Significance
MHC class I molecules present small fragments of proteins from within the cell to alert the immune system to infection and cellular damage. Two protein accessory proteins, tapasin and TAPBPR, assist in the loading and selection of these peptides inside the cell. Here we show that one of these proteins, TAPBPR, surprisingly still works when delivered to the outside of cells and can be used to load peptides from viruses and tumours directly on surface MHC molecules. Therefore, we have found an efficient way to override the peptides naturally presented by cells and can use this to target immune responses against cells. This may prove beneficial to mount immune responses against cancer in the future.
Keywords: TAPBPR/TAPBPL, MHC, HLA, antigen processing, antigen presentation
Abstract
The repertoire of peptides displayed at the cell surface by MHC I molecules is shaped by two intracellular peptide editors, tapasin and TAPBPR. While cell-free assays have proven extremely useful in identifying the function of both of these proteins, here we explored whether a more physiological system could be developed to assess TAPBPR-mediated peptide editing on MHC I. We reveal that membrane-associated TAPBPR targeted to the plasma membrane retains its ability to function as a peptide editor and efficiently catalyzes peptide exchange on surface-expressed MHC I molecules. Additionally, we show that soluble TAPBPR, consisting of the luminal domain alone, added to intact cells, also functions as an effective peptide editor on surface MHC I molecules. Thus, we have established two systems in which TAPBPR-mediated peptide exchange on MHC class I can be interrogated. Furthermore, we could use both plasma membrane-targeted and exogenous soluble TAPBPR to display immunogenic peptides on surface MHC I molecules and consequently induce T cell receptor engagement, IFN-γ secretion, and T cell-mediated killing of target cells. Thus, we have developed an efficient way to by-pass the natural antigen presentation pathway of cells and load immunogenic peptides of choice onto cells. Our findings highlight a potential therapeutic use for TAPBPR in increasing the immunogenicity of tumors in the future.
Optimal peptide selection on MHC I molecules is essential to mount effective antiviral and antitumor immune responses. This process is aided by two intracellular MHC I peptide editors. The first peptide editor identified was tapasin, which works within the peptide-loading complex, which is where peptides are imported into the endoplasmic reticulum (ER) (1–4). Following our initial identification of a role for TAPBPR in the MHC I antigen processing and presentation pathway (5), TAPBPR was more recently shown to function as a second peptide editor for MHC I molecules (6, 7). Molecular insight regarding the mechanisms by which peptide editors can help assist in the selection of high-affinity peptides onto MHC I has recently been provided with the determination of two crystal structures of human TAPBPR in complex with mouse MHC I molecules (8, 9). In contrast to tapasin, TAPBPR is not a component of the peptide-loading complex (5), however it can recruit UDP-glucose:glycoprotein glucosyltransferase 1 to provide a quality-control checkpoint in the process of peptide selection on MHC I (10). Thus, the two MHC I peptide editors work in different environments to shape the peptide repertoire presented to the immune system.
In 2007, two elegant assays were developed to directly explore the ability of tapasin to mediated peptide exchange on MHC class I; one involved artificially zippering tapasin to MHC I and measuring peptide exchange using fluorescent anisotropy in vitro (11), whereas the other used a recombinant tapasin-ERp57 disulphide-linked conjugate and measured its effect on peptide exchange, using iodinated peptides in a cell-free system (12). We and others previously used an approach analogous to the one developed by Chen and Bouvier (11) to demonstrate that TAPBPR enabled efficient peptide exchange on MHC I in vitro; however, as opposed to tapasin, the luminal domain of TAPBPR alone, in the absence of an artificial intermolecular tether, was sufficient to mediate exchange in this system (6, 7).
