 WT mouse cerebral cortex were stained with cTAGE5 (gray), MAP2 (green), and GFAP (red) antibodies (C); or cTAGE5, SAR1-GTP and MAP2 antibodies (D). (Scale bars, 20 μm in C and 2 μm in D.) Nucleus was stained with DAPI.
cTAGE5/MEA6 cKO Impairs Dendritic and Synaptic Structure, as Well as Neuronal Function.
To decipher why cTAGE5/MEA6 cKO led to smaller brain size and also the underlying molecular mechanism regulating this outcome, we inspected whether loss of cTAGE5/MEA6 affected the proliferation of neuronal progenitor cells (NPC). cTAGE5/MEA6 deletion had no effect on the diameter of neurospheres (SI Appendix, Fig. S5 A and B). In addition, pregnant mice were intraperitoneally injected with BrdU at embryonic day 14.5 (E14.5) and inspected 24 h later. We found that cTAGE5/MEA6 cKO had no significant effect on the distribution and density of BrdU+ cell in the cerebral cortex or on the cell cycle exit index (percentage of Ki67−/BrdU+ cells among BrdU+ cells) (SI Appendix, Fig. S5 C–F). Therefore, cTAGE5/MEA6 cKO does not significantly affect the proliferation and differentiation of NPCs. Moreover, immunostaining with the activated form of Caspase 3 antibody showed that cTAGE5/MEA6 cKO did not cause significant apoptosis at P20 (SI Appendix, Fig. S5G).
We analyzed the cell composition of the cerebral cortex and found that there were no significant differences in the numbers of NeuN+ and Parvalbumin+ (PV+) cells in the same width (400 µm) of the cortex between the WT and cKO brains at P20. However, cortical neuronal densities in cKO mice were significantly increased compared with those of WT mice (SI Appendix, Fig. S6 A and B). In addition, astrocytes were dramatically activated in P20 cKO brains (SI Appendix, Fig. S6 C and D). Finally, compared with control brain, the increased protein levels of GFAP in cKO brain at P20 were also confirmed by Western blotting (SI Appendix, Fig. S6E).
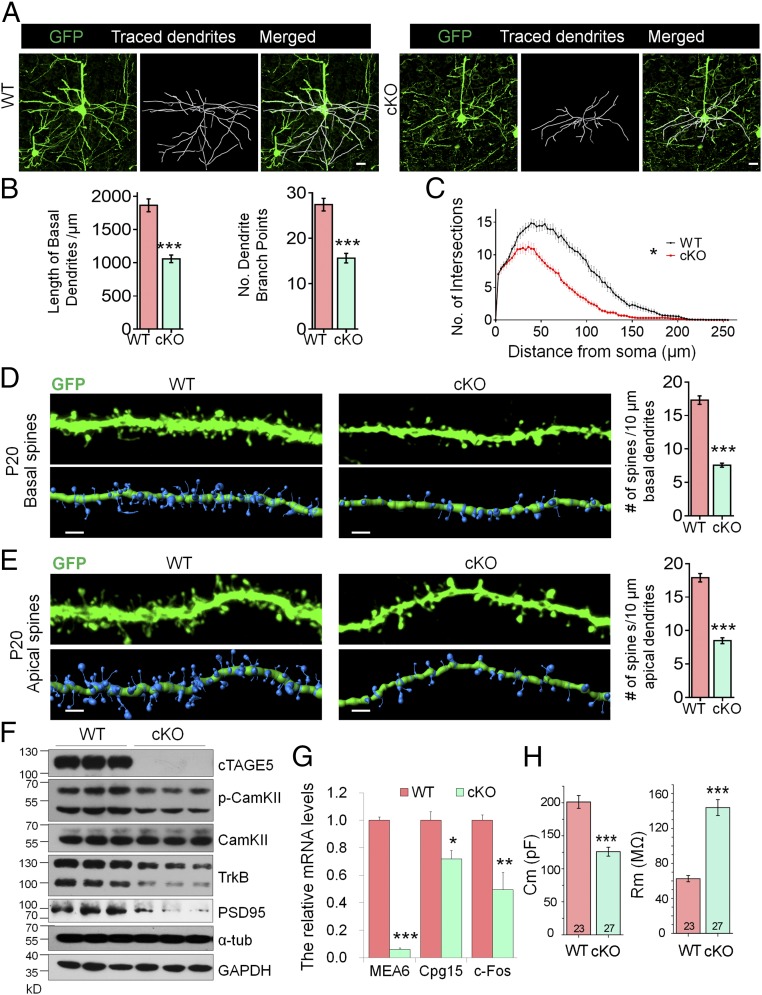
During neural development, dendritic and axonal outgrowth increase both the membrane area and volume of neurons. Thus, the increased neuronal density in the cerebral cortices of cKO mice may be due to defects in dendritic and axonal outgrowth or decrease in overall volume (SI Appendix, Fig. S6 A and B). To validate this hypothesis, we crossed Thy1-GFPm transgenic mice with cTAGE5F, Nestin-Cre//cTAGE5+ mice to enable us to label pyramidal neurons with GFPm. Within the motor cortex of cKO mice at P20, pyramidal neuron basal dendritic outgrowth was impaired compared with that of WT mice (Fig. 3A). Quantification analysis showed that the total length of basal dendrites, and the numbers of dendrite branch points in pyramidal neuron dendrites in cKO brains were significantly reduced at P20 compared with dendrites in WT brains (Fig. 3B). Moreover, Sholl analysis indicated significant decreases in dendritic complexity in cKO brains (Fig. 3C). Similar but less significant defects in basal dendrites of cKO pyramidal neurons were also detected at P14 (SI Appendix, Fig. S7 A and B). In addition, we inspected whether dendritic outgrowth was also affected in cultured primary neurons dissected from the WT and cKO mouse brain, and the similar results indicated that dendritic outgrowth was also impaired in primary cultured cTAGE5/MEA6 cKO neurons at 7 and 14 d in vitro (div), although the difference at 7 div was not as dramatic as that at 14 div (SI Appendix, Fig. S7 C–F). Besides the dendrite growth, we also investigated the effects of cTAGE5/MEA6 cKO in axonal growth, considering that cTAGE5/MEA6 was already knocked out at early developmental stages when axons are growing (SI Appendix, Fig. S8A). However, as shown in SI Appendix, Fig. S8 B and C, cTAGE5/MEA6 cKO did not significantly affect the expression of axonal protein (NF-200) in the P20 cortex and axonal growth in cultured neurons at 4 div. Taken together, these results show that cTAGE5/MEA6 plays a critical role in the outgrowth and maintenance of dendrites. Furthermore, these impaired dendritic structural features lend support to the idea that cTAGE5/MEA6 cKO mice display phenotypes of developmental defects.
