Significance
The polymerase-associated factor 1 (PAF1) complex is a general transcription elongation factor of RNA polymerase II, which not only regulates various stages of the transcription cycle but also broadly influences gene expression through modulating chromatin structure and/or recruiting other transcription-related factors. This study presents a high-resolution crystal structure of the core region of the Paf1-Ctr9-Cdc73 ternary complex, which not only greatly facilitates our understanding of the overall architecture of the Paf1 complex but also provides a structure-based platform for understanding the molecular mechanism underlying the role of the Paf1 complex in regulating gene expression and sheds light toward deciphering the impact of its mutational spectrum on human diseases.
Keywords: Paf1 complex, transcription elongation, crystal structure
Abstract
The polymerase-associated factor 1 (Paf1) complex is a general transcription elongation factor of RNA polymerase II, which is composed of five core subunits, Paf1, Ctr9, Cdc73, Leo1, and Rtf1, and functions as a diverse platform that broadly affects gene expression genome-wide. In this study, we solved the 2.9-Å crystal structure of the core region composed of the Ctr9-Paf1-Cdc73 ternary complex from a thermophilic fungi, which provides a structural perspective of the molecular details of the organization and interactions involving the Paf1 subunits in the core complex. We find that Ctr9 is composed of 21 tetratricopeptide repeat (TPR) motifs that wrap three circular turns in a right-handed superhelical manner around the N-terminal region of an elongated single-polypeptide–chain scaffold of Paf1. The Cdc73 fragment is positioned within the surface groove of Ctr9, where it contacts mainly with Ctr9 and minimally with Paf1. We also identified that the Paf1 complex preferentially binds single-strand–containing DNAs. Our work provides structural insights into the overall architecture of the Paf1 complex and paves the road forward for understanding the molecular mechanisms of the Paf1 complex in transcriptional regulation.
RNA polymerase II (Pol II) mediated transcription is responsible for the expression of most coding genes, a process which can be divided into three major stages: initiation, elongation, and termination. In eukaryotes, transcriptional elongation is a highly regulated and complicated process which affects the integrity of the genome (1). In metazoans, promoter-proximal pausing is a widespread feature of gene regulation (2). Specific transcription elongation factors are needed to help Pol II to overcome obstacles in the elongation process and to release paused Pol II. The polymerase-associated factor I (Paf1) complex is one of these transcription elongation factors, which not only broadly affects gene expression in all three stages of transcription, but also plays a critical role in the release of promoter-proximal paused Pol II (3, 4).
The Paf1 complex is a five-subunit eukaryotes-specific transcription elongation factor, first identified in budding yeast, which is composed of Paf1, Ctr9, Cdc73, Leo1, and Rtf1 (5). In humans, the Paf1 complex contains a sixth subunit, Ski8. Although the Rtf1 subunit is central to the function of the Paf1 complex, it does not associate stably with the Paf1 complex in some species (6). The Paf1 complex is a multifunctional complex, which is closely related to many transcription-related processes and can regulate gene expression through diverse mechanisms. The Paf1 complex can regulate transcription through direct interaction with Pol II or through coordinated interactions with other transcription factors. Both yeast and human Paf1 complexes have been shown to have positive roles in transcription elongation (7, 8). In budding yeast, the Paf1 complex is recruited to the ORFs of actively transcribed genes (9) and has been shown to have extensive genetic and physical links to transcription elongation factors including the Spt4-Spt5 complex and the FACT complex (10–12). The human Paf1 complex can also regulate transcription elongation in synergy with the transcription factor SII (7). The Paf1 complex can regulate transcription through modulating histone modifications, with the Paf1 complex required for histone H3 trimethylation at K36 and monoubiquitylation of histone H2B at position K120 in humans (13). H2B ubiquitylation at K120 is required for the methylation of H3K4 and H3K79 (14–16), which are marks of actively transcribed genes. The Paf1 complex has also been shown to repress small-RNA–mediated epigenetic gene silencing (17), which defines an important role of the Paf1 complex in building up epigenetic memory.
Progress on molecular-based mechanistic understanding of Paf1 complex function has been slow. Structural studies to date revealed that Paf1 and Leo1 form a complex through antiparallel β-sheet interactions (18). Cdc73 contains a GTPase-like domain at the C terminus and a rigid N-terminal domain (19–21). Rtf1 has a plus3 domain, which can bind single-stranded DNA and phosphorylated C-terminal repeats of Spt5 (22, 23). Currently, there is no structural information on the largest subunit Ctr9, nor on the overall architecture of the complex. A cryo-EM structure of the Paf1 complex–Pol II-TFIIS complex reveals that the Paf1 complex has an elongated shape (24), but no structural details could be elucidated due to the lower resolution of the Paf1 component of the complex.
