Significance
The NLRP3 inflammasome is a multimolecular protein complex which is responsible for regulating the processing and secretion of the proinflammatory cytokine interleukin-1β. Understanding the mechanisms regulating the assembly of the inflammasome is of outstanding biological interest and importance, not least because of the contributions of the NLRP3 inflammasome to disease processes. Here we report insights into the dynamic nature of inflammasome oligomerization and its dependence on chloride ions. These studies reveal an additional layer of regulation and priming that can further enhance and drive inflammatory responses.
Keywords: inflammasome, inflammation, caspase-1, interleukin-1, chloride
Abstract
The NLRP3 inflammasome is an important regulator of inflammation and immunity. It is a multimolecular platform formed within cells that facilitates the activation of proinflammatory caspases to drive secretion of cytokines such as interleukin-1β (IL-1β). Knowledge of the mechanisms regulating formation of the NLRP3 inflammasome is incomplete. Here we report Cl− channel-dependent formation of dynamic ASC oligomers and inflammasome specks that remain inactive in the absence of K+ efflux. Formed after Cl− efflux exclusively, ASC specks are NLRP3 dependent, reversible, and inactive, although they further prime inflammatory responses, accelerating and enhancing release of IL-1β in response to a K+ efflux-inducing stimulus. NEK7 is a specific K+ sensor and does not associate with NLRP3 under conditions stimulating exclusively Cl− efflux, but does after K+ efflux, activating the complex driving inflammation. Our investigation delivers mechanistic understanding into inflammasome activation and the regulation of inflammatory responses.
Inflammasomes are cytosolic multimeric protein complexes formed in inflammatory cells in response to pathogenic infection and tissue injury (1). Activated inflammasomes drive the processing of proinflammatory cytokine precursors such as prointerleukin-1β to its mature secreted form (IL-1β) and initiate inflammation (2). The most commonly studied inflammasome is composed of the cytosolic pattern recognition receptor (PRR) NACHT, LRR, and PYD domains containing protein 3 (NLRP3). Aberrant activation of NLRP3 is now associated with the worsening of noncommunicable diseases, including atherosclerosis, Alzheimer’s disease, type II diabetes, arthritis, and others (3), so a research priority has been to understand the mechanism of NLRP3 inflammasome activation.
Specific pathogen- or damage-associated molecular patterns (PAMPs and DAMPs, respectively), or stimuli that disrupt cellular homeostasis or integrity, induce the activation of NLRP3 (1, 4). NLRP3 activation is represented by rapid conformational change of NLRP3 and its physical interaction with never in mitosis A-related kinase 7 (NEK7) (5, 6). Oligomerized NLRP3 then recruits the adaptor protein apoptosis-associated speck-like protein containing a caspase-recruitment domain (CARD) (ASC), which then assembles into large inflammasome specks upon which the CARD of ASC recruits procaspase-1 which undergoes a proximity-dependent autoactivation to produce mature caspase-1 (7, 8). Caspase-1 then cleaves substrate molecules such as pro–IL-1β which are released from the cell to promote inflammation (9). The inflammasome specks released from pyroptotic cells can also further contribute to inflammatory responses (10, 11).
NLRP3 inflammasome activation requires two sequential events for its activation in vitro. Toll-like receptor stimulation and NF-κB signaling induces expression of pro–IL-1β and NLRP3 (12). Posttranslational modifications involving NLRP3 phosphorylation and deubiquitination are crucial for subsequent inflammasome assembly (13–16). In addition to these priming and licensing events, K+ efflux is now widely accepted as an essential second step in the activation of NLRP3 inflammasomes (17). Recent studies also suggest a potential involvement of Cl− channels (18–20), although exactly how Cl− channels contribute to inflammasome formation is unknown. Here we describe the regulation of NLRP3 inflammasomes by Cl− ion fluxes showing that Cl− efflux is the ASC oligomerizing signal, but that a K+ efflux-dependent activation and interaction of NEK7 with NLRP3 is required for activation of caspase-1 and secretion of IL-1β.
Results
Effects of Chloride on IL-1β Secretion.
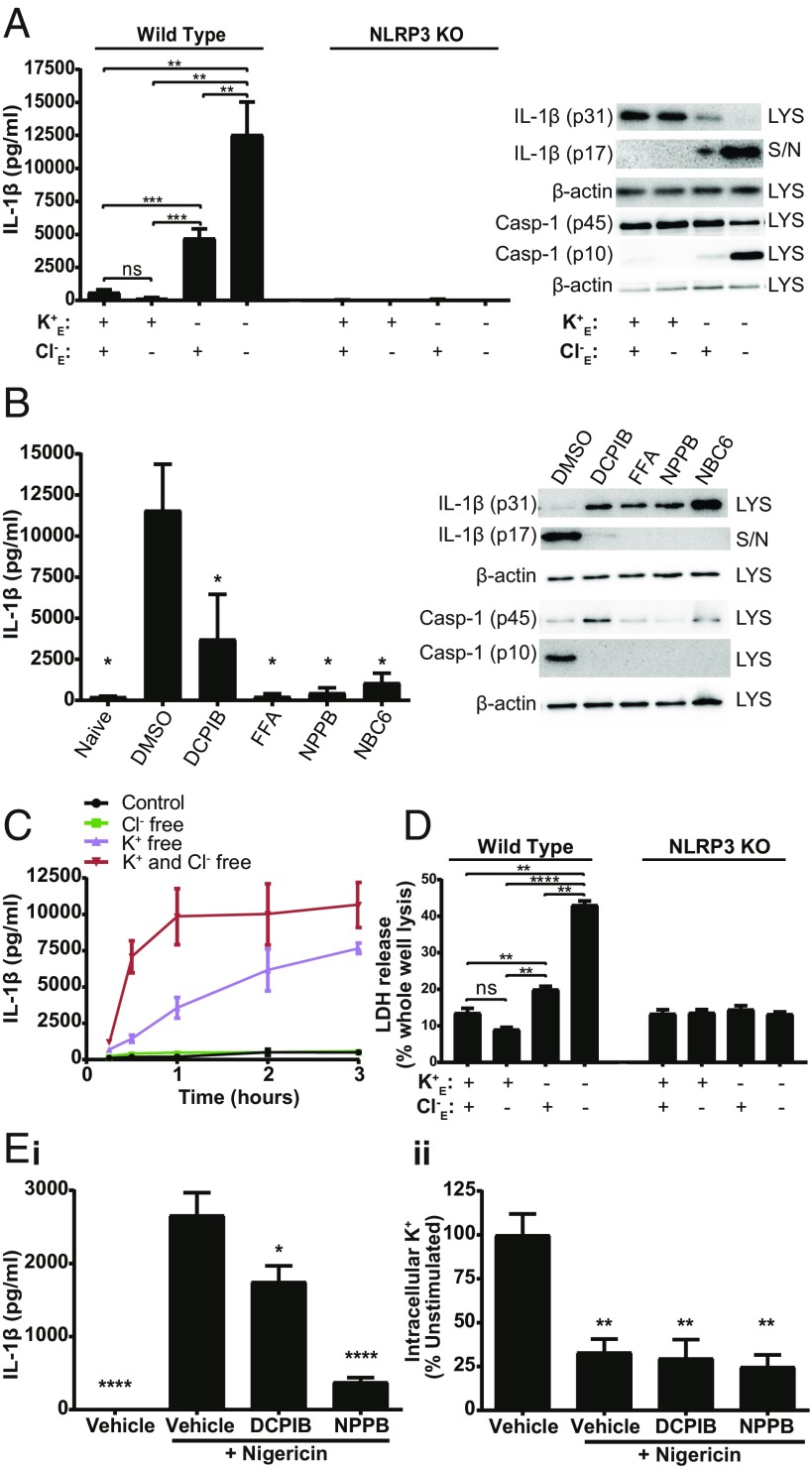
To understand the effect of Cl− fluxes on inflammasome formation, we performed ion substitution experiments and incubated LPS-primed primary mouse bone marrow derived macrophages (BMDMs) in isotonic solutions deficient in K+, Cl−, or both, to drive efflux of K+ or Cl−, respectively. In the absence of additional inflammasome activators, K+ free conditions induced a significant increase in levels of extracellular IL-1β (Fig. 1A). IL-1β secretion was further enhanced in the combined absence of K+ and Cl− (Fig. 1A). Cl− free conditions alone (with normal extracellular 5 mM K+) had no effect on release of IL-1β (Fig. 1A). The effects of K+, and K+ and Cl− free conditions on IL-1β release were NLRP3 dependent, as under the same conditions no IL-1β release was observed from primary BMDMs isolated from NLRP3 knockout (KO) mice (21) (Fig. 1A). IL-1β release under conditions of no extracellular K+ or Cl− was also inhibited by broad spectrum Cl− channel blockers, suggesting that the effect of K+ efflux was sensitive to Cl− channel inhibition (Fig. 1B). The Cl− channel inhibitors used here inhibited Cl− channel activation in HeLa cells under hypotonic stress measured by I− quenching of halide-sensitive YFP H148Q/I152L (22) (SI Appendix, Fig. S1). The recently described NLRP3 inhibitor NBC6 (23) also inhibited IL-1β release under conditions of no K+ and Cl− (Fig. 1B). The Cl− channel inhibitors used here were previously reported to specifically inhibit the NLRP3 inflammasome (18), and do not inhibit production of TNFα as reported previously (19) and as shown in SI Appendix, Fig. S2. A temporal analysis of IL-1β release under K+ free, or K+ and Cl− free conditions up to 3 h revealed that IL-1β release was not only enhanced, but also released more rapidly under conditions of K+ and Cl− free (Fig. 1C). Cl− free solutions in the presence of K+ did not induce any IL-1β release, even with up to 3 h incubation (Fig. 1C). A similar pattern to IL-1β release was observed for cell death in the ion substitution experiments with significant cell death in the K+ free alone, and K+ and Cl− free conditions (Fig. 1D). The cell death was also NLRP3 dependent, suggesting that it could be pyroptotic (Fig. 1D). Cl− channel inhibitors also inhibited LPS and nigericin induced IL-1β release from immortalized (i) mouse BMDMs (iBMDMs) (Fig. 1 E, i) without affecting nigericin-induced K+ efflux from these cells as measured by inductively coupled plasma mass spectrometry (ICP-MS) (Fig. 1 E, ii). Conditions of K+ free, and K+ and Cl− free also induced IL-1β release from LPS-primed human THP-1 monocytes (SI Appendix, Fig. S3A). Cl− channel blockers also prevented K+ and Cl− free solution-induced IL-1β release from LPS-primed THP-1 monocytes (SI Appendix, Fig. S3B). These data suggest that Cl− channel activation is essential for NLRP3 inflammasome activation, consistent with previous observations (18–20), and that the Cl− channel-dependent effect can be blocked without affecting K+ efflux.