Because TAPBPR normally performs peptide editing on glycosylated MHC I molecules within a cellular environment, we wondered whether a more physiological system could be developed to explore TAPBPR-mediated peptide exchange. Although TAPBPR usually resides intracellularly, we previously observed that overexpression of TAPBPR results in a proportion of TAPBPR mislocalizing to the cell surface (5). We speculated that this surface pool of TAPBPR still interacts with MHC I and could thus function as a peptide editor on the plasma membrane (PM). Here, we explore the ability of both PM-targeted and exogenous soluble TAPBPR to function as peptide-exchange catalysts on surface-expressed MHC I molecules. We reveal that both forms of TAPBPR function as efficient peptide-exchange catalysts on surface-expressed MHC I molecules and can be utilized to display immunogenic peptides at the surface of various tumor cell lines to CD8+ cytotoxic T lymphocytes (CTL).
Results
PM-Expressed TAPBPR Promotes Exogenous Peptide Association onto Surface MHC I.
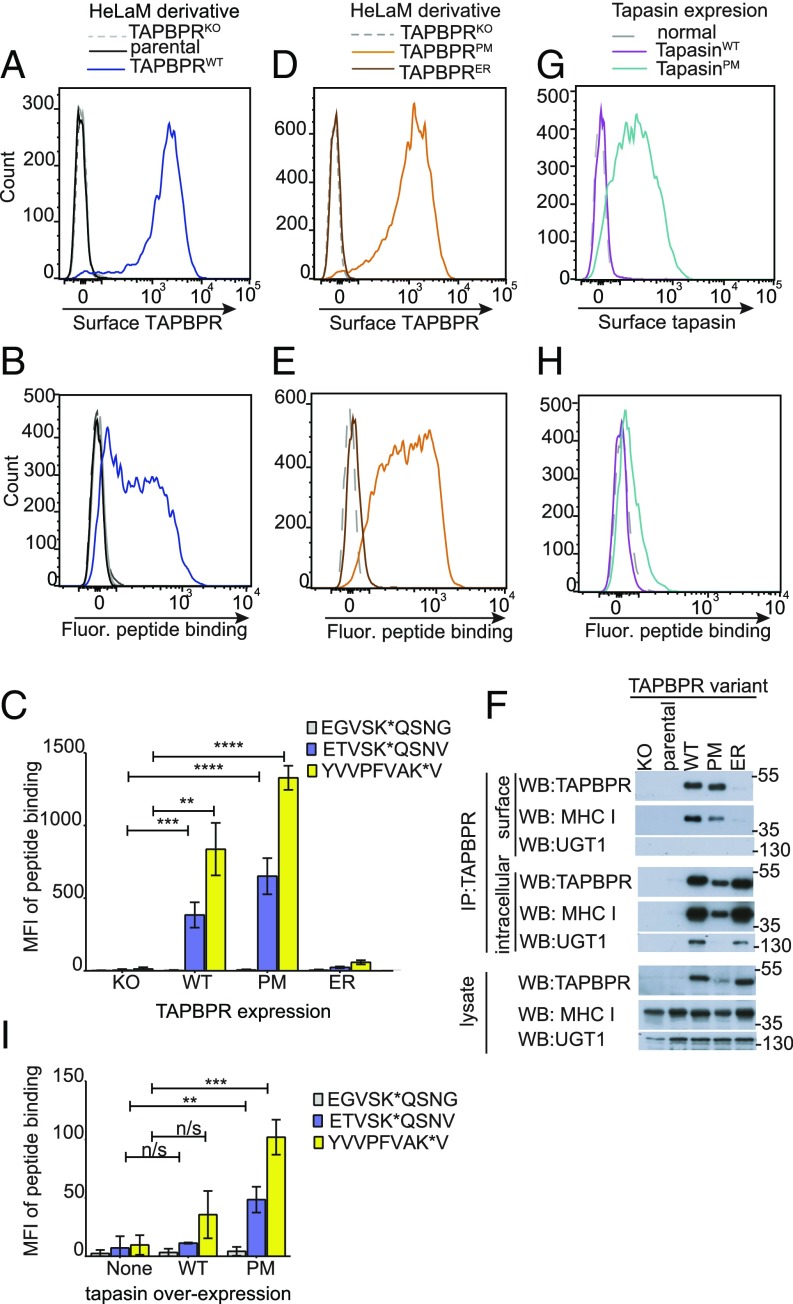
While IFN-γ–treated HeLaM and HeLaM-TAPBPRKO cells do not express TAPBPR on their cell surface, the transduction of TAPBPRWT into HeLaM-TAPBPRKO results in significant expression of TAPBPR at the PM (Fig. 1A). The functionality of surface-expressed TAPBPR was unknown. To explore whether the pool of surface-expressed TAPBPR retains its peptide-editing functionality, we first asked whether a fluorescently labeled exogenous peptide specific for HLA-A*68:02 (an MHC I molecule expressed by HeLaM cells) exhibited increased binding to cells expressing surface TAPBPR compared with cells lacking surface TAPBPR. Upon incubation with a fluorescent variant of the neoantigen ETVSEQSNV, which binds to HLA-A*68:02 with high affinity (13), cells expressing TAPBPRWT became fluorescent, while cells that lacked surface TAPBPR remained nonfluorescent (Fig. 1B). We next investigated the binding of two additional fluorescent peptides: YVVPFVAK*V, which binds to HLA-A*68:02; and EGVSK*QSNG, a nonbinding derivative of ETVSK*QSNV, in which the anchor residues that permit HLA-A*68:02 binding are mutated. While YVVPFVAK*V significantly bound to cells