Fig. 3.
cTAGE5 cKO impairs formation of dendrites and spines, and neuronal function. (A) Representative images of Thy1-GFPm labeled pyramidal neurons in WT and cKO motor cortex at P20. The basal dendrites of pyramidal neurons were reconstructed using the “filament” function of Imaris software. (Scale bars, 20 μm.) (B) The total basal dendrite length and number of dendrite branch points in A were analyzed. (C) The basal dendritic complexity in A was analyzed by Sholl analysis with Imaris software. t test (Kolmogorov–Smirnov test for Sholl analysis); *P < 0.05; ***P < 0.001. WT: n = 21/5; cKO: n = 19/5. (n, no. of neurons analyzed/no. of mice analyzed). (D and E) Representative images of basal (D) and apical (E) dendrites of Thy1-GFPm labeled pyramidal neurons in WT or cKO cerebral cortex at P20. Spine density was defined as the number of spines along 10-μm secondary dendrites and analyzed with the Imaris Software. t test; ***P < 0.001. Basal (WT: n = 40/5; cKO: n = 45/5); apical (WT: n = 24/5; cKO: n = 28/5); (n, no. of dendrites analyzed/no. of mice analyzed). (Scale bars, 2 μm.) (F) The protein levels of cTAGE5, p-CamKII, CamKII, TrkB, and PSD95 in the WT and cKO cerebral cortex at P20 were analyzed by Western blotting. Three independent replicates were performed. α-Tub and GAPDH were used as loading control. (G) The mRNA levels of cTAGE5, Cpg15 and c-Fos in the WT and cKO cerebral cortex at P20 were determined with real-time PCR. β-Actin was used as loading control. n = 4. *P < 0.05; **P < 0.01; ***P < 0.001. (H) The membrane resistance and membrane capacitance of layer II/III pyramidal neurons in the motor cortex of WT and cKO mouse at P17-P23. t test; ***P < 0.001. WT: n = 23/3; cKO: n = 27/3. (n, no. of neurons analyzed/no. of mice analyzed).
In addition to dendrites, we carefully examined the morphology of dendritic spines and found that the density of both basal and apical dendritic spines of pyramidal neurons in the motor cortex of cKO mouse was substantially lower, compared with that of WT mouse at both P20 and P14 (Fig. 3 D and E and SI Appendix, Fig. S9). Consistently, postsynaptic density protein 95 (PSD95) expression levels were dramatically down-regulated in cKO brains compared with those in WT brains (Fig. 3F). In addition, levels of phosphorylated CamKII, which is activated in the calcium signaling pathway, were also significantly reduced in cKO brains at P20. Moreover, both Cpg15 and c-fos mRNA levels, which are markers for neuronal activation (32, 33), were significantly down-regulated in cKO brains at P20 compared with WT brains (Fig. 3G). These results, together with altered neuronal morphology changes, indicate a down-regulation of neuronal activity in cKO brains. This down-regulation was further supported by electrophysiology assays that revealed that the membrane capacitance and membrane resistance of layers II/III pyramidal neurons in the cKO motor cortex were significantly decreased and increased, respectively, compared with that of WT motor cortices at P17–P23 (Fig. 3H). Moreover, compared with the controls, the frequency of mini-excitatory postsynaptic currents (mEPSC) of layer II/III pyramidal neurons in the cKO mouse motor cortex was significantly lower with no change in the amplitude of mEPSC; whereas the mini-inhibitory postsynaptic currents (mIPSC) of these cKO cells was not affected (SI Appendix, Fig. S10).
Transport of Cellular Components in Neuron Is Affected in cTAGE5/MEA6 cKO Mice.
To decipher the molecular mechanisms involved in cTAGE5/MEA6 cKO-induced defects in dendritic outgrowth and spine formation, we labeled proteins from three cKO and three WT P14 brains with tandem mass tag technology and compared the protein levels using quantitative MS. There were significant trends in both the up- and down-regulation of some proteins in cKO brains, especially those related to the ER and Golgi apparatus structure and function (SI Appendix, Table S1). In particular, several component proteins of the COPI complex (COPz1, COPg2, COPa, COPb2, and COPg1), the COPII complex (SEC31a and SEC16a), as well as VIP36, which shuttles between the ER and Golgi apparatus to regulate glycoprotein sorting and trafficking (34), were significantly down-regulated. Surprisingly, the levels of another essential COPII complex component, SAR1, were up-regulated in cKO brains. Taken together, these results suggest that cTAGE5/MEA6 is involved in protein trafficking between the ER and Golgi apparatus in neurons.
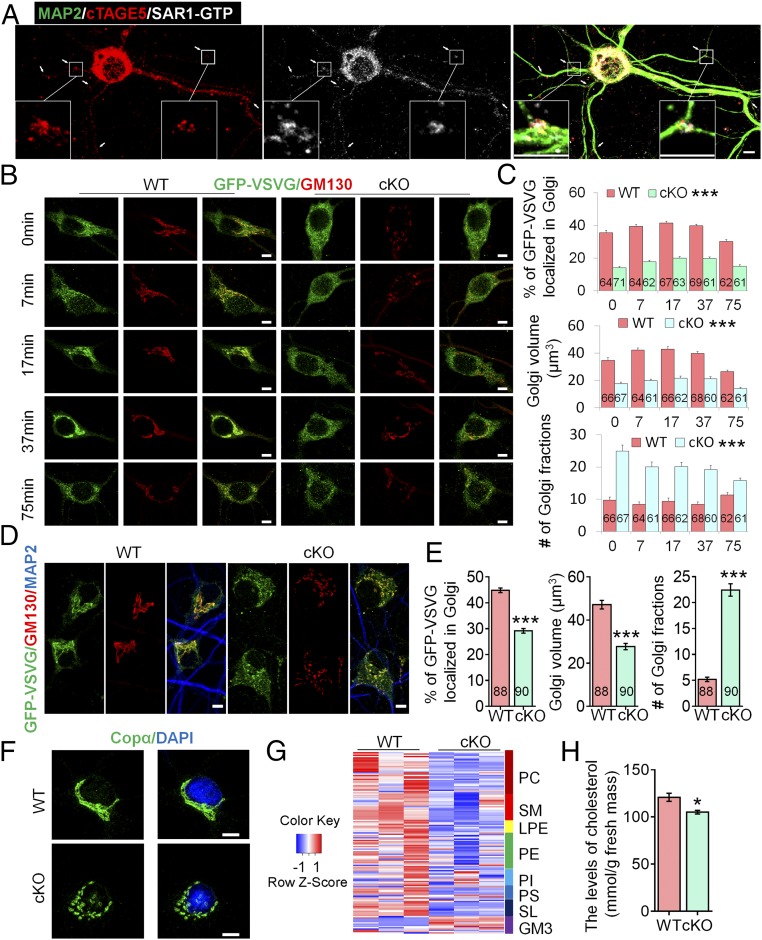
ER complexity is significantly higher in dendritic branch points, where multiple ERES and COPII complexes exist; it is therefore also a site where forward transport of proteins transport is enriched (7, 35). Moreover, dendritic outgrowth were shown to be disrupted when ER complexity was down-regulated by manipulation of CLIMP63, which may affect COPII vesicle formation and thus forward transport of proteins in the dendritic branch points (7). Accordingly, we found that cTAGE5/MEA6, together with SAR1-GTP, were both relatively enriched in the dendritic branch points of pyramidal neurons (Fig. 4A).