Fig. 1.
Chloride efflux modulates NLRP3 activation and IL-1β processing. (A) IL-1β ELISA, caspase-1, and IL-1β immunoblot of cell lysates (LYS) and supernatants (S/N) from LPS-primed WT and NLRP3 KO BMDMs incubated in a control (145 mM NaCl/5 mM KCl), Cl− free (145 mM NaGluconate/5 mM KGluconate), K+ free (150 mM NaCl), or K+ and Cl− free (150 mM NaGluconate) solution for 2 h (n = 6). (B) IL-1β ELISA, caspase-1, and IL-1β immunoblot of LYS and S/N from LPS-primed BMDMs pretreated with a vehicle control, DCPIB (10 µM), flufenamic acid (FFA, 100 µM), NPPB (100 µM), or NBC6 (30 µM) and incubated in a K+ and Cl− free solution for 2 h (n = 5). (C) Time course of released IL-1β in the supernatant of LPS-primed BMDMs incubated in a control, Cl− free, K+ free, or K+ and Cl− free solution, determined by IL-1β ELISA. (D) LDH release from LPS-primed WT or NLRP3 KO BMDMs incubated in a control, Cl− free, K+ free, or K+ and Cl− free solution for 2 h. (E, i) IL-1β ELISA performed on supernatants from LPS-primed iBMDMs pretreated with a vehicle control, DCPIB (10 µM) or NPPB (100 µM) and then stimulated with nigericin (10 µM, 1 h) (n = 4). (E, ii) Intracellular K+ measurements from LPS-primed iBMDM cell lysates pretreated with a vehicle control, DCPIB (10 µM) or NPPB (100 µM) and stimulated with nigericin (10 µM, 30 min) in the presence of Z-VAD-FMK (50 µM) to prevent pyroptosis (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significantly different determined by a one-way ANOVA with Tukey’s (A and D) or Dunett’s (B vs. vehicle control; E, i vs. nigericin; E, ii vs. vehicle) post hoc analysis. Values shown are the mean ± SEM.
Chloride Dependence of ASC Speck Formation.
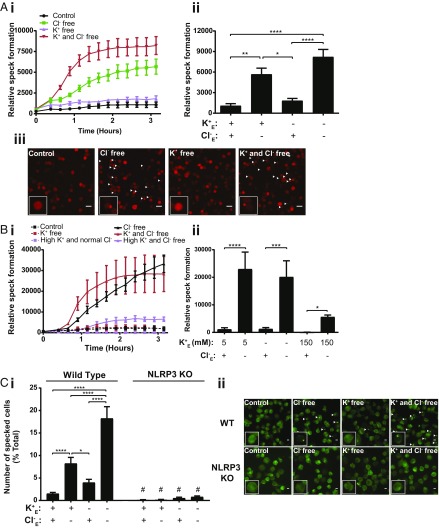
Using iBMDMs stably expressing ASC-mCherry (18), we studied the effects of Cl− channel activation on inflammasome speck formation. Incubation of LPS-primed ASC-mCherry iBMDMs in both Cl− free, and K+ and Cl− free solutions induced formation of ASC specks (Fig. 2A). Thus, Cl− free conditions are sufficient to induce the formation of ASC inflammasome specks without inducing the activation of caspase-1, whereas K+ free alone conditions induced IL-1β release without inducing significant ASC speck formation. We then analyzed ASC speck formation over time in LPS-primed ASC-mCherry expressing iBMDMs using the same solutions, but with the addition of solutions composed of 150 mM KCl, or 150 mM KGluconate, concentrations of K+ far in excess of those required to inhibit NLRP3 inflammasome activation (17). In this experiment we observed significant ASC speck formation in the absence of Cl−, and in the absence of K+ and Cl−, as expected (Fig. 2B). Cells incubated in 150 mM KCl did not form many specks, although incubation in 150 mM KGluconate did induce a modest but significant formation of ASC specks (Fig. 2B). These data reveal speck formation in the presence of inflammasome-inhibiting levels of K+, but in the absence of any Cl−. The NLRP3 dependence of Cl− free solution inducing ASC speck formation was confirmed using wild-type (WT) and NLRP3 KO THP-1 macrophages and staining for ASC specks using immunocytochemistry after incubation in the Cl− free, and K+ and Cl− free solutions described. Here we observed that ASC speck formation under Cl− free conditions was completely NLRP3 dependent (Fig. 2C). We then repeated the experiments on IL-1β release and ASC speck formation with the Cl− and K+ free buffers but added (i) 10 mM Hepes (pH 7.3), (ii) 10 mM glucose, (iii) 2 mM CaCl2, or (iv) Hepes, glucose, and CaCl2 (SI Appendix, Fig. S4). These experiments were conducted in primary BMDMs (for IL-1β release) and iBMDMs expressing ASC-mCherry (for ASC speck formation). Under all these new conditions, Cl− free buffers still failed to cause IL-1β release, but did cause ASC speck formation (SI Appendix, Fig. S4), fully supporting the data presented in Fig. 2.
Fig. 2.
Chloride efflux is sufficient to induce NLRP3-dependent ASC speck formation. (A, i) LPS-primed immortalized BMDMs stably expressing ASC-mCherry (ASC-mCherry iBMDMs) were incubated in a control (145 mM NaCl/5 mM KCl), Cl− free (145 mM NaGluconate/5 mM KGluconate), K+ free (150 mM NaCl), or K+ and Cl− free (150 mM NaGluconate) solution and ASC speck formation was measured in real time (n = 5). (A, ii) ASC speck formation was analyzed after a 3-h incubation in the respective solutions (n = 5). (A, iii) Representative images of ASC-mCherry iBMDM cells after a 3-h incubation in the respective solutions. (Scale bar, 50 µm, arrows denote ASC speck.) (B, i) LPS-primed ASC-mCherry iBMDMs were incubated in a control, Cl− free, K+ free, K+ and Cl− free, high K+ and normal Cl− (150 mM KCl), or high K+ and Cl− free (150 mM KGluconate) solution and ASC speck formation was measured in real time (n = 4). (B, ii) Speck formation was analyzed after a 3-h incubation in the respective solutions (n = 4). (C, i) LPS-primed WT and NLRP3 KO THP-1 macrophages were incubated in a control, Cl− free, K+ free, or K+ and Cl− free solution for 3 h and immunostained for ASC (n = 4). (C, ii) Representative images of WT and NLRP3 KO THP-1 macrophages incubated for 3 h in the respective solutions and immunostained for ASC. (Scale bar, 50 µm, arrows denote ASC speck.) All experiments were performed in the presence of Z-VAD-FMK (50 µM) to prevent pyroptosis and loss of ASC specks. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significantly different determined by a one-way ANOVA with Tukey’s (A and B) post hoc analysis, or two-way ANOVA with Sidak’s post hoc analysis (C, #P < 0.01 vs. WT in the respective solution). Values shown are the mean ± SEM.