expressing TAPBPRWT, but not to HeLaM-TAPBPRKO cells, EGVSK*QSNG did not bind to either cell line (Fig. 1C and SI Appendix, Fig. S1A). These data suggest that the cellular fluorescence observed was due to peptide binding to MHC I, rather than via peptide internalization. Furthermore, when cells expressing surface TAPBPR were incubated at 4 °C to inhibit membrane trafficking (SI Appendix, Fig. S2 A and B), a significant amount of exogenous peptide still bound within 5 min, providing additional evidence that peptide binding occurs directly at the PM.
Fig. 1.
Surface-expressed TAPBPR enhances exogenous peptide association onto MHC I. Surface expression of (A and D) TAPBPR, detected using the mAb PeTe-4 or (G) tapasin, detected using Pasta1 on IFN-γ–treated (A) HeLaM cells and HeLaM-TAPBPRKO ± TAPBPRWT transduction, (D) HeLaM-TAPBPRKO ± TAPBPRPM or TAPBPRER transduction, or (G) HeLaM-TAPBPRKO ± tapasinWT or tapasinPM transduction. Staining with an isotype control (solid black line) is included in A. Note: HeLaM-TAPBPRKOTAPBPRPM cells with low transduction levels were selected to generate cells with similar surface expression as TAPBPRWT. (B, E, and H) Histograms show the typical peptide binding observed when the cells were incubated with the HLA-A*68:02-specific fluorescent peptide ETVSK*QSNV at 10 nM for 15 min at 37 °C. (C and I) Bar charts summarizing the level of exogenous fluorescent peptide binding when cells were incubated with 10 nM EGVSK*QSNG, ETVSK*QSNV, or YVVPFVAK*V for 15 min at 37 °C. Bars show mean fluorescence intensity (MFI) ± SD from three independent experiments. n/s not significant, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 using unpaired two-tailed t test. (F) Immunoprecipitation of the cell surface pool of TAPBPR, by staining intact cells with PeTe-4 before lysis and addition of protein-A Sepharose, and the remaining intracellular TAPBPR pool from cells postsurface TAPBPR preclear, followed by Western blotting for TAPBPR, MHC I (using HC10), or UGT1 on immunoprecipitates and lysates as indicated. Data shown is representative of three independent experiments. For comparison, a classic coimmunoprecipitation from these cells is also provided (SI Appendix, Fig. S1D).