Fig. 4.
cTAGE5 cKO affects the transport of GFP-VSVG from the ER to the Golgi apparatus in neuron and lipid composition in brain. (A) Primary neurons at 14 div dissected from around E15.5 WT cerebral cortex were stained with MAP2, cTAGE5, and SAR1-GTP antibodies. Insets are enlarged images of two of the branch points. (B) Primary neurons at 9 div dissected from around E15.5 WT and cKO cerebral cortex were infected with adenovirus-expressing temperature-sensitive GFP-ts045-VSVG, and cultured at 40 °C for 24 h. Then, after the temperature was switched to 32 °C for 0, 7, 17, 37, and 75 min, cells were fixed and stained with GFP and GM130 antibodies. (C) Quantification analysis of the percentage of GFP-VSVG localized in the Golgi, Golgi volume, and number of Golgi fragments at different time points in B. t test; ***P < 0.001. Indicated No. of neurons of 4 WT and cKO mice were analyzed. (D) Primary neurons as in B were cultured at 37 °C for 24 h, then fixed with PFA and stained with GFP, GM130, and MAP2 antibodies. (E) Quantification analysis of percentage of GFP-VSVG localized in the Golgi, Golgi volume, and number of Golgi of neurons in D. t test; ***P < 0.001. Indicated number of neurons of four WT and cKO mice were analyzed. (F) cTAGE5 cKO leads to fragmentation of the Golgi apparatus in neuron. Primary neurons as those in A stained with COPα antibody. Nucleus was stained with DAPI. (G) Heat map results showing the levels of different lipid components in the cerebral cortex of P14 WT and cKO mouse. Every single line represents one subtype of lipid components indicated by the colorful boxes. GM3, monosialodihexosyl-gangliosides; LPE, lyso-PE; PC, phosphatidylcholines; PE, phosphatidylethanolamines; PI, phosphatidylinositols; PS, phosphatidylserines; SL, sulfatides; SM, sphingomyelins. (H) Levels of cholesterol were significantly reduced in P14 cKO cerebral cortex. t test; *P < 0.05. n = 4. (Scale bars, 5 μm in A, B, D, and F.)
To inspect whether cTAGE5/MEA6 cKO affects forward protein transport in neurons, we took advantage of a temperature-sensitive reporter, GFP-ts045-vesicular stomatitis virus G (VSVG), which is arrested in the ER at 40 °C and begins to translocate to the Golgi apparatus at 32 °C (36). WT and cKO primary neurons infected with adenovirus expressing GFP-ts045-VSVG were cultured at 40 °C for 24 h, and then switched to 32 °C for 0, 7, 17, 37, or 75 min, when these cells were fixed with PFA. At 0 min, 35.7 ± 1.4% of GFP-VSVG was localized in the Golgi apparatus (labeled by GM130) of WT neurons (Fig. 4 A and B), implying that GFP-VSVG might not be fully arrested in the ER. In contrast, only 14.1 ± 0.8% of GFP-VSVG was localized in the Golgi apparatus of cKO neurons, indicating that cTAGE5/MEA6 deletion affects the transport of GFP-VSVG from the ER to the Golgi apparatus (Fig. 4 B and C). However, after switching to 32 °C, GFP-VSVG in WT neurons began to translocate and accumulate in the Golgi apparatus to form a clear Golgi-like distribution pattern within 7–37 min, while significantly less GFP-VSVG was translocated to the Golgi apparatus in cKO neurons and remained in a relatively evenly distributed pattern during that period (Fig. 4 B and C). Another obvious difference between WT and cKO neurons was the overall structure of the Golgi apparatus (Fig. 4B). Compared with WT neurons cultured at 32 °C at different time points, Golgi apparatus volume and the number of fragmented Golgi apparatuses in cKO neurons were significantly reduced and increased, respectively (Fig. 4 B and C).
Because primary neurons are sensitive to culture temperature, to exclude the possibility that the phenotype observed at 40 °C was due to the secondary effect of stress conditions, WT and cKO primary neurons infected with adenovirus-expressing GFP-ts045-VSVG were cultured at 37 °C for 24 h. We found also that significantly less GFP-VSVG (29.2 ± 0.9%) in cKO neurons accumulated in the Golgi apparatus compared with that in WT neurons (44.8 ± 0.9%) (Fig. 4 D and E). In addition, Golgi volume was also significantly smaller and number of fragmented Golgi structures increased significantly in cKO neurons (Fig. 4 D and E). This fragmentation was also confirmed by immunostaining with COPα antibody (Fig. 4F). These results suggest that cTAGE5/MEA6 cKO leads to defects in protein transport from the ER to Golgi apparatus, as well as fragmentation of Golgi structure in neurons.
Membrane receptors and ion channels are critical for neuronal function. In addition to the dramatically reduced levels of the brain-derived neurotrophic factor (BDNF) receptor, TrkB (Fig. 3F), we detected the down-regulation of several other membrane receptors and ion channels in cKO brains, including sodium- and chloride-dependent GABA transporter (SLC6A1), voltage-dependent calcium channels (CACNG8 and CACNA2D3), and sodium/potassium-transporting ATPase subunit β1 (ATP1B1) (SI Appendix, Fig. S11).
Disturbance in forward transport from the ER to the Golgi apparatus may also affect the lipid supply from the ER and therefore the lipid composition of neurons (6). We postulated that cTAGE5/MEA6 cKO may also affect the transport of lipid components to the cell membrane during neural development, and resulting in defects in the expansion of the cell membrane, as well as in dendritic outgrowth. Indeed, when the lipid composition of the cKO brain at P14 was analyzed by MS, we found that the levels of many lipid components were significantly down-regulated, including various species of phosphatidylcholines and phosphatidylethanolamines (the major lipid components of cell membrane), Lyso-PE and phosphatidylinositols (the minor components of cell membrane), as well as cell membrane raft-associated lipids including, sphingomyelins and free cholesterols (Fig. 4 G and H and SI Appendix, Fig. S12) (37–39).
cTAGE5/MEA6 cKO Leads to the Deregulation of SEC23, SEC31, and SAR1-GTP Expression in the Brain.
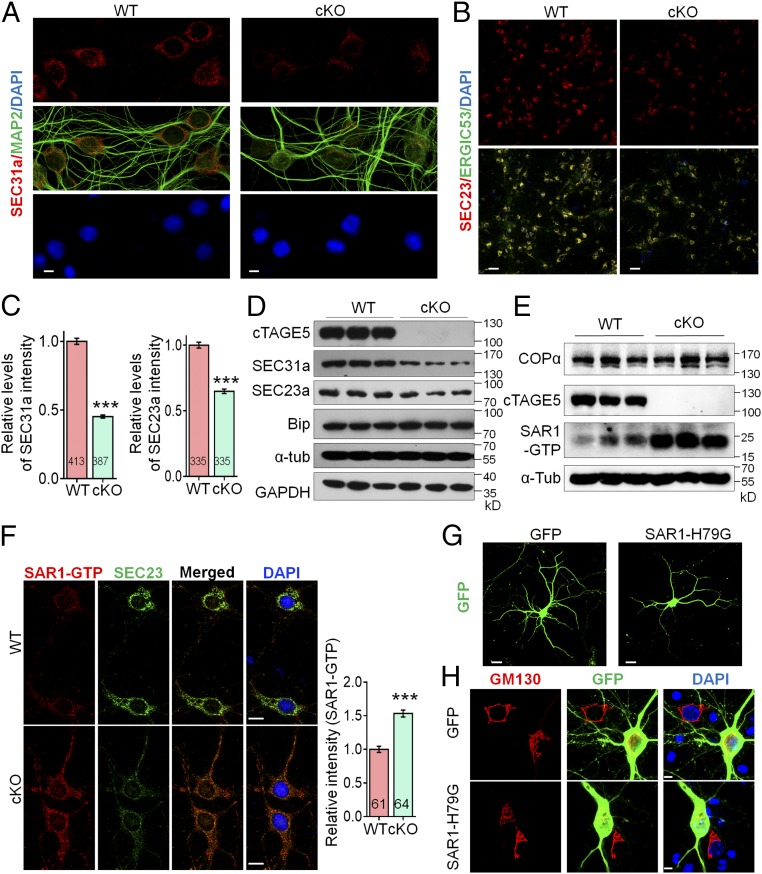
Considering the localization of cTAGE5/MEA6 in the ERES and the down-regulation of COPII components (including SEC31a and SEC16a) (SI Appendix, Table S1) in cKO neurons, the defects in protein transport from the ER to the Golgi apparatus in these neurons may be due to defects in the formation of COPII vesicles. We therefore immunostained markers associated with COPII vesicles, including SEC31a and SEC23a, and found that their intensities in primary cKO neurons were only 45.2% and 64.9% of their respective intensities found in WT neurons (Fig. 5 A–C). The significant down-regulation of SEC31a and SEC23a were confirmed by Western blotting results of P20 cKO cerebral cortices (Fig. 5D). Interestingly, Sec31a mRNA levels in cKO brains were significantly up-regulated (SI Appendix, Fig. S13A), which may be in response to a feedback loop to compensate for decreased numbers of COPII vesicles.