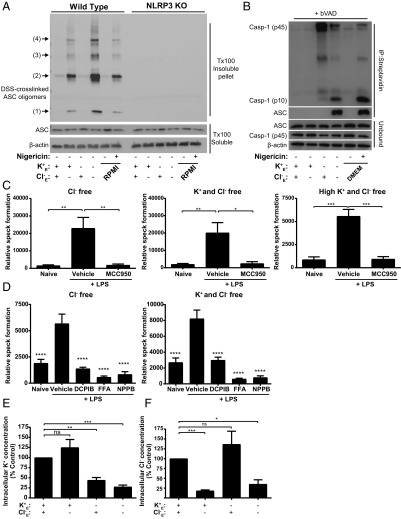
Using WT and NLRP3 KO THP-1 macrophages, we crosslinked ASC using disuccinimidyl suberate (DSS) and detected ASC oligomerization using immunoblotting (24). In Cl− free conditions in LPS-primed THP-1 cells there was ASC oligomerization and this was absent in NLRP3 KO cells, further confirming the NLRP3 dependence of Cl− channel-dependent ASC oligomerization (Fig. 3A). Given that the specks that formed under Cl− free conditions were inactive (i.e., there was no active caspase-1 or mature IL-1β produced) (Fig. 1) we investigated whether caspase-1 was recruited to these inactive ASC specks. Recent research has shown that biotin-VAD-fmk (bVAD-fmk) is a useful probe for activated caspases (25). We utilized bVAD-fmk to measure the recruitment of active caspase-1 to the ASC specks under Cl− or K+ free conditions. Primary BMDM cells were treated with LPS and then incubated in isotonic solutions deficient in K+, Cl−, or both. After 1 h, cell supernatants containing excess bVAD were removed, and active caspase-1 bound to bVAD/streptavidin beads was pulled down for immunoblotting. Similar to previous research (25) we detected active 45 and 10 kDa caspase-1 after nigericin treatment (Fig. 3B). Under these conditions we also blotted for ASC, confirming an association of the activated caspase-1 to ASC (Fig. 3B). When we analyzed the pulldown following incubation with the different buffers, we found significant pulldown of caspase-1 and ASC after incubations in buffers that were K+ and Cl− free (Fig. 3B). In control or only Cl− free conditions, there was no caspase-1 and ASC pulldown (Fig. 3B). These data suggest that under conditions of Cl− free driving formation of inactive ASC specks there is no caspase-1 activation because it is not recruited.
Fig. 3.
NLRP3 dependence and caspase-1 recruitment to the chloride-dependent ASC complex. (A) Immunoblot of DSS-crosslinked ASC oligomers in lysates from LPS-primed WT and NLRP3 KO THP-1 macrophages incubated in a control (145 mM NaCl/5 mM KCl), Cl− free (145 mM NaGluconate/5 mM KGluconate), K+ free (150 mM NaCl), K+ and Cl− free (150 mM NaGluconate) solution, or RPMI ± nigericin (10 µM) for 2 h (n = 3). (B) LPS-primed WT primary BMDMs were incubated in a control, Cl− free, K+ free, K+ and Cl− free solution, or DMEM ± nigericin (10 µM) in the presence of biotin-VAD-FMK (bVAD, 10 µM) for 2 h (n = 7). Cell supernatants were removed and lysates were pulled down with streptavidin T1 beads and immunoblotted for caspase-1 and ASC. (C) Analysis of ASC speck formation in naïve or LPS-primed ASC-mCherry iBMDMs pretreated with a vehicle control or the NLRP3 inflammasome inhibitor MCC950 (10 µM) and incubated in a Cl− free, K+ and Cl− free, or high K+ and Cl− free solution for 3 h (n = 4). (D) Analysis of ASC speck formation in naïve or LPS-primed ASC-mCherry iBMDMs pretreated with a vehicle control or the chloride channel inhibitors DCPIB (10 µM), flufenamic acid (FFA, 100 µM), or NPPB (100 µM) and incubated in a Cl− free or K+ and Cl− free solution for 3 h (n = 5). (E) Intracellular K+ measurements from LPS-primed BMDMs incubated in a control, Cl− free, K+ free, or K+ and Cl− free solution for 2 h, in the presence of Z-VAD-FMK (50 µM) to prevent pyroptosis (n = 4). (F) Intracellular Cl− measurements from LPS-primed NLRP3 KO BMDMs incubated in a control, Cl− free, K+ free, or K+ and Cl− free solution for 2 h (n = 4). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significantly different determined by a one-way ANOVA with Dunett’s (C and D, vs. vehicle control) post hoc analysis, or a one-sample t test versus hypothetical value of 100% (E and F). Values shown are the mean ± SEM.
We then looked at ASC speck formation in the ASC-mCherry iBMDMs in Cl− free conditions in the absence and presence of the NLRP3 inflammasome inhibitor MCC950 (26). MCC950 effectively inhibited ASC speck formation in Cl− free, K+ and Cl− free, and in high K+ and Cl− free (150 mM KGluconate)-containing solutions (Fig. 3C). ASC speck formation induced by Cl− free, and K+ and Cl− free conditions was also inhibited by Cl− channel inhibitors (Fig. 3D). Incubation of primary mouse BMDMs with the respective solutions revealed that only K+ free solutions induced K+ efflux, and that the Cl− free solution (which contains K+) did not, as measured by ICP-MS (Fig. 3E). Using ion chromatography we were also able to determine that Cl− loss occurred specifically from cells incubated with the Cl− free buffers (Fig. 3F). Further, pharmacologically induced Cl− efflux in LPS-primed ASC-mCherry iBMDMs using Eact, a selective agonist of the calcium-activated chloride channel ANO1 (27), dose dependently induced ASC speck formation (SI Appendix, Fig. S5A). Formation of Eact-dependent ASC specks was blocked using the selective ANO1 antagonist Ani9 (28), the NLRP3 inhibitor MCC950, and the Cl− channel inhibitor NPPB (SI Appendix, Fig. S5B). Similar to the Cl− free conditions, Eact did not induce IL-1β processing and release (SI Appendix, Fig. S5C). The ANO1 inhibitor Ani9 did not inhibit nigericin-induced IL-1β release (SI Appendix, Fig. S5D) or cell death (SI Appendix, Fig. S5E) in iBMDM cells. These data suggest that Cl− channel activation and Cl− efflux are essential for ASC speck formation and that Cl− efflux can occur independently of K+ efflux. In the absence of K+ efflux, the ASC specks do not activate caspase-1.