To understand the architecture and structural details of the Paf1 complex, we reconstituted the Paf1 complex from both budding yeast Saccharomyces cerevisiae (designated as scPaf1 complex) and a thermophilic fungi Myceliophthora thermophila (designated as mtPaf1 complex). We mapped interactions among subunits of the Paf1 complex. We reconstituted Paf1 subcomplexes in vitro and solved the crystal structure of the three-subunit ternary core complex of mtPaf1 at 2.9-Å resolution. In the structure of the complex, Ctr9 is composed of 21 tetratricopeptide repeat (TPR) motifs, which tightly wrap around the Paf1 fragment, spanning three circular turns in a right-handed superhelical manner. Cdc73 binds the surface groove of Ctr9 where it makes major contacts with Ctr9 and minor contacts with Paf1. We further identified that both the scPaf1 and the mtPaf1 complexes prefer to bind single-strand–containing DNAs. Our work presents the structural details of intermolecular alignments within the three-subunit mtPaf1 complex core components and sheds light onto the overall architecture of the full-length Paf1 complex.
Results
In Vitro Reconstitution of the Paf1 Complex.
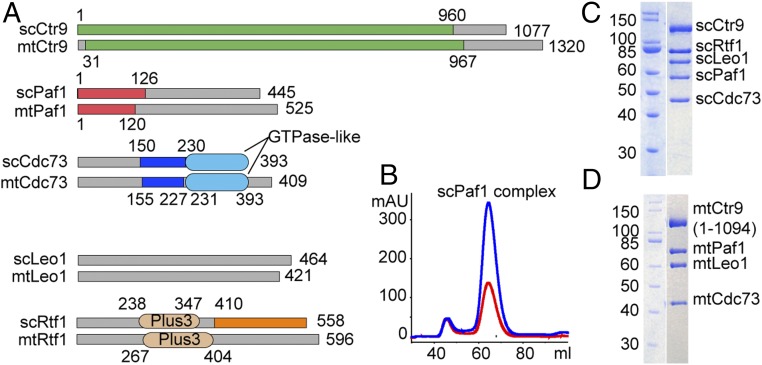
To understand the architecture of the Paf1 complex, we reconstituted the full-length five-subunit yeast scPaf1 complex (Fig. 1 A–C), and several combinations of three- or four-subunit subcomplexes in Escherichia coli by a coexpression protocol or through in vitro reconstitution of the complexes with purified subunits. We found that Paf1 and Ctr9 are two key members of the complex, as in the absence of either, the other three- or four-subunit subcomplexes did not stably form in vitro. We also reconstituted the Paf1 complex from a thermophilic fungi (Myceliophthora thermophile). The five-subunit mtPaf1 complex could be obtained through a one-step affinity purification protocol. However, the mtRtf1 subunit dissociated from the complex during further purification steps, especially when bound DNAs were removed (SI Appendix, Fig. S1A). Addition of the purified mtRtf1 back to the remaining four-subunit complex could not restore a five-subunit complex. This indicates that for the mtPaf1 complex, the Rtf1 subunit may need other partners, such as some type of nucleic acid or protein factors, to stably associate with the other components. This observation is consistent with the controversial behavior of Rtf1 in other species (6). Without Rtf1, the other four subunits of the mtPaf1 complex associate with one another in a 1:1:1:1 stoichiometry (Fig. 1D).
Fig. 1.
Reconstitution of both scPaf1 and mtPaf1 complexes. (A) Schematic drawing of the subunits of the Paf1 complex. Known domains in Cdc73 and Rtf1 are labeled. Crystallized regions in this study for both scPaf1 and mtPaf1 subunits are color-coded. (B) Gel-filtration chromatogram of the scPaf1 complex. (C) SDS/PAGE analysis of the full-length five-subunit scPaf1 complex. (D) SDS/PAGE analysis of the Ctr9-truncated four-subunit mtPaf1 complex.
Identification of the Rigid Structural Core of the Paf1 Complex.