Fig. 5.
cTAGE5 cKO leads to aberrant expression of COPII components. (A and B) Primary neurons at 14 div dissected from around E15.5 WT and cKO brains were stained with SEC31a and MAP2 antibodies (A) or SEC23a and ERGIC53 (B). (C) Quantification relative staining intensity of SEC31a in A and SEC23a in B. t test; ***P < 0.001. Indicated number of neurons from five (for SEC31a) or three (for SEC23a) WT and cKO mice were analyzed. (Scale bars, 5 μm in A, 50 μm in B.) (D and E) The protein levels of SEC31a, SEC23a, and Bip (D) or COPα and SAR1-GTP (E) in the cerebral cortex of P20 WT and cKO mice were determined with Western blotting. α-Tub or GAPDH were used as loading control. Three independent replicates were performed. (F) Neurons as those in A and B were stained with SAR1-GTP and SEC23a antibodies. (Right) Quantification of relative staining intensity of SAR1-GTP. t test; ***P < 0.001. Indicated number of neurons from five WT and cKO mice were analyzed. (Scale bars, 10 μm.) (G and H) Primary neurons dissected from around E15.5 WT mouse brain were transfected with SAR1-H79G or GFP cDNA at 8 div, fixed at 14 div, and stained with GFP (G) or GFP and GM130 antibodies (H). (Scale bars, 20 μm in G, 5 μm in H.) Three WT mice were analyzed. The nucleus was stained with DAPI.
Surprisingly, based on the up-regulated SAR1 expression in cKO brains (SI Appendix, Table S1), we found that the levels of SAR1-GTP, the activated form of SAR1, were also significantly enhanced in cKO brains (Fig. 5E). Consistently, immunostaining intensities of both SAR1-GTP and SAR1 in cKO neurons were also significantly higher than those in WT neurons (Fig. 5F and SI Appendix, Fig. S13B). SAR1 is a critical regulator of COPII vesicle formation (17, 18). To verify whether enhanced SAR1-GTP levels caused defects in dendritic outgrowth and fragmentation of Golgi structures, similar to those caused by cTAGE5/MEA6 cKO, we expressed SAR1-H79G to increase SAR1-GTP levels; we detected defects in dendritic outgrowth, including significant decreases in total dendrite length, in the number of dendrite branch points, and in dendritic complexity compared with WT neurons (Fig. 5G and SI Appendix, Fig. S14 A and B). In addition, SAR1-H79G overexpression also led to significantly decreased Golgi apparatus volume and increased fragmentation of Golgi structure, mimicking the phenotype found in cTAGE5/MEA6 cKO mice (Fig. 5H and SI Appendix, Fig. S14C).
cTAGE5/MEA6 cKO Disturbs the Interaction Between SEC23 and SAR1.
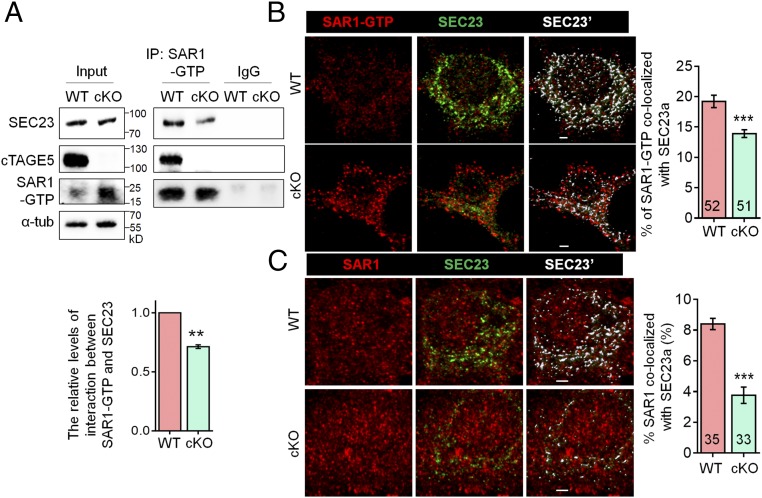
Considering that SEC23 activates SAR1 GTPase activity, and SAR1-H79G expression enhances SAR1-GTP levels through alternating the interaction between SAR1 and SEC23, we hypothesized that enhanced SAR1-GTP levels in cKO brains may be caused by disturbed interactions between SAR1 and SEC23. Endogenous cTAGE5/MEA6 interactions with SAR1-GTP and SEC23 were confirmed by colocalization of cTAGE5/MEA6 and SAR1-GTP, as well with coimmunoprecipitation (co-IP) of both cTAGE5/MEA6 and SEC23 with SAR1-GTP from brain lysates (Figs. 4A and 6A). In addition, significantly less SEC23 was detected in complexes immunoprecipitated with SAR1-GTP antibody from cKO brain lysates (Fig. 6A). Moreover, colocalization of SEC23 with both SAR1-GTP and SAR1 in cKO neurons was also significantly reduced (Fig. 6 B and C). These results indicate that cTAGE5/MEA6 cKO affects the interaction of SEC23 with SAR1, which leads to persistent enhanced SAR1-GTP levels and associated defects in COPII vesicle formation.
Fig. 6.
cTAGE5 cKO disturbs the interaction between SEC23 and SAR1. (A) Tissue lysates of P20 WT and cKO mouse cortex were immunoprecipitated with SAR1-GTP antibody and probed with SEC23, cTAGE5, and SAR1-GTP antibodies. The relative levels of interaction between SAR1-GTP and SEC23 was defined as the level of SEC23 in the immunocomplex, normalized with the level of SEC23 in the input and SAR1-GTP in immunocomplex. Paired t test; **P < 0.01. n = 3. (B and C) Representative images of primary neurons at 14 div, which were dissected from around E15.5 WT and cKO mouse brains and stained with SAR1-GTP (B) or SAR1 (C) and SEC23 antibodies. (Right) Quantification of SAR1-GTP (B) or SAR1 (C) colocalized with SEC23. The “Surface” function of Imaris software was used to simulate the distribution of SEC23 (white SEC23′), and the portion of SAR1-GTP or SAR1 localized on the SEC23 surface was determined as colocalized with SEC23. t test; ***P < 0.001. Indicated number of neurons from five (for SAR1-GTP) and four (for SAR1) WT and cKO mice were analyzed. (Scale bars, 2 μm.)