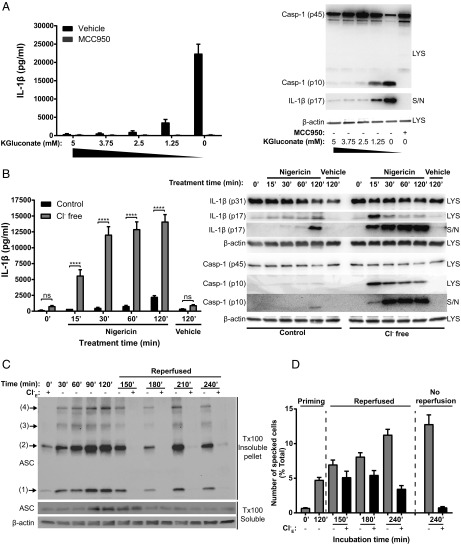
We next incubated LPS-primed primary BMDMs in K+ and Cl− free conditions to induce NLRP3 inflammasome activation, caspase-1 activation, and IL-1β release and added back up to 5 mM extracellular K+. Addition of K+ dose dependently inhibited the activation of caspase-1 and IL-1β secretion (Fig. 4A). Thus, we are observing two events combining to facilitate the activation of caspase-1 and IL-1β secretion: a Cl− efflux-dependent formation of an ASC speck and a K+ efflux-dependent activation of caspase-1. We tested whether specks formed under Cl− free conditions could “prime” or enhance inflammasome-dependent responses. LPS-primed BMDMs were stimulated in Cl− free solution to form inactive ASC specks as above and were then stimulated with nigericin to induce K+ efflux and IL-1β release and were compared with cells incubated under normal K+ and Cl− conditions (Fig. 4B). There was an accelerated and enhanced activation of caspase-1 and release of IL-1β in BMDMs preincubated in Cl− free solution within the first 15 min and subsequent time points, suggesting that the specks formed under Cl− free conditions are competent for subsequent inflammasome activation and IL-1β processing. Currently the formation of the inflammasome speck is considered to be a terminal event dependent upon prion like properties of the ASC molecule (10, 11). The formation of the ASC speck under Cl− free conditions did not result in caspase-1 activation and there was no pyroptotic cell death (Fig. 1) and so we were able to test whether ASC oligomerization is dynamically regulated by extracellular Cl−. LPS-primed THP-1 cells were incubated in Cl− free solution before ASC oligomerization was measured by DSS crosslinking and immunoblotting as described above. Incubation of the cells in Cl− free (but K+ containing) solution resulted in ASC oligomerization as indicated by high molecular weight ASC bands in the Triton X-100 insoluble fraction (Fig. 4C). Reintroducing Cl− containing solution back to the cells at this point caused a reversal of ASC oligomerization when measured 30 min later which was sustained for the duration of the experiment (Fig. 4C). We then used ASC-mCherry iBMDMs to determine whether Cl− free dependent ASC speck formation was reversible. LPS-primed ASC-mCherry iBMDM cells were incubated in Cl− free solution for 120 min and numbers of cells with ASC specks were quantified (Fig. 4D). In one experimental group, extracellular Cl− levels were returned to normal (reperfused), while the other group remained extracellular Cl− free, and specks continued to be monitored. Here we observed that Cl− free dependent speck formation was also dynamic and reversible, albeit over a slower time course than oligomerization measured above (Fig. 4D). The kinetics of oligomerization measured by immunoblot, and of speck formation measured by microscopy are different supporting the concept that oligomerization to soluble oligomers precedes the formation of more stable specks. These data suggest that ASC oligomerization and speck formation are dynamic and reversible processes regulated by Cl−.
Fig. 4.
Chloride efflux-induced ASC specks are readily activated by subsequent potassium efflux. (A) LPS-primed BMDMs were incubated in a K+ and Cl− free solution (150 mM NaGluconate) and K+ was isosmotically added back with the indicated concentration of KGluconate. Involvement of the NLPR3 inflammasome was assessed by pretreatment with the NLRP3 inhibitor MCC950 (10 μM). Caspase-1 processing and IL-1β release was determined by ELISA and immunoblotting of cell lysates (LYS) and supernatants (S/N) (n = 4). (B) LPS-primed BMDMs were incubated in a control (145 mM NaCl/5 mM KCl) or Cl− free (145 mM NaGluconate/5 mM KGluconate) solution for 1 h before stimulation with nigericin (10 µM) for the indicated time (n = 3). Caspase-1 processing and IL-1β release was determined by ELISA and immunoblotting of cell LYS and S/N (n = 3). (C) LPS-primed THP-1 cells were incubated in a Cl− free (145 mM NaGluconate/5 mM KGluconate) solution for the indicated time points for up to 120 min, then reperfused with a Cl− containing (145 mM NaCl/5 mM KCl) or Cl− free solution, and ASC oligomerization was determined by immunoblotting (n = 3). (D) LPS-primed mCherry-ASC iBMDMs were incubated for 120 min in a Cl− free solutions before reperfusion with a Cl− containing or Cl− free solution and ASC speck-containing cells were quantified by immunofluorescence (n = 3). ****P < 0.0001; ns, not significantly different determined by a two-way ANOVA with Sidak’s post hoc analysis. Values shown are the mean ± SEM.
Effects of Chloride on NLRP3 Oligomerization.
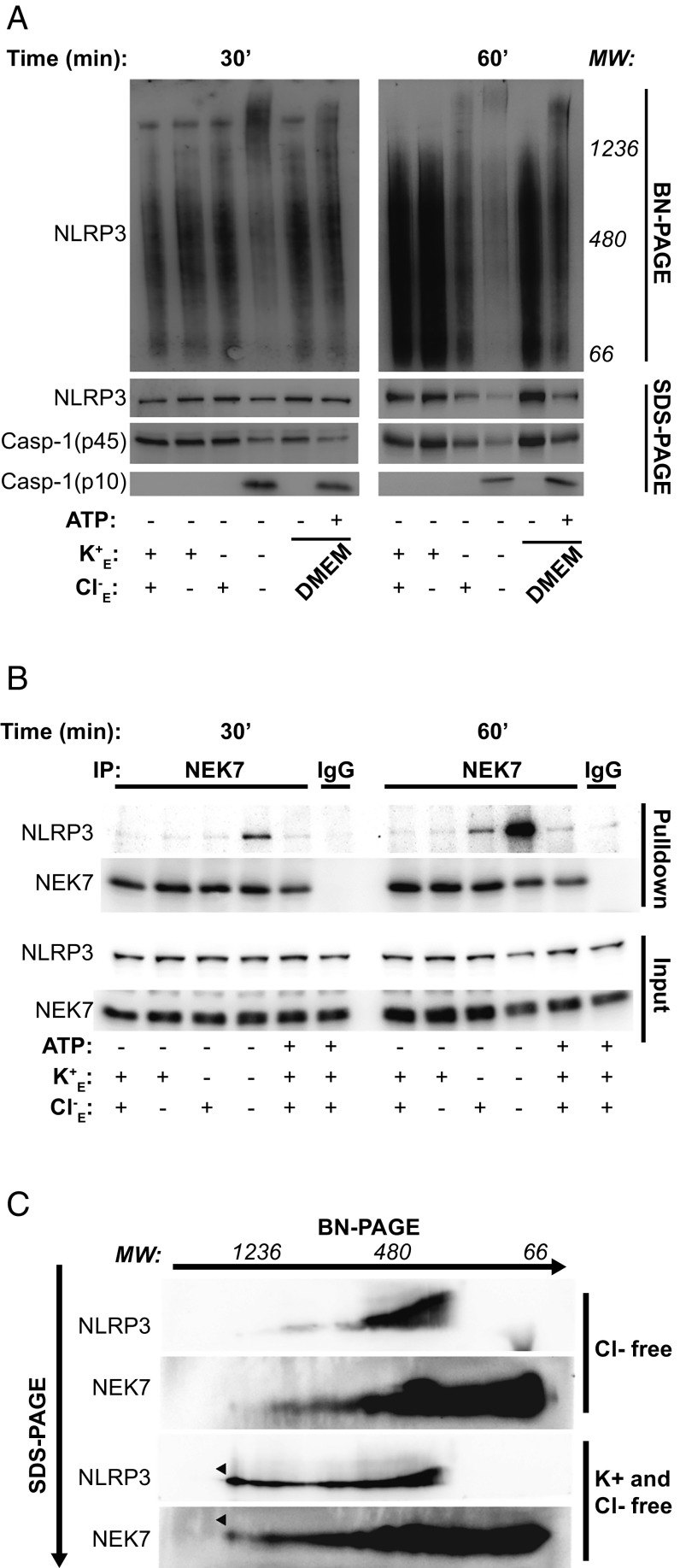
Although Cl− free ASC speck formation was shown to be NLRP3 dependent (Figs. 2 and 3), the fact that the specks were not recruiting active caspase-1 suggested that NLRP3 may not exist in an oligomerized active state under these conditions. To test the activation state of NLRP3, we assessed its oligomerization under conditions of Cl−, K+, or K+ and Cl− free. Using native gel electrophoresis as described previously (5, 29) and immunoblotting for NLRP3, we observed an increased shift in molecular weight of NLRP3 under conditions of K+ free, or K+ and Cl− free, but not of Cl− free alone (Fig. 5A). These data suggest that the formation of high molecular weight complexes containing oligomeric NLRP3 requires K+ efflux (Fig. 5A). We subsequently performed SDS/PAGE under denaturing conditions with these samples to examine processing of caspase-1. Caspase-1 cleavage occurred exclusively in conditions where NLRP3 oligomerization was present, suggesting NLRP3 oligomerization is necessary for caspase-1 cleavage (Fig. 5A). To confirm that NLRP3 oligomerization was different upon selective K+ and Cl− efflux, we performed bioluminescence resonance energy transfer (BRET) experiments using the NLRP3 BRET sensor as described previously (30). HEK293T cells expressing Luc Ct-NALP3-Nt YFP were incubated with different solutions and NLRP3 conformation was monitored over time. We found that both K+ free and Cl− free solutions were able to induce modest but different changes in the BRET signal (SI Appendix, Fig. S6A). Microscopy of the YFP label on the NLRP3 construct revealed that Cl− efflux did not induce NLRP3 speck formation, but when cells were incubated in K+ free solutions NLRP3 specks were observed (SI Appendix, Fig. S6B). These data are consistent with the data presented in Fig. 5A, suggesting that K+ efflux is necessary for NLRP3 oligomerization. The association between NEK7 and NLRP3 is reported to be dependent upon K+ efflux (5). Thus, we performed coimmunoprecipitations of NEK7 and NLRP3 under Cl− free, and K+ and Cl− free conditions. After 30 min in K+ and Cl− free solution we could detect a clear interaction between NEK7 and NLRP3 (Fig. 5B). NLRP3–NEK7 interaction was also apparent in K+ free (Cl− containing) conditions at 60 min, at which time the interaction was even more apparent in the K+ and Cl− free solution (Fig. 5B). To determine whether NEK7 was present in the oligomerized NLRP3 complexes formed under K+ and Cl− free conditions, we first separated samples in one dimension by native gel electrophoresis and then in a second dimension by SDS/PAGE. Immunoblotting revealed that under conditions of K+ and Cl− free, but not Cl− free alone, the formation of a high molecular weight complex including NEK7 and NLRP3 was induced (Fig. 5C). These data suggest that the K+ sensor, which is required for NLRP3 oligomerization and a functional caspase-1–activating inflammasome, is NEK7. Thus, while Cl− free conditions drive NLRP3-dependent ASC oligomerization and speck formation, in the absence of NEK7 binding and NLRP3 oligomerization, these specks remain inactive and do not recruit active caspase-1.