To provide further evidence that the surface pool of TAPBPR, rather than its overexpression, was responsible for loading exogenous peptide onto surface MHC I, we generated two chimeric TAPBPR constructs that target TAPBPR to different subcellular sites. PM targeting of the luminal portion of TAPBPR was achieved by replacing the cytoplasmic tail of TAPBPR with that of CD8 (TAPBPRPM) (14), while TAPBPR was retained within the ER by replacing its transmembrane domain and cytoplasmic tail with those of tapasin (TAPBPRER) (2, 3). In contrast to TAPBPRPM, which was expressed at high levels on the cell surface, TAPBPRER was not found on the PM (Fig. 1D). Only cells expressing TAPBPRPM were able to bind to exogenous fluorescent peptides specific for HLA-A*68:02 molecules, YVVPFVAK*V and ETVSK*QSNV (Fig. 1 C and E and SI Appendix, Fig. S1B). No significant fluorescent peptide binding was detectable on cells expressing TAPBPRER (Fig. 1 C and E and SI Appendix, Fig. S1B). These results suggest that surface TAPBPR promotes the loading of exogenous peptide onto surface expressed MHC I.
PM-Expressed TAPBPR Associates with Surface MHC I.
Given the results above, we next confirmed that cell surface-expressed TAPBPR physically associated with surface MHC I. Immunoprecipitation of the surface pool of TAPBPR from both TAPBPRWT- and TAPBPRPM-expressing cells confirmed it was associated with MHC I, but not with UGT1 (Fig. 1F). As expected, negligible levels of TAPBPRER were detectable using this technique, verifying the lack of significant cross-contamination of intracellular TAPBPR in the cell surface immunoprecipitates (Fig. 1F). Isolation of the intracellular TAPBPR pool confirmed that all TAPBPR variants were expressed and associated with intracellular MHC I (Fig. 1F). In contrast to TAPBPRPM, we observed that UGT1 associates with both TAPBPRWT and TAPBPRER, supporting the predicted subcellular localization of the chimeric proteins (Fig. 1F). Taken together, these results suggest that surface localization of TAPBPR, rather than its intracellular overexpression, is responsible for the loading of exogenous peptide onto MHC I at the PM.
Surface-Expressed Tapasin Does Not Promote Substantial Exogenous Peptide Association onto Surface MHC I.
Because tapasin is also an MHC I peptide editor, we next asked whether tapasin could similarly load exogenous peptide onto MHC I when expressed at the cell surface. The overexpression of tapasinWT did not result in tapasin expression at the cell surface (Fig. 1G), most probably because of the ER retention motif found in its cytoplasmic tail (2, 3). We therefore replaced the cytoplasmic tail of tapasin with that of CD8 (tapasinPM), which led to tapasin expression at the cell surface (Fig. 1G). When the ability of cells expressing surface tapasin to bind to exogenous fluorescent peptides was tested, there was a significant but very slight increase in peptide binding of both ETVSK*QSNV (Fig. 1 H and I) and YVVPFVAK*V (Fig. 1I and SI Appendix, Fig. S1C). Our results suggest that TAPBPRPM is at least 10-fold more efficient at promoting peptide binding in this situation, compared with tapasinPM. This finding is consistent with the differences observed in the ability of TAPBPR and tapasin to facilitate peptide exchange using other reported assays, in which TAPBPR alone functions as an efficient peptide editor without the need for zippering to MHC I (6, 7). In contrast, tapasin requires other association partners or artificial zippering to MHC I (11, 12). We speculate that this low level of peptide binding observed to cells expressing surface tapasin is due the export of peptide-receptive MHC I with tapasin to the cell surface, rather than the surface tapasin efficiently facilitating peptide exchange.
TAPBPR Mediates Exogenous Peptide Binding Quickly and at Low Peptide Concentration.