As the full-length scPaf1 complex could not be crystallized, we tested removal of some flexible regions or domains from each subunit so as to generate a rigid structural core of the complex. We tried many combinations of complexes bearing various deletions and by chance found that only one batch of samples could be crystallized. Unexpectedly, some components of that batch of scPaf1 complex samples degraded into smaller pieces during crystallization. By N/C terminus analysis and Edman sequencing analysis, we identified three fragments of the Paf1 complex, namely, Ctr9(1-960), Paf1(1-126), and Rtf1(410-558), in the degraded sample. We also found that Cdc73 was cleaved at the position of residue 150 (SI Appendix, Fig. S1B). Further experiments verified that the above three fragments plus a middle region of Cdc73(150-230) represent the rigid structural core of the scPaf1 complex and are amenable to crystallization. However, the crystals of the four-component scPaf1 complex did not diffract well, so we further purified and crystallized the three-component mtPaf1 complex composed of Ctr9(31-967)/Paf1(1-120)/Cdc73(155-227) without the Rtf1 subunit based on our knowledge of scPaf1 complex and solved the ternary complex structure to 2.9-Å resolution by the single anomalous dispersion (SAD) method using a selenomethionine-labeled sample of the complex (SI Appendix, Table S1).
Overall Structure of the Ternary mtPaf1 Complex.
The ternary mtPaf1 complex has the overall shape of a thick solenoid (Fig. 2 A and B), with the Paf1 component positioned in the middle as the central core (Fig. 2 A and C) and the Ctr9 component wrapped around the Paf1 as the outer wire, with the Cdc73 fragment decorated in the surface groove of Ctr9 (Fig. 2 A and D). The Paf1 component of the complex displays a stretched conformation that has the shape of the letter “J,” which is composed mainly of loops except for four short helices (Fig. 2C) and makes extensive interactions with the Ctr9 component. Paf1 can be separated into three segments, each of which contacts a different surface of Ctr9 (Fig. 2 B and C). Segment 1 of Paf1 has the shape of a long curved line that spans a distance of 140 Å. Segment 2 extends in a direction that is almost perpendicular to segment 1, while segment 3 bends another 90° and extends antiparallel to segment 1. Except for two short β-strands located at the N terminus, the remaining portion of Ctr9 is composed of 42 α-helices, which organize into 21 consecutive TPR (25). All 21 TPR motifs of Ctr9 fold into a right-handed superhelix which wraps antiparallel to segment 1 of Paf1, spanning three complete circular turns. The solenoidal topology is a general feature of most TPR domain-containing proteins, as shown in several subunits of the anaphase-promoting complex (26). These contiguously stacked TPR motifs form two surfaces of Ctr9. Segment 1 of Paf1 contacts only the inner surface of the Ctr9 spiral, while segments 2 and 3 of Paf1 contact one end and the outer surface of Ctr9, respectively. The solenoid-shaped Ctr9 creates a large groove between the spirals, where segments of the inner wrapped Paf1 are exposed and can be accessed by other proteins. Cdc73 just contacts both Ctr9 and Paf1 through this groove. In the structure of the complex, the Cdc73 fragment is composed of three α-helices linked by two loops (Fig. 2D). Cdc73 traverses the Ctr9 groove, where it makes multiple interactions with the Ctr9 component, while it makes minimal contacts with Paf1, restricted to their intersection site.
Fig. 2.
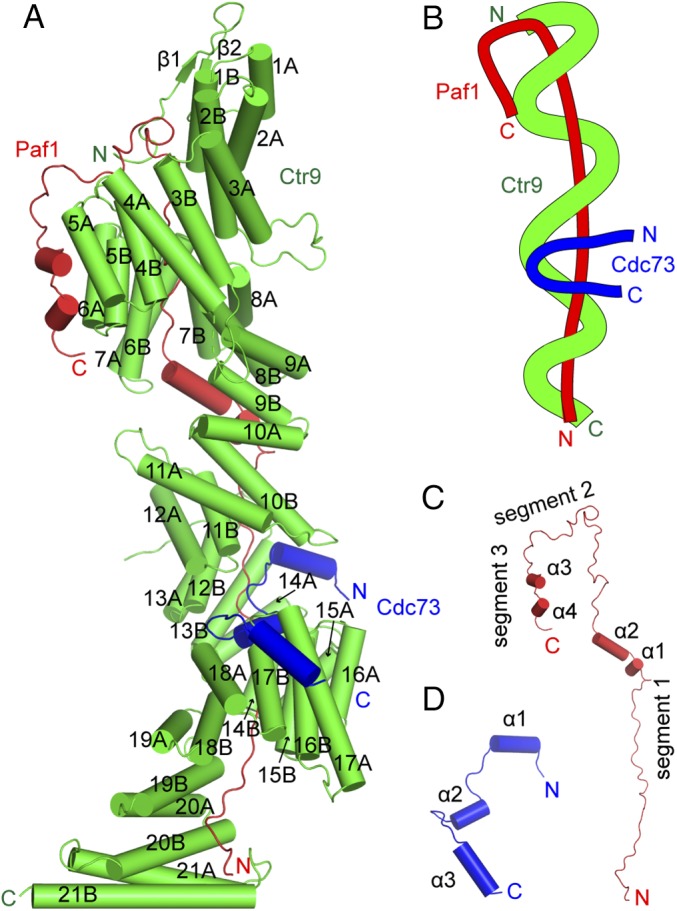
Overall structure of the ternary mtPaf1 complex. (A) Cartoon view of the overall structure of the ternary mtPaf1 complex. The α-helices are shown as cylinders. Ctr9 is colored in green, Paf1 is colored in red, and Cdc73 is colored in blue. (B) A schematic illustration of the ternary mtPaf1 complex. (C) Overall structure of the Paf1 component of the complex. Three segments and four α-helices are labeled. (D) Overall structure of the Cdc73 fragment of the complex. Three α-helices are labeled.