Fig. 5.
Chloride efflux-induced ASC specks do not require NLRP3 oligomerization, while active inflammasomes require NEK7-dependent NLRP3 oligomerization. (A) LPS-primed BMDMs were incubated for 30 or 60 min in a control (145 mM NaCl/5 mM KCl), Cl− free (145 mM NaGluconate/5 mM KGluconate), K+ free (150 mM NaCl), K+ and Cl− free (150 mM NaGluconate) solution, or in DMEM ± ATP (5 mM). Cell lysates were resolved using a blue-native PAGE or SDS/PAGE and immunoblotted for NLRP3 and caspase-1 (n = 3). (B) LPS-primed BMDMs were incubated for 30 or 60 min in a control, Cl− free, K+ free, K+ and Cl− free solution, or treated with ATP (5 mM) in the control solution. Cell lysates were immunoprecipitated (IP) with NEK7 or IgG antibodies and immunoblotted for NLRP3 and NEK7 (n = 2). (C) LPS-primed BMDMs were incubated for 30 min in a Cl− free or K+ and Cl− free solution. Cell lysates were resolved by 2D-PAGE and immunoblotted for NLRP3 and NEK7 (n = 2).
Effects of Chloride on the Alternative NLRP3 Inflammasome in Human Peripheral Blood Mononuclear Cells.
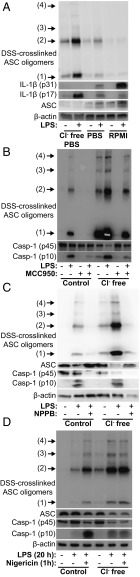
It has been known for some time that LPS can directly induce the release of IL-1β from freshly isolated human monocytes without the need for a secondary stimulus (31). More recent research has characterized this as the “alternative” NLRP3 inflammasome which is less dependent on K+ efflux and does not rely on ASC oligomerization (32). We therefore tested whether Cl− free conditions could influence IL-1β production by the alternative NLRP3 inflammasome in peripheral blood mononuclear cells (PBMCs) freshly isolated from healthy donors. When the cells were stimulated with LPS in RPMI media, there was no ASC oligomerization, although there was caspase-1 activation and mature IL-1β production (Fig. 6A). We repeated this experiment in Cl− containing and Cl− free PBS (supplemented with 10 mM glucose, 1% FBS, and 2 mM l-glutamine) and found that under these conditions there was some ASC oligomerization in response to LPS in PBS (Fig. 6A). Intriguingly, Cl− free PBS alone induced ASC oligomerization, caspase-1 cleavage, and IL-1β processing, which was markedly enhanced by the addition of LPS (Fig. 6A). The formation of these Cl− free dependent ASC oligomers was reduced by MCC950, confirming NLRP3 dependence (Fig. 6B). The Cl− channel inhibitor NPPB also inhibited Cl− free and LPS-induced ASC oligomerization in human PBMCs (Fig. 6C). We then repeated the experiment in PBMCs in PBS and Cl− free PBS plus and minus LPS and this time, nigericin. This experiment showed that nigericin induced ASC oligomerization and caspase-1 activation in LPS-treated PBMCs comparable to Cl− free conditions (Fig. 6D). These data suggest that Cl− free conditions in PBMCs do not activate the alternative pathway, but rather, activate the canonical pathway. Thus, the alternative NLRP3 inflammasome forms independently of ASC oligomerization, and thus independently of the Cl− dependent effect reported here.
Fig. 6.
Effects of chloride on LPS-induced inflammasome activation in human PBMCs. (A) Immunoblot of DSS-crosslinked ASC oligomers and IL-1β from human PBMCs incubated in RPMI, PBS, or Cl− free PBS ± LPS (1 µg⋅mL−1) for 20 h (n = 5). (B) Immunoblot of ASC oligomers and caspase-1 from PBMCs incubated in PBS or Cl− free PBS ± LPS (1 µg⋅mL−1) and the NLRP3 inhibitor MCC950 (5 µM) for 20 h (n = 3). (C) Immunoblot of ASC oligomers and caspase-1 from PBMCs incubated in PBS or Cl− free PBS ± LPS (1 µg⋅mL−1) and the chloride channel inhibitor NPPB (100 µM) for 24 h (n = 2). (D) PBMCs were treated as in A ± nigericin (10 µM) added for the last 1 h of treatment (n = 2).
Discussion
These data reveal a mechanism regulating the assembly of the NLRP3 inflammasome and allow us to propose a model that opens future studies on an additional priming and regulatory step that may further amplify inflammatory responses. Here we show that there is a Cl− channel-dependent step whereby NLRP3 nucleates the oligomerization of an ASC speck. This complex can remain inactive; however, K+ efflux permits NEK7 to bind to NLRP3 causing its oligomerization and activation of the complex. Altered ion balance and transport is a well-established contributor to inflammatory disease (33), and we recently reported that the fenamate nonsteroidal antiinflammatory drugs (NSAIDs) inhibit inflammation by virtue of their ability to inhibit Cl− channels on macrophages (18). We can thus envisage situations where cells are primed to contain fully formed inflammasome specks which will allow a heightened and rapid inflammatory response to subsequent inflammatory situations. It is therefore possible to consider cellular regulation of Cl− as a therapeutic target in inflammatory disease.
Following our publication describing the fenamate NSAIDs as NLRP3 inhibitors through their ability to inhibit Cl− channels (18), several subsequent studies have further reported the importance of Cl− channel activation, and they point to the importance of chloride intracellular channel (CLIC) proteins (19, 20). Interestingly there is evidence to suggest that the NLRP3 inhibitor MCC950 could be a CLIC inhibitor as a related analog was shown to bind CLIC1 (19, 34), and more recently MCC950 was reported to inhibit Cl− flux from cells (35), although we did not observe this after volume-regulated anion channel activation (SI Appendix, Fig. S1). LPS activation of macrophages drives CLICs to the plasma membrane where they can facilitate Cl− flux after NLRP3 inflammasome-activating stimuli, and knockdown of CLICs reduces NLRP3 inflammasome-dependent release of IL-1β (19, 20). Furthermore, previous research examining the effects of Cl− on P2x7-dependent IL-1β release attributed enhanced responses in Cl− free conditions to allosteric effects on the P2x7 receptor, and the potential of increased intracellular Cl− to negatively regulate the activation of caspase-1 (36). These studies, our previous research (18), and several other studies from the literature showing that Cl− channel inhibitors block the release of IL-1β (37–39), strongly support an important role for Cl− efflux in inflammasome assembly and IL-1β secretion.