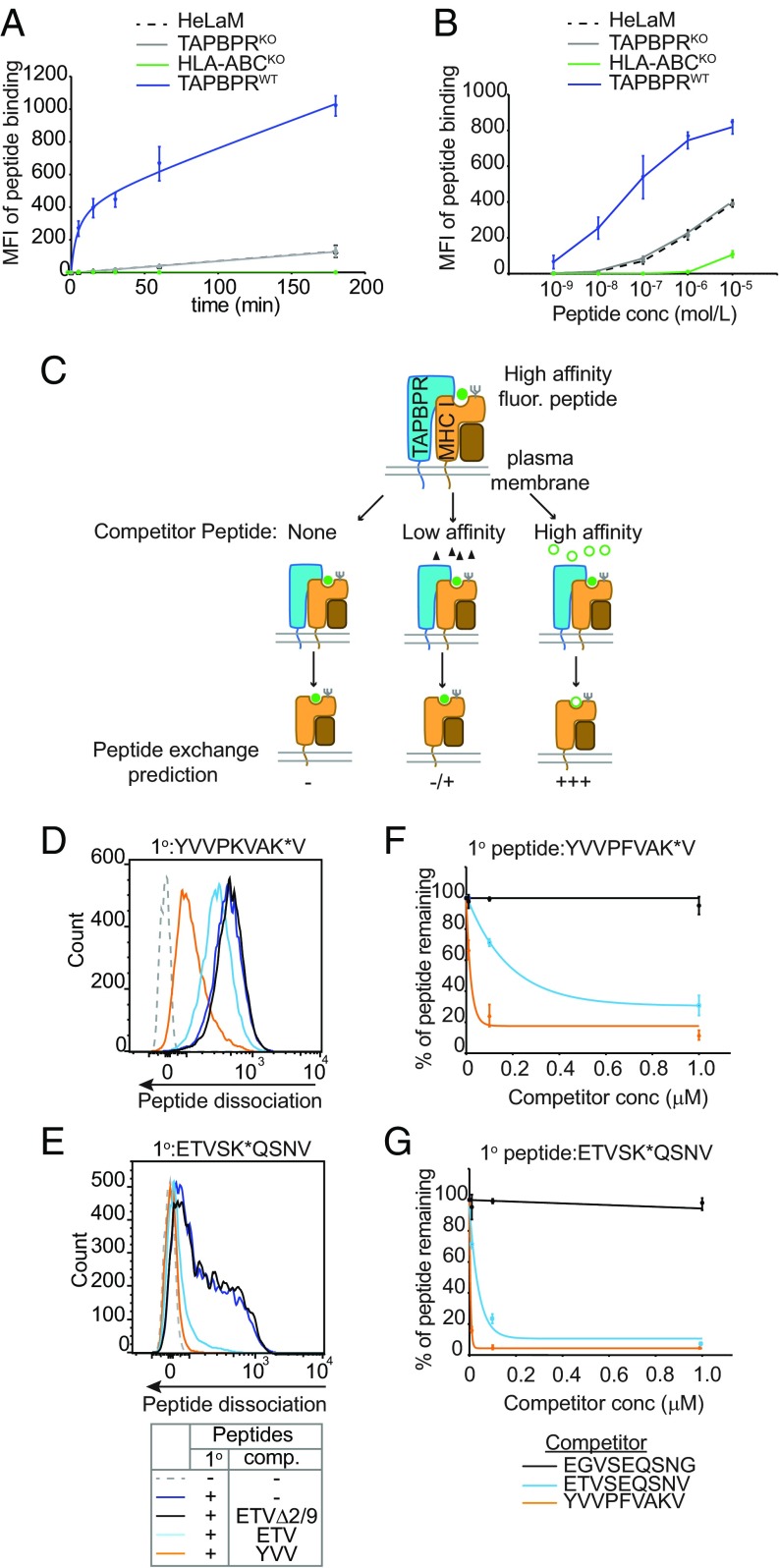
When we explored the kinetics of TAPBPR-mediated peptide binding to cells over time, we observed a striking increase in both the magnitude and speed at which this occurred (Fig. 2A and SI Appendix, Fig. S2C). Within 5 min we observed a >200-fold increase in the level of exogenous peptide binding in cells expressing surface TAPBPR compared with the HeLaM and HeLaM-TAPBPRKO controls (Fig. 2A). Furthermore, TAPBPR-mediated peptide binding occurred at extremely low concentrations of peptide compared with cells lacking surface TAPBPR expression (Fig. 2B and SI Appendix, Fig. S2D). TAPBPR-mediated peptide binding required ∼100-fold less peptide to obtain equivalent peptide binding, compared with cells lacking surface TAPBPR expression (Fig. 2B). These findings demonstrate that surface TAPBPR mediates peptide association onto surface MHC I molecules extremely rapidly and at very low concentrations of peptide.