Structural Details of Paf1-Ctr9 Intermolecular Contacts.
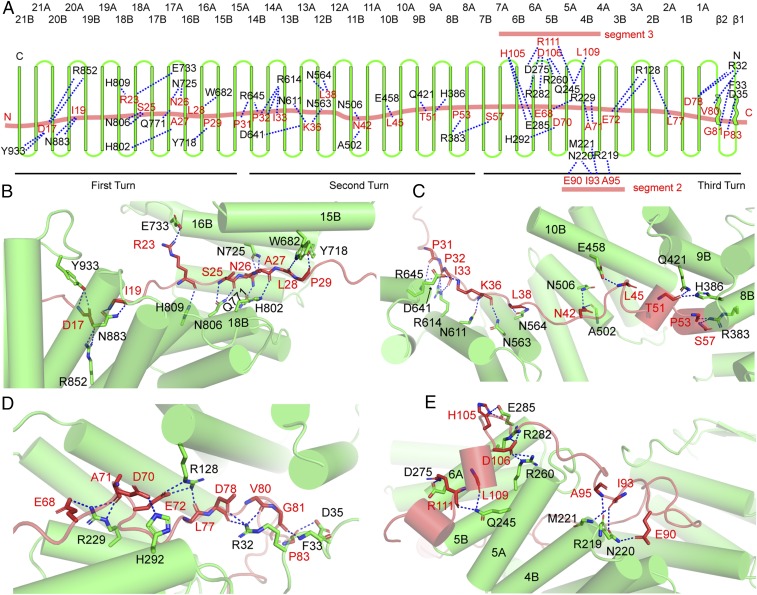
A typical TPR motif is composed of a pair of α-helices, which are named helix A and helix B, respectively. The 21 contiguously stacked TPR motifs of Ctr9 are separated into two layers that form two surfaces, with all of the B-helices forming the inner surface and with all of the A-helices forming the outer surface (Fig. 3A). The 140-Å-long segment 1 of Paf1, which is composed of both hydrophobic and hydrophilic residues, makes extensive contacts with the inner surface of the Ctr9 spiral, which is also enriched in hydrophobic and hydrophilic residues. The intermolecular contacts are stabilized by multiple hydrophobic, hydrogen-bonding, and charge interactions (Fig. 3A). From the C terminus to the N terminus, Ctr9 wraps around Paf1 segment 1, spanning three complete circular turns. We thus divide Ctr9 into three turn segments to illustrate its interactions with Paf1. The first turn of Ctr9 contains TPR motifs 15–21, which cover Paf1 residues 12–30. The wrapped Paf1 fragment makes sequence-specific interactions with the left half of this turn of Ctr9 mainly through residues D17 and I19, whose main chains form hydrogen bonds with the side chains of Y933 and N883 of Ctr9, respectively (Fig. 3B). In addition, a pair of salt bridges between D17 of Paf1 and R852 of Ctr9 further stabilize this interaction. Paf1 makes more interactions with the right half of this turn of Ctr9, as this fragment of Paf1 extends almost antiparallel with the Ctr9 helices, with this region of Paf1 sandwiched between the B-helices of TPR motifs 16 and 18 of Ctr9. A salt bridge between Paf1 R23 and Ctr9 E733 and multiple hydrogen bonds between both proteins further strengthen the interaction (Fig. 3 A and B).
Fig. 3.