In this study, we have dissociated Cl− dependent effects from those caused by K+ efflux and report that NLRP3-dependent ASC oligomerization is Cl− efflux dependent, dynamic, and reversible. ASC speck formation is generally considered an essential and sufficient event for NLRP3-dependent inflammasome activation. We report here formation of ASC specks without the downstream activation of caspase-1 and release of IL-1β. We show that Cl− efflux-dependent preformed inflammasome specks may represent a further phase of priming to accelerate and amplify inflammatory responses, potentially in a positive feedback loop as recently described for the amplification of AIM2 oligomerization (40). When there is a concomitant efflux of K+ we report that NEK7 binds to NLRP3 leading to caspase-1 recruitment and the formation of an active inflammasome (Fig. 5). Cl− efflux-dependent ASC specks can also undergo a rapid disassembly following restoration of intracellular Cl− levels providing insight into the dynamic nature of these complexes (Fig. 4). These insights now open the door for studies of inflammasome assembly in the absence of pyroptotic cell death and reveal further levels of regulation for the NLRP3 inflammasome. Further understanding of the effects of Cl− will reveal and refine future and current therapeutic targets, respectively. Given that there are Cl− channel inhibiting drugs available and that the well-characterized inhibitor MCC950 may be a CLIC inhibitor suggests that there is considerable therapeutic potential in understanding the contribution of Cl− channels to inflammation. Our data also highlight the importance of understanding the regulation of inflammasome complexes when considering therapeutic strategies. In conditions where alternative NLRP3 inflammasomes contribute to disease, Cl− channel inhibition is unlikely to be a viable therapeutic strategy (Fig. 6). Rather, diseases dependent upon ASC speck formation, such as Alzheimer’s disease (41), may be more likely to respond specifically to therapies targeting formation of the ASC speck. Indeed in rodent models of Alzheimer’s disease, Cl− channel inhibitors are protective (18). Further understanding of inflammasome activation in disease models will therefore enable a more refined approach to inflammasome targeting.
Methods
Antibodies and Reagents.
Details are in SI Appendix.
Cell Culture.
Primary BMDMs were prepared by flushing femurs of male and female WT C57BL/6 mice (Envigo) or NLRP3 KO mice. All procedures were performed with appropriate personal and project licenses in place, in accordance with the Home Office (Animals) Scientific Procedures Act (1986), and approved by the Home Office and the local Animal Ethical Review Group, University of Manchester. Red blood cells were lysed and BMDMs were generated by culturing the resulting marrow cells in 70% DMEM [containing 10% (vol/vol) FBS, 100 U⋅mL−1 penicillin, 100 μg⋅mL−1 streptomycin] supplemented with 30% L929 mouse fibroblast-conditioned media for 7–10 d. Before experiments, BMDMs were seeded overnight at a density of 1 × 106 mL−1. Human WT and NLRP3 KO THP-1 cells were cultured in RPMI-1640 medium supplemented with 10% (vol/vol) FBS, 100 U⋅mL−1 penicillin, and 100 μg⋅mL−1 streptomycin, 2 mM l-glutamine, and 55 μM 2-mercaptoethanol. Before experiments, THP-1 cells were seeded overnight at a density of 1 × 106 mL−1 in media supplemented with 500 nM phorbol 12-myristate 13-acetate (PMA). iBMDMs and iBMDMs stably expressing ASC-mCherry (18) were cultured in DMEM supplemented with 10% (vol/vol) FBS, 100 U⋅mL−1 penicillin, and 100 μg⋅mL−1 streptomycin. Before experiments, iBMDMs were seeded overnight at a density of 0.75 × 106 mL−1. HeLa cells were cultured in DMEM supplemented with 10% (vol/vol) FBS, 100 U⋅mL−1 penicillin, and 100 μg⋅mL−1 streptomycin. HeLa cells were seeded out at a density of 1.2 × 105 mL−1. HEK293 constitutively expressing Luc Ct-Nlrp3-Nt YFP (30) were maintained in DMEM: F12 (1:1) supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, and 0.3 mg⋅mL−1 G418.
Human PBMCs were obtained with consent from healthy human donors (National Health Service Blood and Transplant, Manchester, UK), with full ethical approval from the Research Governance, Ethics, and Integrity Committee at the University of Manchester (ref. 2018-2696-5711). Briefly, PBMCs were separated from leukocyte cones using Ficoll reagent (Thermo Fisher) at 400 × g for 40 min at room temperature (RT). The layers containing PBMCs were gently isolated using a Pasteur pipette and washed at least three times in sterile PBS supplemented with 1% FBS and 2 mM EDTA to remove debris and platelets. PBMCs were counted and seeded out in RPMI-1640 medium supplemented with 1% (vol/vol) FBS and 2 mM l-glutamine in 12-well plates at 5 × 10−6 density per well. PBMCs were then stimulated with or without 1 µg⋅mL−1 LPS for 20 h. For the buffer stimulations, PBMCs were directly resuspended in PBS or Cl− free PBS supplemented with 10 mM d-glucose, 1% FBS, and 2 mM l-glutamine, and stimulated with or without 1 µg⋅mL−1 LPS for 20 h. Both cell supernatants and lysates were collected for immunoblotting.
Inflammasome Activation Assays.
Primary BMDMs and THP-1 cells were primed with 1 μg⋅mL−1 LPS (from Escherichia coli O26:B6; Sigma) for 4 h. iBMDMs were primed with 1 μg⋅mL−1 LPS for 2 h. After priming, cells were reperfused with the indicated isotonic salt solution: control (145 mM NaCl/5 mM KCl), Cl− free (145 mM NaGluconate/5 mM KGluconate), K+ free (150 mM NaCl), or K+ and Cl− free (150 mM NaGluconate). When specified, solutions were supplemented with CaCl2 (2 mM), glucose (10 mM), or Hepes (10 mM, pH 7.3). When used, chloride channel and NLRP3 inhibitors were added to the solutions just before reperfusion. For experiments where NLRP3 agonists were used, cells were incubated in either serum-free DMEM (primary BMDMs, iBMDMs) or RPMI-1640 (THP-1) media and stimulated with nigericin (10 μM, 1 h) or ATP (5 mM, 1 h). When used, chloride channel and NLRP3 inhibitors were pretreated for 15 min before inflammasome stimulation. Supernatants were collected and cells were lysed in RIPA buffer [150 mM NaCl, 1% (vol/vol) Nonidet P-40, 0.5% (wt/vol) sodium deoxycholate, 0.1% (wt/vol) SDS, 50 mM Tris, pH 8], supplemented with a protease inhibitor mixture.
IL-1β release was determined in supernatants by ELISA (DuoSet, R&D Systems) according to the manufacturer’s instructions. IL-1β and caspase-1 processing was determined by immunoblotting. When necessary, both cell supernatant and lysates were collected and precipitated in deoxycholate containing 20% trichloroacetic acid (20% TCA) and washed three times with 100% acetone, before concentration in 1× Laemmli buffer. All supernatants or cell lysates were separated by Tris-glycine SDS/PAGE and then transferred onto nitrocellulose or PVDF membranes at 25 V using a semidry Trans-Blot Turbo system (Bio-Rad). Membranes were blocked in 5% (wt/vol) milk in PBS, 1% (vol/vol) Tween 20 (PBST) before being incubated with indicated primary antibodies at 4 °C overnight. Membranes were then labeled with HRP-tagged secondary antibodies and visualized with Amersham ECL detection reagent (GE Healthcare). Immunoblot images were captured digitally using a G:Box Chemi XX6 (Syngene) or C-digit scanner (Li-Cor). Cell supernatants were also assayed for lactate dehydrogenase (LDH) release using CytoTox 96 nonradioactive cytotoxicity assay (Promega) according to manufacturer’s instructions.
Intracellular K+ and Cl− Determination.
Intracellular K+ measurements were performed as previously described (42). Full details on K+ and Cl− measurements are in SI Appendix.
H148Q/I152L YFP Quenching Assay.
A cell-based assay for measuring chloride channel activity was performed as previously described with pcDNA3.1 EYFP H148Q/I152L (22), a gift from Peter Haggie, University of California, San Francisco (Addgene plasmid 25872). Further details are in SI Appendix.
ASC Speck Imaging.