Fig. 2.
Surface TAPBPR functions as a MHC I peptide exchange catalyst. (A) Time course and (B) dose–response curves showing the level of exogenous ETVSK*QSNV binding to IFN-γ–treated HeLaM, HeLaM-TAPBPRKO ± TAPBPRWT, and to HeLaM-HLA-ABCKO cells treated with (A) 10 nM ETVSK*QSNV from 0 to 180 min at 37 °C or (B) increasing concentration of ETVSK*QSNV for 15 min. Line graphs show MFI ± SD from three independent experiments. Histograms displaying the typical fluorescent peptide binding observed on HeLaMKO-TAPBPRWT–expressing cells for both the time course and dose–response experiment are provided in SI Appendix, Fig. S2 C and D, respectively. Note: in B ETVSK*QSNV binding using 1 nM–1 µM peptide was dependent on MHC I, given that no exogenous peptide association was observed on HeLaM-HLA-ABCKO cells at these concentrations. (C) Schematic representation of experimental workflow used to measure peptide exchange by PM-bound TAPBPR. (D–G) IFN-γ–treated HeLaKOTAPBPRWT cells were incubated with 10 nM (D and F) YVVPKVAK*V or (E and G) ETVSK*QSNV for 15 min at 37 °C, then washed to remove unbound peptide. Dissociation of the fluorescent peptides was subsequently monitored in the absence or presence of increasing concentrations of the unlabeled competitor peptides YVVPFVAKV (YVV), ETVSEQSNV (ETV), or EGVSEQSNG (ETVΔ2/9) for 15 min at 37 °C. (D and E) Histograms show the typical dissociation of fluorescent peptide observed following incubation with 100-nM competitor peptide. (F and G) Line graphs show the percentage of fluorescent peptide remaining ±SD following treatment with increasing concentrations of unlabeled peptide from (F) four and (G) three independent experiments.
Surface TAPBPR Functions as a Peptide-Exchange Catalyst on Surface MHC I.
There are two conceivable mechanisms by which surface-expressed TAPBPR could promote the loading of exogenous peptides onto MHC I; it may drag peptide-receptive MHC I molecules with it through the secretory pathway to the cell surface or it may retain its ability to function as a peptide-exchange catalyst in this atypical location. Given that enhanced peptide binding continued over a prolonged period on cells expressing surface TAPBPR (Fig. 2A), it was probable that TAPBPR retained its ability to function as a peptide-exchange catalyst at this atypical location. To explore this, we developed an assay to directly assess whether TAPBPR actively mediated peptide dissociation from MHC I at the cell surface (Fig. 2C). First, cells expressing surface TAPBPR were incubated with fluorescently labeled peptide for 15 min to enable surface HLA-A*68:02 molecules to bind to labeled peptides. Subsequently, cells were extensively washed to remove any unbound peptide, thus removing any excess fluorescent peptide from the system. We then tested the ability of cells to dissociate the labeled peptide in the presence of a vast excess of unlabeled competitor peptides. Using this method, we observed dissociation of both YVVPKVAK*V (Fig. 2D) and ETVSK*QSNV (Fig. 2E) in the presence of a high-affinity unlabeled competitor peptide (ETVSEQSNV or YVVPFVAKV), but not in the presence of a nonbinding competitor peptide (EGVSEQSNG). This suggests that surface TAPBPR is capable of promoting peptide exchange from MHC I molecules in a peptide-affinity (YVVPFVAKV > ETVSEQSNV > EGVSEQSNQ) and peptide-concentration–dependent manner (Fig. 2 F and G).