Discussion
cTAGE5/MEA6 has been shown to be important for the secretion of proteins, including collagen, VLDL, and insulin (22–25). In the present study, we generated a mouse model with cKO of cTAGE5/MEA6 to investigate the function of cTAGE5/MEA6 in the brain. We showed that cTAGE5/MEA6 plays a critical role in the transport of nonsecretory cellular components in neurons and regulates brain development.
cTAGE5/MEA6 Is Critical for Brain Development.
During brain development, neurons generate dendrites and axons to construct neural circuits and networks (1–4). cTAGE5/MEA6 is expressed in the mouse brain at different developmental stages and is highly expressed in neurons. Interestingly, as opposed to other tissues, cTAGE5/MEA6 mainly exists as a short splicing isoform in neurons (Fig. 2) and implies that cTAGE5/MEA6 may have a unique function in the brain. cTAGE5/MEA6 cKO leads to severe developmental retardation, smaller brain size, and lethality before P25. Moreover, these mice also exhibit phenotypes of developmental defects, including abnormal limb-clasping reflexes in the tail suspension test, inability to stay on the rotating rod in the rotarod test, dramatic activation of astrocytes, defects in dendritic and spine morphology, and decreased electrophysiological activities in the brain (Figs. 1 and 3 and SI Appendix, Figs. S2, S6, S7, S9, and S10). Interestingly, cTAGE5/MEA6 and its upstream gene, Mia2, were reported to form a hybrid gene, Tali, and the hybrid protein TALI plays a role in VLDL secretion (27, 28). However, Tali is not expressed in the brain (SI Appendix, Fig. S2C) (28). More importantly, transgenic expression of human cTAGE5/MEA6 largely rescued the phenotypes caused by cTAGE5/MEA6 cKO in the brain, including developmental retardation, the dramatic activation of astrocytes, and lethality (SI Appendix, Figs. S2 and S6). This indicates that cTAGE5/MEA6 itself plays an important role in brain development.
cTAGE5/MEA6 Is Critically Involved in the Transport of Cellular Components and Neuronal Development.
cTAGE5/MEA6 has been shown to be important for the secretion of collagen, VLDL, and insulin (22–25). Here, we took advantage of a temperature-sensitive reporter, GFP-ts045-VSVG, to demonstrate that cTAGE5/MEA6 cKO leads to defects in protein transport from the ER to the Golgi apparatus and the fragmentation of the Golgi structure in neurons (Fig. 4). More importantly, we detected down-regulation of several membrane receptors and ion channels in cKO brain (Fig. 3F and SI Appendix, Fig. S11). In addition to protein levels, the levels of lipid rafts where membrane proteins are localized, might be also affected based on the reduced levels of essential lipid components, such as sphingomyelins and cholesterol (38, 39) that were found in cKO brains (Fig. 4 G and H and SI Appendix, Fig. S12). Therefore, cTAGE5/MEA6 is involved in not only the transport of secretory proteins, but also the transport of nonsecretory cellular components in neurons.
We hypothesized that these defects in membrane protein transport and lipid raft assembly would likely result in abnormal dendrite and spine development. We detected significant abnormalities in dendritic outgrowth and spine formation and maintenance in cKO neurons in vivo. Consistently, the postsynaptic protein, PSD95, was dramatically down-regulated in cKO brains (Fig. 3F). In addition, levels of TrkB—the receptor for the neurotropic factors BDNF and NT4/5, which play important roles in neuronal development (40, 41)—were also significantly down-regulated (Fig. 3F). Moreover, levels of phosphorylated CamKII, and mRNA levels of Cpg15 and c-fos, markers for neuronal activation (32, 33), were all significantly reduced in cKO brains (Fig. 3 F and G). Taken together, these results explain how cTAGE5/MEA6 plays critical roles in dendritic outgrowth and synaptic formation or maintenance in neurons; they also help to explain the aberrant electrophysiological activities detected in cKO neurons (Fig. 3H and SI Appendix, Fig. S10).
cTAGE5/MEA6 Plays an Important Role in ER/Golgi Apparatus Trafficking Through the Regulation of COPII Complex Formation in Neurons.
COPII—which is composed of a small GTPase (SAR1) and the COPII coat proteins, SEC23, -24, -13, and -31—plays a crucial role in transporting ER cargo to the Golgi apparatus (8–10). ER complexity is significantly higher in the dendritic branch points of neurons, where ERES sites and COPII complexes are enriched, and is therefore an active area for forward protein transport (7, 35). Accordingly, we found that cTAGE5/MEA6, together with SAR1-GTP, were both relatively enriched in the dendritic branch points of neurons (Fig. 4A). Importantly, we show that cTAGE5/MEA6 cKO in neurons affected the interaction between SAR1 and SEC23. This would lead to inefficient down-regulation of SAR1 activity by SEC23 and subsequently persistent elevated levels of SAR1-GTP (Figs. 5 and 6) and, thus, the disruption of the cycling of SAR1 activity. Saito et al. (25) showed that cTAGE5/MEA6 interacts with the GEF of SAR1, SEC12, and regulates collagen transport. However, in contrast to results from the present study, where significant down-regulation of SEC31a protein levels and up-regulation of Sec31a mRNA levels were found (Fig. 5), Saito et al. (25) showed that cTAGE5/MEA6 knockdown had no effect on SEC31a expression. This inconsistency may be due to the different cell/systems used in each study. In addition, cTAGE5/MEA6 may control both the GEF and GAP of SAR1, and act as a key regulator of SAR1 activity to adapt to different cellular functions. Interestingly, cTAGE5/MEA6 expression is also under tight control and transgenic expression of hcTAGE5/MEA6 dramatically suppressed endogenous cTAGE5/MEA6 protein expression in the brain to maintain stable total cTAGE5/MEA6 levels (SI Appendix, Fig. S2).
In addition to the effects of cTAGE5/MEA6 cKO on the COPII complex, five components of COPI complex—including COPz1, COPg2, COPa, COPb2, and COPg1—were also significantly down-regulated (SI Appendix, Table S1), implying that cTAGE5/MEA6 may also be important for retrograde transport from the Golgi apparatus to the ER (42, 43).
cTAGE5/MEA6 plays an essential role in the development and maintenance of dendrites and spines, but unexpectedly, cTAGE5/MEA6 cKO in the brain had no significant effect on the proliferation and differentiation of NPCs, although cTAGE5/MEA6 is also expressed in these cells. In addition, cTAGE5/MEA6 is evolutionarily conserved only in vertebrates and not in yeast, in which the function of COPII vesicles is well conserved (24, 44, 45). This raises the possibility that cTAGE5/MEA6 may be dispensable for the formation of COPII vesicles at the basal level but becomes essential when transport of bulk or large amounts of cargo are needed. The forward transport of proteins and lipids in neurons during dendritic outgrowth is likely greater than that of transport in NPCs and might help to explain why cTAGE5/MEA6 is essential for neuronal development, but not NPC development. Similarly, cTAGE5/MEA6 is also essential for liver hepatocytes and pancreatic β cells in the bulk transport of VLDL and insulin, respectively (22, 23).