Detailed interactions between Ctr9 and Paf1 in the mtPAF1 complex. (A) A diagrammatic view of the interactions between Ctr9 and Paf1. (B) Detailed interactions between the first turn of Ctr9 and Paf1. (C) Detailed interactions between the second turn of Ctr9 and Paf1. (D) Interactions between the third turn of Ctr9 and Paf1. (E) Detailed interactions between segments 2 and 3 of Paf1 and Ctr9.
The second turn of Ctr9 contains TPR motifs 8–14 that span Paf1 residues 31–66. This turn of Ctr9 displays a more stretched conformation, which covers around half the length of segment 1 of Paf1 (Fig. 2C). The wrapped fragment of Paf1 makes sequence-specific contacts with one or two residues from each helix it traverses through hydrogen-bonding or salt-bridge interactions (Fig. 3 A and C). Paf1 fragment 45–66 contains two short α-helices and its flanking loops, which extend antiparallel with the B-helices of TPR motifs 8–10 of Ctr9. E458 from TPR motif 10, Q421 from TPR motif 9, and H386 and R383 from TPR motif 8 hold this segment of Paf1 by forming hydrogen bonds with the main-chain atoms of Paf1 through their long side chains (Fig. 3C).
The third turn of the Ctr9 spiral contains TPR motifs 1–7 and two β-strands located at the N terminus. This turn of Ctr9 wraps Paf1 in a more compact manner and makes extensive contacts with a short segment (67-87) of Paf1. Multiple salt bridges and hydrogen bonds are formed between Ctr9 and this fragment of Paf1 (Fig. 3 A and D). Three arginines (R229, R128, and R32) of Ctr9 form multiple hydrogen bonds and salt bridges with this fragment of Paf1. In addition, H292 and F33 also form hydrogen bonds with the residues of Paf1. These close contacts result in burying this segment of Paf1 deep inside the Ctr9 spiral.
Segment 2 of Paf1 contains residues 88–101. Starting from G88, Paf1 reaches the N-terminal end of the Ctr9 spiral and extends almost perpendicular to segment 1. This segment of Paf1 traverses along the edge of the Ctr9 N-terminal end and makes sequence-specific interactions with several residues at the end of helix 4B of Ctr9. A95, E90, and I93 of Paf1 form several hydrogen bonds with the R219-N220-M221 segment of Ctr9 (Fig. 3 A and E).
Segment 3 of Paf1 contains residues 102–119. This segment contains two short α-helices, which contact only the outer surface of Ctr9 and extend antiparallel with segment 1 of Paf1 (Fig. 3E). This segment of Paf1 is sandwiched between helices 5A and 6A of Ctr9. The hydrophobic residues of this segment are positioned at the bottom and pack against the hydrophobic surface formed by helices 5A, 5B, and 6A of Ctr9. Sequence-specific interactions include salt bridges between D106 of Paf1 and both R260 and R282 of Ctr9 and between R111 of Paf1 and D275 of Ctr9. In addition, several hydrogen bonds between both proteins also contribute to the interaction (Fig. 3E).
Structural Details of Cdc73-Paf1 and Cdc73-Ctr9 Intermolecular Contacts.
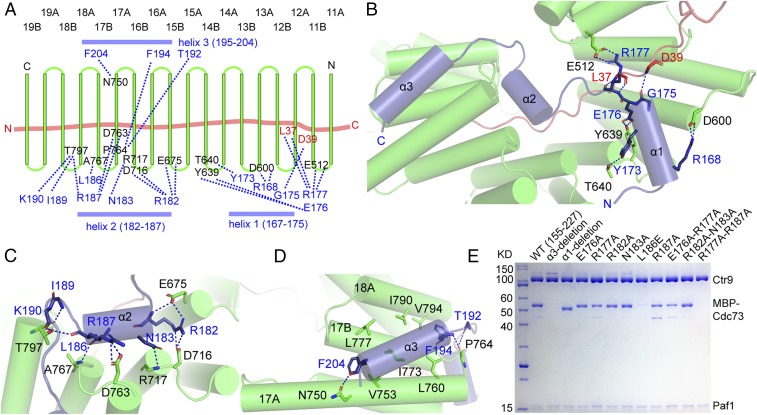
Our in vitro analysis identified that a short fragment in the middle region of Cdc73 is sufficient for forming a complex with the other components of the Paf1 complex. In the structure of the mtPaf1 ternary complex, the traceable fragment of Cdc73 (residues 164–205) contains three α-helices, which are connected by two loops (Figs. 2D and 4 A and B). The three helices are located on both ends and in the middle of the Cdc73 fragment, respectively. The Cdc73 N-terminal helix wedges inside the Ctr9 groove till it meets the central segment of Paf1. This helix makes sequence-specific contacts with Ctr9 through a pair of salt bridges and a hydrogen bond (Fig. 4B). The loop connecting the N-terminal helix and the middle helix of Cdc73 extends antiparallel with the central Paf1 segment, where it contacts residues from both Paf1 and Ctr9. A short segment of Cdc73 that contains only three residues, G175, E176, and R177, constitutes the only region that contacts both Paf1 and Ctr9 directly. The main chains of G175 and R177 of Cdc73 form three hydrogen bonds with the main-chain atoms of Paf1, which are the only hydrogen bonds observed between Paf1 and Cdc73 in this complex (Fig. 4B). The long side chains of R177 and E176 of Cdc73 form a pair of salt bridges and hydrogen bonds with E512 and Y639 of Ctr9, respectively, which further stabilize the ternary complex (Fig. 4B).