In keeping with this, the binding of YVVPFVAK*V to cells expressing surface TAPBPR appeared as a single sharp peak (e.g., Fig. 2D and SI Appendix, Fig. S1A), while the binding of ETVSK*QSNV appeared bimodal in comparison (e.g., Figs. 1B and 2E). Although in our standard assay conditions (10 nM peptide for 15 min at 37 °C) ETVSK*QSNV was not at saturation, the binding of this peptide to cells expressing surface TAPBPR was improved by either increasing the time of peptide incubation (SI Appendix, Fig. S2C) or by increasing the concentration of peptide used (SI Appendix, Fig. S2D). The binding of ETVSK*QSNV could be brought up to comparable levels and distribution as observed with 10 nM YVVPFVAK*V by increasing ETVSK*QSNV concentration to 1 µM (SI Appendix, Fig. S2D). Thus, the differences observed in the pattern of peptide binding to cells appears to be due to the different affinity of the two peptides for HLA-A*68:02 (YVVPFVAK*V > ETVSK*QSNV), rather than heterogeneity of the cells.
Surprisingly, TAPBPR also retained this catalytic activity at 4 °C, albeit at a slower rate and in the presence of a higher concentration of competitor peptide (SI Appendix, Fig. S3 A and B). To rule out the possibility that peptide was simply binding to empty MHC I expressed on the surface, cells were incubated at 4 °C to inhibit further membrane trafficking and a reversed assay to measure peptide exchange was performed. Cells at 4 °C were first incubated with an excess of unlabeled high-affinity peptide to occupy any potentially empty MHC I molecules with peptide. Then, after extensive washing to remove any excess unbound peptide, cells were subsequently incubated with a fluorescent competitor peptide (SI Appendix, Fig. S3C). We still observed high levels of fluorescent peptide binding to cells in the presence of surface TAPBPR, but not in its absence (SI Appendix, Fig. S3D). Thus, for TAPBPR to promote fluorescent peptide loading observed in this assay, it must have dissociated peptide from MHC I first.