Real-time ASC speck assays were performed using iBMDMs stably expressing ASC-mCherry (ASC-mCherry iBMDMs). ASC-mCherry iBMDMs were seeded overnight into 96-well plates at a density of 0.75 × 105 mL−1 and subsequently primed for 2 h with 1 μg⋅mL−1 LPS. To prevent pyroptosis and detachment during the experiment, iBMDMs were pretreated with the pan-caspase inhibitor Z-VAD-FMK (50 µM) for 30 min before microscopy. After priming, cells were reperfused with the indicated isotonic salt solution: control, Cl− free, K+ free, K+ and Cl− free, high K+ and normal Cl−, or high K+ and Cl− free. When appropriate, cells were treated with either a vehicle control, DCPIB (10 µM), flufenamic acid (100 µM), NPPB (100 µM), or the NLRP3 inflammasome inhibitor MCC950 (10 µM). Images were captured using a 20×/0.61 S Plan Fluor objective at 15-min intervals and quantified using an IncuCyte ZOOM System (Essen Bioscience). Comparison of ASC speck formation between treatments was analyzed after stimulation for 3 h. All experiments involving the ANO1 agonist Eact were performed in OptiMEM.
WT and NLRP3 KO THP-1 cells were seeded overnight onto sterilized 10-mm coverslips in 24-well plates at a density of 1 × 106 mL−1 cells, supplemented with PMA (500 nM). THP-1 macrophages were primed with LPS (1 µg⋅mL−1, 4 h) before reperfusion with the indicated isotonic salt solution: control, Cl− free, K+ free, or K+ and Cl− free and incubated for 3 h. All solutions contained Z-VAD-FMK (50 µM) to prevent pyroptosis and therefore loss of ASC specks. Cells were washed with cold PBS and then fixed with 4% (wt/vol) PFA for 20 min. Fixed cells were blocked and permeabilized with 1% (wt/vol) BSA, 0.1% (vol/vol) Triton X-100, and 0.05% (vol/vol) Tween 20 in PBS. Anti-ASC antibodies were incubated in 1% (wt/vol) BSA, 0.3% (vol/vol) Triton X-100 in PBS overnight at 4 °C. Coverslips were then washed three times with PBS before incubation with anti-rabbit AlexaFluor 488 secondary antibodies (Thermo Fisher) for 1 h at RT. After a further three washes with PBS, cells were stained with 1 μg⋅mL−1 DAPI. Samples were washed three times before being mounted in Prolong Diamond (Thermo Fisher). For experiments testing reversal of ASC specks, ASC-mCherry iBMDMs were seeded overnight onto sterilized 10-mm coverslips at a density of 0.75 × 106 mL−1. ASC-mCherry iBMDMs were primed with LPS (1 µg⋅mL−1, 2 h) and stimulated as indicated in the presence of Z-VAD-FMK (50 μM). ASC-mCherry iBMDMs were washed with cold PBS and fixed with 4% (wt/vol) PFA for 20 min. Fixed cells were stained with 1 μg⋅mL−1 DAPI and washed a further three times in PBS before being mounted in Prolong Diamond. Images were captured using a 20× objective on a Zeiss Axioimager D2 upright microscope and subsequently processed using ImageJ software.
ASC Oligomerization Assay.
WT and NLRP3 KO THP-1 cells were seeded into 12-well plates at a density of 1 × 106 mL−1 and incubated overnight with PMA (500 nM). Following LPS priming (1 µg⋅mL−1, 4 h), cells were incubated in the indicated isotonic salt solution: control, Cl− free, K+ free, or K+ and Cl− free, for 2 h. Cells were directly lysed in the well by the addition of 1% (vol/vol) Triton X-100 and protease inhibitor mixture. Cell lysates were separated into Triton X-100 soluble fraction and insoluble fraction using differential centrifugation at 6,800 × g for 20 min at 4 °C. The soluble fraction of cell lysates was subsequently used for immunoblotting, whereas the Triton X-100 insoluble pellets were chemically crosslinked with 2 mM DSS (Thermo Fisher) for 30 min at RT. Crosslinked pellets were further spun down at 6,800 × g for 20 min and eluted in Laemmli buffer for SDS/PAGE.
Active Caspase-1 Pulldown.
For active caspase-1 pulldown, a cell-permeable pan-caspase activity probe bVAD-fmk (bVAD) (Santa Cruz: sc-311290) was used as previously described (25). Briefly, 1.5 × 106 WT BMDMs were seeded into 12-well plates and incubated overnight. A total of 10 µM bVAD was premixed in stimulating conditions before reperfused onto LPS-primed (1 µg⋅mL−1, 4 h) BMDMs for 1 h. Cell supernatant containing excess bVAD was removed. For caspase-1 pulldown, cells were lysed overnight in 1% Triton X-100 lysis buffer supplemented with EDTA-free protease inhibitor mixture and 1 mM PMSF. Cell lysates were then incubated in magnetic streptavidin T1 Dynabeads (Thermo Fisher) for 1 h. The proteins bound to streptavidin were washed three times using 1% Triton X-100 lysis buffer before being resuspended in 1× Laemmli buffer and boiled before immunoblotting.
Blue Native PAGE and 2D PAGE.
For 1D Blue Native-PAGE (BN-PAGE), 2 × 106 primary WT and NLRP3 KO BMDMs were seeded overnight into six-well plates. LPS-primed BMDMs (1 µg⋅mL−1, 4 h) were incubated in a control, Cl− free, K+ free, or K+ and Cl− free solution for 30 or 60 min. Alternatively, BMDMs were incubated in serum-free DMEM and stimulated with ATP (5 mM, 30 or 60 min). For 2D-PAGE, Z-VAD-FMK (50 µM) was added simultaneously to prevent loss of protein. Cells were lysed in a native lysis buffer [20 mM Bis-Tris, 500 mM 6-aminocaproic acid, 20 mM NaCl, 10% (vol/vol) glycerol, 0.25% (wt/vol) digitonin, 0.25% (vol/vol) Triton X-100, 0.5 mM Na3VO4, 0.5 mM NaF, 1 mM PMSF, EDTA-free protease inhibitor mixture, pH 7.0] for 30 min at 4 °C. Insoluble debris was pelleted by centrifugation at 20,000 × g for 30 min at 4 °C. Soluble lysates were separated by isoelectric focusing using Coomassie Brilliant Blue G-250 (Fluka) on 3–12% NativePAGE BisTris gels (Thermo Fisher) as described previously (5, 43). Proteins were then denatured by incubating the gel in 10% SDS for 10 min before semidry transfer and immunoblotting with indicated antibodies. For second dimension PAGE, 6.5 cm of BN-PAGE gel slices was cut and denatured in Laemmli buffer. The gel slices were rotated 90° counterclockwise and transferred into a precast SDS/PAGE for immunoblotting.
Immunoprecipitation.
The 1 × 106 LPS-primed BMDMs (1 µg⋅mL−1, 4 h) were incubated in a control, Cl− free, K+ free, K+ and Cl− free solution, or stimulated with ATP (5 mM) for 30 or 60 min. Cells were lysed in a Nonidet P-40 lysis buffer [0.5% (vol/vol) Nonidet P-40, 50 mM Tris pH 7.4, 150 mM NaCl, 2 mM EDTA, protease inhibitor mixture]. Lysates were precleared by incubation with protein G magnetic beads (Thermo Fisher) for 1 h at 4 °C. Precleared lysates were immunoprecipitated with anti-NEK7 or anti-rabbit IgG (EPR25A, Abcam) overnight. The proteins bound to the antibodies were then pulled down by protein G magnetic beads. Beads were then washed three times in Nonidet P-40 lysis buffer and proteins were eluted from the beads by boiling in Laemmli buffer at 95 °C for 10 min. Proteins were then detected by immunoblotting.
BRET Assay.
BRET experiments were performed as described previously (30). Further details are in SI Appendix.
Quantification and Statistical Analysis.
Data are presented as mean values plus the SEM. Accepted levels of significance were *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Statistical analyses were carried out using GraphPad Prism (version 7). Data with multiple groups were analyzed using a one-way ANOVA followed by Tukey’s corrected post hoc comparisons. Alternatively, in the case where comparisons were made against a vehicle control, a Dunnett’s post hoc comparison was used. Experiments with two independent variables were analyzed using a two-way ANOVA followed by a Sidak’s post hoc corrected analysis. Percentage control data were analyzed with Holm–Sidak corrected one-sample t test against the value of 100%. Equal variance and normality were assessed with Levene’s test and the Shapiro–Wilk test, respectively, and appropriate transformations were applied when necessary. n represents experiments performed on individual animals (in the case of primary BMDMs) or different passage (in the case of iBMDMs and THP-1s).