Fig. 4.
Detailed interactions between Cdc73 and Ctr9, as well as between Cdc73 and Paf1 in the mtPAF1 complex. (A) Schematic representation of the interactions between Cdc73 and Ctr9, as well as between Cdc73 and Paf1. (B) Cdc73 α1 and its flanking loop interact with both Ctr9 and Paf1. (C) Detailed interactions between Cdc73 α2 and its flanking loops with Ctr9. (D) Interactions between Cdc73 α3 and Ctr9. (E) In vitro reconstitution of MBP-tagged Cdc73(155-227) or its mutants with the binary complex of Ctr9(31-967)-Paf1(1-120).
The middle helix of Cdc73 extends perpendicular to Paf1, which directs Cdc73 toward the outside of the Ctr9 groove. As the middle helix and its flanking loops of Cdc73 are positioned deep inside the Ctr9 groove, they make extensive contacts with the residues located on the edge of the Ctr9 groove (Fig. 4 A and C). Intermolecular contacts between both proteins are mainly mediated by salt bridges and hydrogen bonds. R182 and R187 of Cdc73 form multiple salt bridges with neighboring acidic residues of Ctr9. In addition, N183 and L186 of Cdc73 form several hydrogen bonds with residues from Ctr9. Residues I189 and K190, which are located on a sharp turn of the loop of Cdc73, are stabilized in position by forming hydrogen bonds with the main chain and side chain of T797 of Ctr9, respectively. Residues of Cdc73 after K190 extend outside of the Ctr9 groove and make contacts with the outer surface of Ctr9 (Fig. 4C).
The hydrophobic side of the C-terminal helix of the Cdc73 fragment packs against a hydrophobic groove formed by helices 17A, 17B, and 18A of TPR motifs 17–18 of Ctr9, which stabilizes the contact by hydrophobic interactions (Fig. 4D). Besides, hydrogen bonds between the side chain of N750 of Ctr9 and the main chain of F204 of Cdc73 and between the main chain of P764 of Ctr9 and both main chains of T192 and F194 of Cdc73 further strengthen their interactions.
To detect residues or regions of the Cdc73 fragment important for forming a complex with both Ctr9 and Paf1, we made a series of mutations and deletions of this fragment of Cdc73 and then checked their association with the binary complex of Ctr9-Paf1 through an in vitro reconstitution procedure. We find that the C-terminal helix of Cdc73 is important for complex formation, as its deletion resulted in a complete loss of the Cdc73 fragment in the complex (Fig. 4E). By contrast, deletion of the N-terminal helix would not affect complex formation (Fig. 4E). We also identified that Cdc73 fragments bearing R177A single mutation or E176A-R177A double mutations showed reduced binding to the Ctr9-Paf1 binary complex, while R177A-R187A double mutations and L186E single mutation showed a complete loss of binding to the Ctr9-Paf1 binary complex. As a control, several other single or double mutations in the middle region of Cdc73 could not disrupt complex formation (Fig. 4E). This indicates that strong mutations are needed to disrupt the interaction network between Cdc73 and the Ctr9-Paf1 binary complex.
ssDNA-Binding Properties.