A point mutation-P521A in cTAGE5/MEA6 was reported to be associated with Fahr’s syndrome (46, 47). Fahr’s syndrome, also known as idiopathic basal ganglion calcification, is a rare dominant inheritance neurological disorder in humans. The patients showed calcification in brain regions controlling movement, including the basal ganglion and cerebral cortex; these patients also exhibited unsteady gaits and seizures (48–50). However, whether the P521A point mutation in cTAGE5/MEA6 is a contributing factor in the onset of Fahr’s syndrome is still unclear. It should be noted that the transgenic human cTAGE5/MEA6 used for rescue study here contains this P521A mutation and could largely rescue the developmental defects caused by knockout of cTAGE5/Mea6, indicating that this mutation does not have significant effects, at least, in young mice. This is consistent with the fact that the average onset age of Fahr’s syndrome in humans is around 40 y of age. Therefore, whether the P521A mutation in cTAGE5/MEA6 has a causal link with Fahr’s syndrome needs further investigation in older mice.
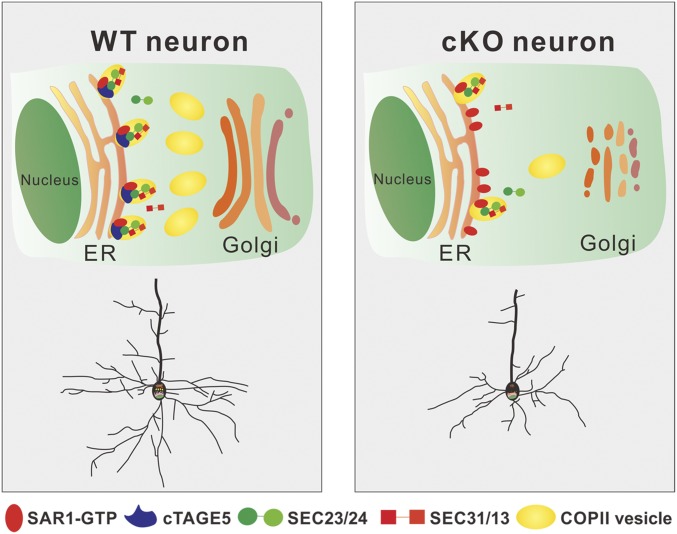
In conclusion, as described in Fig. 7, we found that cTAGE5/MEA6 cKO in the brain affects the interaction between SAR1 and SEC23 in neurons. This would lead to the disruption of the cycling of SAR1 activity, which is replaced by a persistent elevated level of SAR1-GTP and, thus, defects in COPII vesicle formation and transport from the ER to Golgi. This will result in fragmentation of the Golgi structure and disturb the trafficking of both proteins and lipids in neurons. These defects not only affect the transport of materials required for the development and maintenance of dendrites and spines (plasma membrane expansion), but also the signaling pathways required for neuronal development. Therefore, cTAGE5/MEA6 is critical for brain development; and when absent, severe phenotypes of developmental retardation will develop.
Fig. 7.
Model for the roles of cTAGE5/MEA6 in brain development. cTAGE5/MEA6 cKO in the brain affects the interaction between SAR1 and SEC23 in neurons. This would lead to the disruption of the cycling of SAR1 activity, which is replaced by persistent elevated level of SAR1-GTP and, thus, defects in COPII vesicle formation and transport from the ER to the Golgi. This will result in fragmentation of the Golgi structure, disturbing the trafficking of both proteins and lipids in neurons and defects in brain development.
Materials and Methods
Animals.
cTAGE5F/F mice, with insertion of two loxp sites flanking the 11th exon of cTAGE5 gene, were constructed by Yaqing Wang (22). Nestin-Cre [B6.Cg(SJL)-TgN(Nes-cre)1Kln)/J] transgenic mice were bought from Model animal research center of Nanjing University. Thy1-GFPm transgenic mice were generously provided by Yi Zuo, University of California, Santa Cruz, CA. Human FLAG-cTAGE5 (P521A) transgenic mice were generated by Beijing Biocytogen through methods of homologous recombination based on CRISPR/Cas9. The targeting vector contains 5′ homologous arm (2 kb), CAG promoter (CAG-Pr), LoxP-Stop-LoxP element (for conditional expression under the control of Cre), 3x-FLAG–tagged human cTAGE5 containing P521A point mutation, WPRE-Probe (for Southern blot), poly-A signal (pA), and 3′ homologous arms (2 kb). sgRNAs were designed in the site of Rosa26 gene on the sixth Chromosome and the CAG-Pr-loxP-Stop-loxP-cTAGE5-(P521A)-3xFLAG-WPRE-pA fragments were inserted in mouse Rosa26 site through homologous recombination. The experimental procedures on mice were performed based on the protocols which were approved by the Institutional Animal Care and Use Committee at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences.
Immunostaining.
Immunostaining was performed as described previously, with minor modifications (51). Briefly, the dissected brains were fixed with PFA (4%) and dehydrated in sucrose (30%), and then sectioned into slices in 40- or 100-μm width. The primary neurons were fixed with PFA (4%) at room temperature or cold methanol at 4 °C for 15 min. The brain slices or primary neurons were first incubated in the blocking buffer (10% FBS, 5% BSA, 0.3% Triton X-100, 0.01% NaN3 dissolved in PBS) at room temperature for 1 h, and then incubated in the primary antibody dissolved in the blocking buffer at 4 °C for overnight, and finally incubated in the secondary antibody conjugated with Alexa Fluor 488, 568, or 647 dyes dissolved in the blocking buffer at room temperature for 1 h. The antibodies used for immunostaining: Bip (ab21685, 1:2,000; Abcam), BrdU (ab6326, 1:500; Abcam), Caspase3 (ab2302, 1:800; Abcam), COPα (HPA028024, 1:400; Sigma), ERGIC53 (E6782, 1:1,000; Sigma), GFAP (ab7260, 1:1,000; Abcam), GFP (ab290, 1:1,000; Abcam), GFP (ab13970, 1:1,000; Abcam), GM130 (610822, 1:1,000; BD), Ki67 (ab15580, 1:1,000; Abcam), MAP2 (ab5392, 1:1,000; Abcam), cTAGE5/MEA6 (HPA000387, 1:400; Sigma), NeuN (ab104224, 1:1,000; Abcam), Parvalbumin (ab11427, 1:1,000; Abcam), SAR1 (ab77029, 1:400; Abcam), SAR1-GTP (26916, 1:200; Neweast), SEC23a (8162s, 1:400; CST), SEC31a (3466s, 1:400; CST), γ-Tubulin (ab11316, 1:1,000; Abcam).
Co-IP and Western Blotting.