Supplementary Material
Acknowledgments
We thank Dr. Vishva Dixit (Genentech) for providing the NLRP3 KO mice; Dr. Veit Hornung (Ludwig Maximilian University, Munich) for providing the NLRP3 KO THP-1 cells; and Mr. Alistair Bewsher and Dr. Richard Unwin (both University of Manchester) for performing the ion chromatography analysis on intracellular Cl− levels and the ICP-MS to measure K+. This work was supported by Medical Research Council Grant MR/N029992/1 (to D.B. and C.B.L.) and European Research Council ERC-2013-CoG Grant red 614578 (to P.P.). S.Y. is funded by the Faculty at the University of Manchester on a Joint China Council Scholarship and Presidential Doctoral Scholar award. G.L.-C. is funded by Wellcome Trust and the Royal Society (Grant 104192/Z/14/Z).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. I.E.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812744115/-/DCSupplemental.
References
- 1.Broz P, Dixit VM. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 2.Dinarello CA, Simon A, van der Meer JW. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11:633–652. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Netea MG, et al. A guiding map for inflammation. Nat Immunol. 2017;18:826–831. doi: 10.1038/ni.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liston A, Masters SL. Homeostasis-altering molecular processes as mechanisms of inflammasome activation. Nat Rev Immunol. 2017;17:208–214. doi: 10.1038/nri.2016.151. [DOI] [PubMed] [Google Scholar]
- 5.He Y, Zeng MY, Yang D, Motro B, Núñez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530:354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi H, et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol. 2016;17:250–258. doi: 10.1038/ni.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dick MS, Sborgi L, Rühl S, Hiller S, Broz P. ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat Commun. 2016;7:11929. doi: 10.1038/ncomms11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brough D, Pelegrin P, Nickel W. An emerging case for membrane pore formation as a common mechanism for the unconventional secretion of FGF2 and IL-1β. J Cell Sci. 2017;130:3197–3202. doi: 10.1242/jcs.204206. [DOI] [PubMed] [Google Scholar]
- 10.Franklin BS, et al. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat Immunol. 2014;15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baroja-Mazo A, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol. 2014;15:738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- 12.Hornung V, Latz E. Critical functions of priming and lysosomal damage for NLRP3 activation. Eur J Immunol. 2010;40:620–623. doi: 10.1002/eji.200940185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stutz A, et al. NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J Exp Med. 2017;214:1725–1736. doi: 10.1084/jem.20160933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juliana C, et al. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem. 2012;287:36617–36622. doi: 10.1074/jbc.M112.407130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Castejon G, et al. Deubiquitinases regulate the activity of caspase-1 and interleukin-1β secretion via assembly of the inflammasome. J Biol Chem. 2013;288:2721–2733. doi: 10.1074/jbc.M112.422238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song N, et al. NLRP3 phosphorylation is an essential priming event for inflammasome activation. Mol Cell. 2017;68:185–197.e186. doi: 10.1016/j.molcel.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Muñoz-Planillo R, et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniels MJ, et al. Fenamate NSAIDs inhibit the NLRP3 inflammasome and protect against Alzheimer’s disease in rodent models. Nat Commun. 2016;7:12504. doi: 10.1038/ncomms12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domingo-Fernández R, Coll RC, Kearney J, Breit S, O’Neill LAJ. The intracellular chloride channel proteins CLIC1 and CLIC4 induce IL-1β transcription and activate the NLRP3 inflammasome. J Biol Chem. 2017;292:12077–12087. doi: 10.1074/jbc.M117.797126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang T, et al. CLICs-dependent chloride efflux is an essential and proximal upstream event for NLRP3 inflammasome activation. Nat Commun. 2017;8:202. doi: 10.1038/s41467-017-00227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 22.Galietta LJ, Haggie PM, Verkman AS. Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS Lett. 2001;499:220–224. doi: 10.1016/s0014-5793(01)02561-3. [DOI] [PubMed] [Google Scholar]
- 23.Baldwin AG, et al. Boron-based inhibitors of the NLRP3 inflammasome. Cell Chem Biol. 2017;24:1321–1335.e1325. doi: 10.1016/j.chembiol.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandes-Alnemri T, Alnemri ES. Assembly, purification, and assay of the activity of the ASC pyroptosome. Methods Enzymol. 2008;442:251–270. doi: 10.1016/S0076-6879(08)01413-4. [DOI] [PubMed] [Google Scholar]
- 25.Boucher D, et al. Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J Exp Med. 2018;215:827–840. doi: 10.1084/jem.20172222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coll RC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Namkung W, Yao Z, Finkbeiner WE, Verkman AS. Small-molecule activators of TMEM16A, a calcium-activated chloride channel, stimulate epithelial chloride secretion and intestinal contraction. FASEB J. 2011;25:4048–4062. doi: 10.1096/fj.11-191627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo Y, et al. Ani9, a novel potent small-molecule ANO1 inhibitor with negligible effect on ANO2. PLoS One. 2016;11:e0155771. doi: 10.1371/journal.pone.0155771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swamy M, Siegers GM, Minguet S, Wollscheid B, Schamel WW. Blue native polyacrylamide gel electrophoresis (BN-PAGE) for the identification and analysis of multiprotein complexes. Sci STKE. 2006;2006:pl4. doi: 10.1126/stke.3452006pl4. [DOI] [PubMed] [Google Scholar]
- 30.Martín-Sánchez F, Compan V, Pelegrín P. Measuring NLR oligomerization III: Detection of NLRP3 complex by bioluminescence resonance energy transfer. Methods Mol Biol. 2016;1417:159–168. doi: 10.1007/978-1-4939-3566-6_10. [DOI] [PubMed] [Google Scholar]
- 31.Perregaux DG, Laliberte RE, Gabel CA. Human monocyte interleukin-1beta posttranslational processing. Evidence of a volume-regulated response. J Biol Chem. 1996;271:29830–29838. doi: 10.1074/jbc.271.47.29830. [DOI] [PubMed] [Google Scholar]
- 32.Gaidt MM, et al. Human monocytes engage an alternative inflammasome pathway. Immunity. 2016;44:833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 33.Eisenhut M. Changes in ion transport in inflammatory disease. J Inflamm (Lond) 2006;3:5. doi: 10.1186/1476-9255-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laliberte RE, et al. Glutathione s-transferase omega 1-1 is a target of cytokine release inhibitory drugs and may be responsible for their effect on interleukin-1beta posttranslational processing. J Biol Chem. 2003;278:16567–16578. doi: 10.1074/jbc.M211596200. [DOI] [PubMed] [Google Scholar]
- 35.Jiang H, et al. Identification of a selective and direct NLRP3 inhibitor to treat inflammatory disorders. J Exp Med. 2017;214:3219–3238. doi: 10.1084/jem.20171419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhoef PA, Kertesy SB, Lundberg K, Kahlenberg JM, Dubyak GR. Inhibitory effects of chloride on the activation of caspase-1, IL-1beta secretion, and cytolysis by the P2X7 receptor. J Immunol. 2005;175:7623–7634. doi: 10.4049/jimmunol.175.11.7623. [DOI] [PubMed] [Google Scholar]
- 37.Laliberte R, Perregaux D, Svensson L, Pazoles CJ, Gabel CA. Tenidap modulates cytoplasmic pH and inhibits anion transport in vitro. II. Inhibition of IL-1 beta production from ATP-treated monocytes and macrophages. J Immunol. 1994;153:2168–2179. [PubMed] [Google Scholar]
- 38.Perregaux DG, Svensson L, Gabel CA. Tenidap and other anion transport inhibitors disrupt cytolytic T lymphocyte-mediated IL-1 beta post-translational processing. J Immunol. 1996;157:57–64. [PubMed] [Google Scholar]
- 39.Compan V, et al. Cell volume regulation modulates NLRP3 inflammasome activation. Immunity. 2012;37:487–500. doi: 10.1016/j.immuni.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Matyszewski M, Morrone SR, Sohn J. Digital signaling network drives the assembly of the AIM2-ASC inflammasome. Proc Natl Acad Sci USA. 2018;115:E1963–E1972. doi: 10.1073/pnas.1712860115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venegas C, et al. Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature. 2017;552:355–361. doi: 10.1038/nature25158. [DOI] [PubMed] [Google Scholar]
- 42.Church SJ, et al. Deficient copper concentrations in dried-defatted hepatic tissue from ob/ob mice: A potential model for study of defective copper regulation in metabolic liver disease. Biochem Biophys Res Commun. 2015;460:549–554. doi: 10.1016/j.bbrc.2015.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.