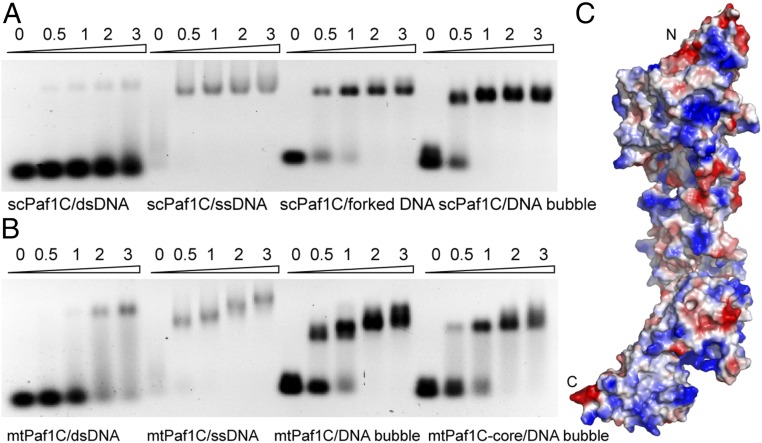
As most transcription factors can bind DNA, we hypothesized that the Paf1 complex may also bind some type of DNA. We designed several forms of DNA and tested their binding properties for the scPaf1 complex based on electrophoretic mobility shift assay (EMSA). The full-length scPaf1 complex showed almost no binding to double-stranded DNA (dsDNA), but exhibited a strong affinity for various single-strand–containing DNAs tested, such as single-stranded DNA (ssDNA), forked DNA, and dsDNA containing an internal single-stranded bubble (Fig. 5A). We further undertook tests to identify components that contribute to DNA binding and found that Rtf1, Cdc73, and the Paf1-Ctr9-Leo1 ternary complex all bind single-strand–containing DNAs, but with reduced affinity (SI Appendix, Fig. S2). Therefore we propose that the DNAs may be recognized by several components of the scPaf1 complex simultaneously. Similarly, the four-subunit mtPaf1 complex that does not contain the Rtf1 subunit also showed a preference for the above single-strand–containing DNAs (Fig. 5B). Furthermore, the crystallized solenoid-shaped rigid structural core of the mtPaf1 ternary complex also retains a strong binding affinity to single-strand–containing DNAs (Fig. 5B). Several positively charged patches could be identified on the electrostatic surface of the structure of the mtPaf1 ternary complex structure (Fig. 5C), which may serve as potential ssDNA-binding sites. However, a detailed mechanism of ssDNA recognition by the Paf1 complex and its function in vivo remain to be explored.
Fig. 5.
The Paf1 complex prefers to bind single-strand–containing DNAs. (A) EMSA analysis of the full-length scPaf1 complex with various DNAs. dsDNA, double-stranded DNA; Paf1C, Paf1 complex; ssDNA, single-stranded DNA. One hundred picomoles of ssDNA or 20 picomoles of all of the other forms of DNAs were used as input. Protein-to-DNA molar ratios are listed above gel lanes. (B) EMSA analysis of the mtPaf1 complex with various DNAs. mtPaf1C-core represents the crystallized ternary complex of the mtPaf1 complex. DNA concentrations are the same as above. Protein-to-DNA molar ratios are listed above gel lanes. (C) Surface electrostatic representation of the structure of the mtPaf1 core complex solved in this study. Positively charged regions are colored in blue. Negatively charged regions are colored in red. Neutral regions are colored in white.
Discussion
The Paf1 complex is conserved throughout eukaryotes and functions as a general RNA polymerase II transcription elongation factor. In this study, we present the 2.9-Å crystal structure of the rigid core of the mtPaf1 ternary complex, allowing an analysis of the interactions among different subunits/domains, which provides a much clearer overview of the architecture of the Paf1 complex. The largest subunit Ctr9 is a TPR protein that folds into a right-handed superhelical structure, which shapes the overall architecture of the Paf1 complex. The Paf1 N terminus folds into an extended linear conformation, which is deeply buried inside the Ctr9 spirals, so that both Ctr9 and Paf1 fold together into an inseparable structural unit. Other subunits join the complex mainly through direct association with these two subunits. Our structure showed that Cdc73 joins the complex mainly through the Ctr9 subunit. The Leo1 subunit interacts with the complex mainly through a direct association with the Paf1 subunit (18), but not through contacts with the other subunits. In addition, in vitro reconstitution studies showed that removing either the Paf1 or the Ctr9 subunit, but not the other subunits of the Paf1 complex, would disrupt complex formation. These evidences establish the Paf1-Ctr9 binary complex as the central core of the complex. The Rtf1 subunit behaves differently among different species. In budding yeast, results from both ourselves and others clearly verified that the C-terminal region of Rtf1 is enough for its stable association with the remaining subunits (27). By contrast, in Myceliophthora thermophile, in vitro purified Rtf1 does not join the other four-subunit complex to form a five-subunit full complex. Other factors may be needed for mtRtf1 to associate stably with the remaining subunits of the mtPaf1 complex, which remains to be clarified.