Co-IP and Western blotting were performed as described previously with minor modifications (52). In brief, protein lysates of mouse brain were prepared in the cell lysis buffer [20 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 10 mM Hepes (pH 7.4), 0.5% Nonidet P-40, 0.5 mM EDTA (pH 8.0), Phosphatase Inhibitor Mixture (Roche), Protease Inhibitor Mixture (Roche), 0.2 mM PMSF]. For Co-IP of endogenous proteins, 2–3 μg of primary antibodies (SAR1-GTP) was first incubated with Protein A/G agarose beads (15 μL + 15 μL) at 4 °C for 2 h, and then the agarose beads were incubated in the 1-mg protein lysates of mouse brain at 4 °C for overnight. After being washed with cell lysis buffer at least three times, the proteins bound on the beads were eluted with SDS sample buffer and analyzed through Western blotting. For Western blotting, the proteins in the brain lysates were separated through SDS/PAGE and transferred into the nitrocellulose (NC) membranes. Then, the NC membranes were incubated in the primary antibodies at 4 °C for overnight and subsequently in the secondary antibodies or heavy-chain specific secondary antibodies (for the co-IP samples) conjugated with horseradish peroxidase. Finally, enhanced chemiluminescence substrate was used to visualize the protein bands in the NC membrane. The antibodies used for Western blotting: Bip (ab21685, 1:5,000; Abcam), COPα (HPA028024, 1:1,000; Sigma), FLAG (M185, 1:2,000; MBL), GAPDH (2118s, 1:2,000; CST), GFAP (3670s, 1:1,000; CST), cTAGE5/MEA6 (HPA000387, 1:1,000; Sigma), NeuN (ab104224, 1:1,000; Abcam), p-CamKII (ab124880, 1:1,000; Abcam), SAR1-GTP (26916, 1:1,000; Neweast), SEC23a (8162s, 1:1,000; CST), SEC31a (3466s, 1:1,000; CST), TrkB (4603s, 1:1,000; CST), α-Tubulin (3873s, 1:2,000; CST).
Quantitative MS.
The protein sample preparation of brain tissues for MS analysis and protocols of the MS analysis were performed as described previously with some modifications (53, 54). Three WT and cKO mouse brains at P14 were dissected and lysed in the lysis buffer [0.4 M sucrose, 50 mM 3-(N-morpholino) propanesulfonic acid (pH 7.0), 10 mM NaCl, 5 mM EDTA, and 0.5 mM PMSF]. The protein samples were digested with the filter-aided sample preparation method, in which the 100 μg lysates of mouse brain were reduced with 10 mM DTT at 37 °C for 1 h, and alkylated with 55 mM iodoacetamide. After digested with sequencing grade trypsin, the collected peptides of three biological replicates of the WT and cKO mouse brain were labeled with 6-plex TMT (Tandem Mass Tag; Thermo Fisher Scientific) and the labeling reaction was inactivated by addition of 5% hydroxylamine. The labeled samples were mixed together with equal ratios of these six samples and fractionated with reversed-phase (RP)-high performance liquid chromatography (HPLC). The fractioned peptides were resuspended in 0.1% formic acid (FA) and analyzed with a LTQ Orbitrap Elite mass spectrometer. For data analysis, raw mass spectrometric files were analyzed with the software MaxQuant (55), in which the Mus musculus (including canonical and isoforms) proteome sequence database used was downloaded from Uniprot. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010386 (56).
RNA Extraction and Real-Time PCR.
RNA extraction from the brain tissue and real-time PCR were performed as described previously (51, 57). Briefly, 1 mL TRIZOL was used to dissolve the brain tissues and 200 μL chloroform was subsequently added. After precipitation with isopropanol and washed with 70% (vol/vol) cold ethanol, the mRNA was collected and reversely transcribed into cDNA by using the reverse transcriptase (Promega). A 2× SYBR Green PCR mixture (Bio-Rad) was used to perform real-time PCR and analyzed with Bio-Rad C1000 Thermal Cycler. The primers used in PCR are listed in SI Appendix, Table S2.
Imaging and Data Analysis.
The images of mouse body and brain were taken by Canon camera, and confocal images were achieved through Zeiss LSM700 confocal microscopy. The images of neurospheres were achieved through Nikon fluorescence microscopy. All of the images were analyzed with software of Photoshop, ImageJ, or Imaris as described previously (51), and the process of digitalization of the data are described in SI Appendix, Supplemental Methods and Materials. For the quantification analysis, which was not performed blinded to the experimental condition, t test (Kolmogorov–Smirnov test for the Sholl analysis) or one-way ANOVA followed by Tukey’ multiple comparison test were used through GraphPad Prism or Excel software to analyze where there was significant difference, and the significant difference was indicated as *P < 0.05, **P < 0.01, and ***P < 0.001.
Other methods and materials are shown in SI Appendix, Supplemental Methods and Materials.
Supplementary Material
Acknowledgments
We thank Drs. Xiaowei Chen, Weixiang Guo, and Qingfeng Wu for their constructive advice, and Dr. Yi Zuo for providing Thy1-GFPm transgenic mice. This work was supported by National Natural Science Foundation of China Grants 31730108, 31430037, 31471132, and 31571038, Strategic Priority Research Program and Innovation Program of the Chinese Academy of Sciences Grants XDBS1020100, XDA16010306, and QYZDJ-SSW-SMC007, Shanghai brain-intelligence project from Science and Technology Commission of Shanghai Municipality Grant 16JC1420500, Beijing Brain Project Grant Z161100002616004, and National Science and Technology Major Project of China Grant 2014CB942801.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (accession no. PXD010386).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804083115/-/DCSupplemental.