The Paf1 complex is a versatile platform that associates with multiple transcription factors and chromatin-modifying enzymes and displays diverse functions in transcriptional and epigenetic regulation (6, 13). We here showed that the Paf1 complex prefers to bind single-strand–containing DNAs, which would add another layer of complexity to its function. The Rtf1 subunit has been shown to bind ssDNAs (22). Our data indicate that in budding yeast, other subunits also contribute to ssDNA binding. ssDNAs are generated during transcription, but how they are recognized and interpreted by the Paf1 complex remains to be studied.
In humans, mutations of subunits in the Paf1 complex are closely related to various cancers. Mutations in Cdc73 are directly associated with hyperparathyroidism/jaw tumor syndrome (28), while mutations in CTR9 gene predispose individuals to Wilms tumor (29). CTR9 gene mutation results in the deletion of 78 amino acids in the CTR9 protein, which correspond roughly to helices 17–20 (TPR motifs 9–10) of mtCtr9. Deletion of these helices would result in a register shift of all of the following helices, which could likely disrupt the interaction between Ctr9 and Paf1, thereby leading to instability of the Paf1 complex. Our structure provides a molecular basis for deciphering the impact of these mutations in human cancers in the future.
Materials and Methods
Details of the methods, including protein expression and purification, crystallization and structure determination, EMSA analysis, N/C terminus analysis and Edman sequencing, and in vitro reconstitution of the complexes, are presented in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff of BL17U1, BL18U1, and BL19U1 beamlines at Shanghai Synchrotron Radiation Facility (SSRF) in China for assistance during data collection. This work was supported by the National Natural Science Foundation of China (31570729, 31870725, and 31370719), Beijing Natural Science Foundation (5152015), starting funds from “the Thousand Talents Plan” for young scientists, and the Fundamental Research Funds for the Central Universities (2017EYT19) (to Z.W.), funds from the Leukemia and Lymphoma Society and Memorial Sloan-Kettering Cancer Center Core Grant P30CA008748, as well as from the Office of the President, SUSTech (to D.J.P.), and NIH Grants DK071900 and CA129325 (to R.G.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 6AF0).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812256115/-/DCSupplemental.
References
- 1.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript elongation by RNA polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 2.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen FX, et al. PAF1 regulation of promoter-proximal pause release via enhancer activation. Science. 2017;357:1294–1298. doi: 10.1126/science.aan3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu M, et al. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 2015;350:1383–1386. doi: 10.1126/science.aad2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wade PA, et al. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr Purif. 1996;8:85–90. doi: 10.1006/prep.1996.0077. [DOI] [PubMed] [Google Scholar]
- 6.Jaehning JA. The Paf1 complex: Platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rondón AG, Gallardo M, García-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 10.Costa PJ, Arndt KM. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics. 2000;156:535–547. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogan NJ, et al. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Squazzo SL, et al. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Oss SB, Cucinotta CE, Arndt KM. Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem Sci. 2017;42:788–798. doi: 10.1016/j.tibs.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogan NJ, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 15.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 16.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 17.Kowalik KM, et al. The Paf1 complex represses small-RNA-mediated epigenetic gene silencing. Nature. 2015;520:248–252. doi: 10.1038/nature14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu X, et al. Structural insights into Paf1 complex assembly and histone binding. Nucleic Acids Res. 2013;41:10619–10629. doi: 10.1093/nar/gkt819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amrich CG, et al. Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin. J Biol Chem. 2012;287:10863–10875. doi: 10.1074/jbc.M111.325647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, et al. Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold. Acta Crystallogr D Biol Crystallogr. 2012;68:953–959. doi: 10.1107/S0907444912017325. [DOI] [PubMed] [Google Scholar]
- 21.Sun W, et al. Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome. Sci Rep. 2017;7:15638. doi: 10.1038/s41598-017-15715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jong RN, et al. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure. 2008;16:149–159. doi: 10.1016/j.str.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Wier AD, Mayekar MK, Héroux A, Arndt KM, VanDemark AP. Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci USA. 2013;110:17290–17295. doi: 10.1073/pnas.1314754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, et al. Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat Commun. 2017;8:15741. doi: 10.1038/ncomms15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sikorski RS, Boguski MS, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 26.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warner MH, Roinick KL, Arndt KM. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol. 2007;27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carpten JD, et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat Genet. 2002;32:676–680. doi: 10.1038/ng1048. [DOI] [PubMed] [Google Scholar]
- 29.Hanks S, et al. Germline mutations in the PAF1 complex gene CTR9 predispose to Wilms tumour. Nat Commun. 2014;5:4398. doi: 10.1038/ncomms5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.