References
- 1.Calabrese B, Wilson MS, Halpain S. Development and regulation of dendritic spine synapses. Physiology (Bethesda) 2006;21:38–47. doi: 10.1152/physiol.00042.2005. [DOI] [PubMed] [Google Scholar]
- 2.Spruston N. Pyramidal neurons: Dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- 3.Arikkath J. Molecular mechanisms of dendrite morphogenesis. Front Cell Neurosci. 2012;6:61. doi: 10.3389/fncel.2012.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horton AC, Ehlers MD. Secretory trafficking in neuronal dendrites. Nat Cell Biol. 2004;6:585–591. doi: 10.1038/ncb0704-585. [DOI] [PubMed] [Google Scholar]
- 5.Ramírez OA, Couve A. The endoplasmic reticulum and protein trafficking in dendrites and axons. Trends Cell Biol. 2011;21:219–227. doi: 10.1016/j.tcb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Ye B, et al. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui-Wang T, et al. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012;148:309–321. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonny B, Schekman R. ER export: Public transportation by the COPII coach. Curr Opin Cell Biol. 2001;13:438–443. doi: 10.1016/s0955-0674(00)00234-9. [DOI] [PubMed] [Google Scholar]
- 9.Lee MC, Miller EA. Molecular mechanisms of COPII vesicle formation. Semin Cell Dev Biol. 2007;18:424–434. doi: 10.1016/j.semcdb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Miller EA, Schekman R. COPII—A flexible vesicle formation system. Curr Opin Cell Biol. 2013;25:420–427. doi: 10.1016/j.ceb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano A, Brada D, Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakańo A, Muramatsu M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuoka K, et al. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 14.Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol. 2001;3:531–537. doi: 10.1038/35078500. [DOI] [PubMed] [Google Scholar]
- 15.Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 16.Miller E, Antonny B, Hamamoto S, Schekman R. Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 2002;21:6105–6113. doi: 10.1093/emboj/cdf605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storrie B, et al. Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuberth CE, Tängemo C, Coneva C, Tischer C, Pepperkok R. Self-organization of core Golgi material is independent of COPII-mediated endoplasmic reticulum export. J Cell Sci. 2015;128:1279–1293. doi: 10.1242/jcs.154443. [DOI] [PubMed] [Google Scholar]
- 19.Heckel D, et al. cDNA cloning and chromosomal mapping of a predicted coiled-coil proline-rich protein immunogenic in meningioma patients. Hum Mol Genet. 1997;6:2031–2041. doi: 10.1093/hmg/6.12.2031. [DOI] [PubMed] [Google Scholar]
- 20.Comtesse N, et al. MGEA6 is tumor-specific overexpressed and frequently recognized by patient-serum antibodies. Oncogene. 2002;21:239–247. doi: 10.1038/sj.onc.1205005. [DOI] [PubMed] [Google Scholar]
- 21.Kalniņa Z, et al. Evaluation of T7 and lambda phage display systems for survey of autoantibody profiles in cancer patients. J Immunol Methods. 2008;334:37–50. doi: 10.1016/j.jim.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Mea6 controls VLDL transport through the coordinated regulation of COPII assembly. Cell Res. 2016;26:787–804. doi: 10.1038/cr.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan J, et al. cTAGE5 deletion in pancreatic β cells impairs proinsulin trafficking and insulin biogenesis in mice. J Cell Biol. 2017;216:4153–4164. doi: 10.1083/jcb.201705027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito K, et al. cTAGE5 mediates collagen secretion through interaction with TANGO1 at endoplasmic reticulum exit sites. Mol Biol Cell. 2011;22:2301–2308. doi: 10.1091/mbc.E11-02-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito K, et al. Concentration of Sec12 at ER exit sites via interaction with cTAGE5 is required for collagen export. J Cell Biol. 2014;206:751–762. doi: 10.1083/jcb.201312062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petkovic M, et al. The SNARE Sec22b has a non-fusogenic function in plasma membrane expansion. Nat Cell Biol. 2014;16:434–444. doi: 10.1038/ncb2937. [DOI] [PubMed] [Google Scholar]
- 27.Pitman JL, Bonnet DJ, Curtiss LK, Gekakis N. Reduced cholesterol and triglycerides in mice with a mutation in Mia2, a liver protein that localizes to ER exit sites. J Lipid Res. 2011;52:1775–1786. doi: 10.1194/jlr.M017277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos AJ, Nogueira C, Ortega-Bellido M, Malhotra V. TANGO1 and Mia2/cTAGE5 (TALI) cooperate to export bulky pre-chylomicrons/VLDLs from the endoplasmic reticulum. J Cell Biol. 2016;213:343–354. doi: 10.1083/jcb.201603072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 30.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 31.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 32.Nedivi E, Wu GY, Cline HT. Promotion of dendritic growth by CPG15, an activity-induced signaling molecule. Science. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramirez S, et al. Creating a false memory in the hippocampus. Science. 2013;341:387–391. doi: 10.1126/science.1239073. [DOI] [PubMed] [Google Scholar]
- 34.Kamiya Y, et al. Sugar-binding properties of VIP36, an intracellular animal lectin operating as a cargo receptor. J Biol Chem. 2005;280:37178–37182. doi: 10.1074/jbc.M505757200. [DOI] [PubMed] [Google Scholar]
- 35.Hanus C, et al. Synaptic control of secretory trafficking in dendrites. Cell Rep. 2014;7:1771–1778. doi: 10.1016/j.celrep.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zilberstein A, Snider MD, Porter M, Lodish HF. Mutants of vesicular stomatitis virus blocked at different stages in maturation of the viral glycoprotein. Cell. 1980;21:417–427. doi: 10.1016/0092-8674(80)90478-x. [DOI] [PubMed] [Google Scholar]
- 37.Silvius JR. Role of cholesterol in lipid raft formation: Lessons from lipid model systems. Biochim Biophys Acta. 2003;1610:174–183. doi: 10.1016/s0005-2736(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 38.de Almeida RF, Fedorov A, Prieto M. Sphingomyelin/phosphatidylcholine/cholesterol phase diagram: Boundaries and composition of lipid rafts. Biophys J. 2003;85:2406–2416. doi: 10.1016/s0006-3495(03)74664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niemelä PS, Ollila S, Hyvönen MT, Karttunen M, Vattulainen I. Assessing the nature of lipid raft membranes. PLoS Comput Biol. 2007;3:e34. doi: 10.1371/journal.pcbi.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yacoubian TA, Lo DC. Truncated and full-length TrkB receptors regulate distinct modes of dendritic growth. Nat Neurosci. 2000;3:342–349. doi: 10.1038/73911. [DOI] [PubMed] [Google Scholar]
- 41.Yoshii A, Constantine-Paton M. Postsynaptic BDNF-TrkB signaling in synapse maturation, plasticity, and disease. Dev Neurobiol. 2010;70:304–322. doi: 10.1002/dneu.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee C, Goldberg J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010;142:123–132. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson LP. Structure and mechanism of COPI vesicle biogenesis. Curr Opin Cell Biol. 2014;29:67–73. doi: 10.1016/j.ceb.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Bednarek SY, et al. COPI- and COPII-coated vesicles bud directly from the endoplasmic reticulum in yeast. Cell. 1995;83:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- 45.Bickford LC, Mossessova E, Goldberg J. A structural view of the COPII vesicle coat. Curr Opin Struct Biol. 2004;14:147–153. doi: 10.1016/j.sbi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Oliveira JR, et al. Analysis of candidate genes at the IBGC1 locus associated with idiopathic basal ganglia calcification (“Fahr’s disease”) J Mol Neurosci. 2007;33:151–154. [PubMed] [Google Scholar]
- 47.Lemos RR, Oliveira DF, Zatz M, Oliveira JR. Population and computational analysis of the MGEA6 P521A variation as a risk factor for familial idiopathic basal ganglia calcification (Fahr’s disease) J Mol Neurosci. 2011;43:333–336. doi: 10.1007/s12031-010-9445-7. [DOI] [PubMed] [Google Scholar]
- 48.Moskowitz MA, Winickoff RN, Heinz ER. Familial calcification of the basal ganglions: A metabolic and genetic study. N Engl J Med. 1971;285:72–77. doi: 10.1056/NEJM197107082850202. [DOI] [PubMed] [Google Scholar]
- 49.Geschwind DH, Loginov M, Stern JM. Identification of a locus on chromosome 14q for idiopathic basal ganglia calcification (Fahr disease) Am J Hum Genet. 1999;65:764–772. doi: 10.1086/302558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saleem S, et al. Fahr’s syndrome: Literature review of current evidence. Orphanet J Rare Dis. 2013;8:156. doi: 10.1186/1750-1172-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F, Xu D, Yuan L, Sun Y, Xu Z. Epigenetic regulation of Atrophin1 by lysine-specific demethylase 1 is required for cortical progenitor maintenance. Nat Commun. 2014;5:5815. doi: 10.1038/ncomms6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang F, et al. A novel c-Jun N-terminal Kinase (JNK) signaling complex involved in neuronal migration during brain development. J Biol Chem. 2016;291:11466–11475. doi: 10.1074/jbc.M116.716811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiśniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 54.Ge H, et al. Translating divergent environmental stresses into a common proteome response through the histidine kinase 33 (Hik33) in a model cyanobacterium. Mol Cell Proteomics. 2017;16:1258–1274. doi: 10.1074/mcp.M116.068080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 56.Vizcaíno JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.