Taken together, we find that although these findings do not exclude the possibility that surface TAPBPR carries some peptide-receptive MHC I molecules en route, they do demonstrate that TAPBPR retains its ability to function as a peptide-exchange catalyst when expressed on the cell surface. Thus, membrane-bound TAPBPR expressed on the surface of a cell can be used as an assay to interrogate TAPBPR-mediated peptide exchange on MHC I on a cellular membrane.
Soluble TAPBPR Facilitates Peptide Exchange on Surface HLA-A*68:02 Molecules.
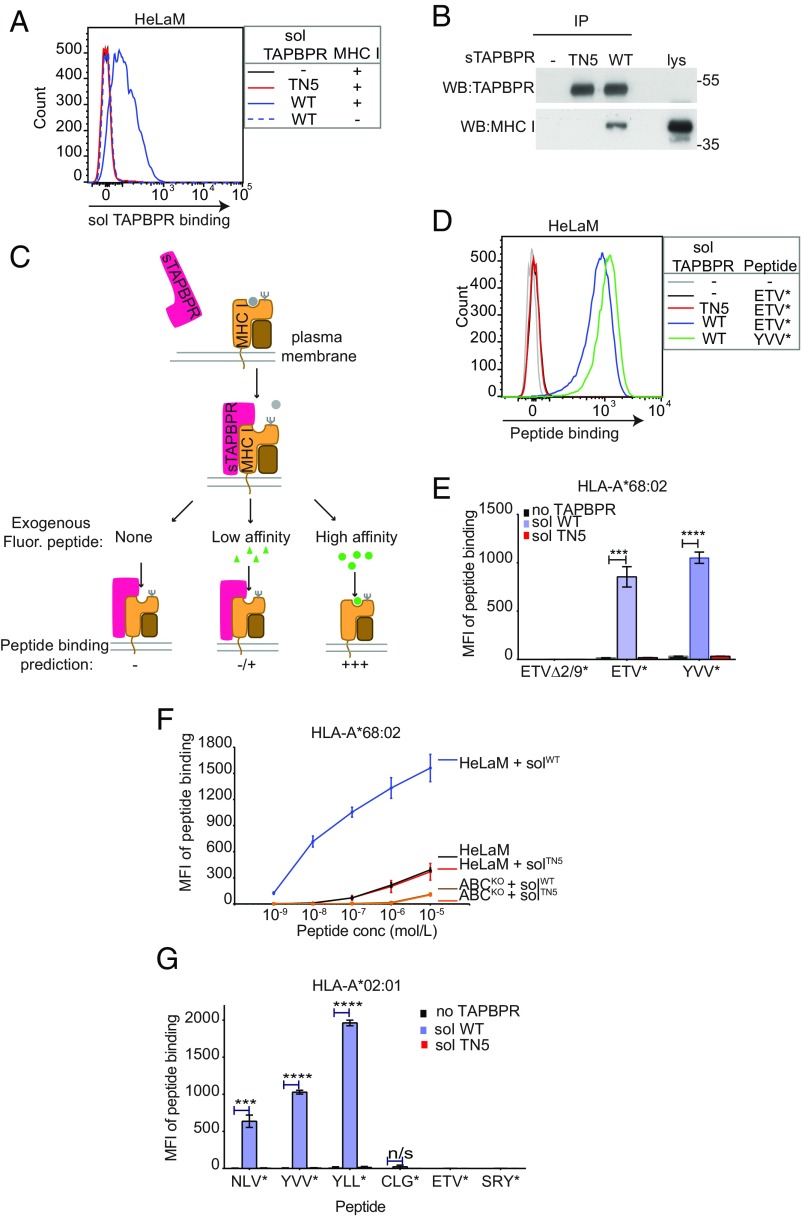
Following on from this, we were curious whether soluble exogenous TAPBPR added to cells was also capable of functioning as a peptide-exchange catalyst on surface MHC I molecules. First, we tested whether soluble TAPBPR, consisting only of its luminal domain (i.e., lacking its transmembrane domain and cytoplasmic tail) could bind to surface MHC I molecules. When we incubated HeLaM cells with soluble TAPBPRWT for 15 min, TAPBPR was clearly detectable on the cell surface (Fig. 3A). The binding of TAPBPR to cells was dependent on its association with MHC I because soluble TAPBPRTN5, a mutant that cannot bind to MHC I (15), did not bind to HeLaM cells (Fig. 3A) and since soluble TAPBPRWT could no longer bind to HeLaM cells in which HLA-A, -B, and -C molecules had been knocked out (Fig. 3A). Furthermore, the binding of soluble TAPBPRWT to HLA-ABC deficient HeLaM cells was restored when HLA-A*68:02 expression was reconstituted (SI Appendix, Fig. S4A). In addition, soluble TAPBPRWT, but not TAPBPRTN5, bound to MHC I in TAPBPR pulldown experiments (Fig. 3B).
Fig. 3.
Soluble TAPBPR enhances exogenous peptide association onto surface MHC I. (A) IFN-γ–treated HeLaM cells were incubated ±100 nM soluble TAPBPRWT or TAPBPRTN5 for 15 min at 37 °C, followed by detection of surface-bound TAPBPR using PeTe-4. Soluble TAPBPRWT binding to HeLaM-HLA-ABCKO cells (MHC−) is included as a control. (B) TAPBPR pulldowns on lysates from IFN-γ–treated HeLaM-TAPBPRKO cells incubated ±soluble TAPBPRTN5 or TAPBPRWT demonstrate that TAPBPRWT binds to MHC I. Data are representative of three independent experiments. (C) Schematic representation of experimental workflow used to measure peptide exchange by soluble TAPBPR. (D and E) IFN-γ–treated HeLaM cells were incubated ±100 nM soluble TAPBPRWT or TAPBPRTN5 for 15 min at 37 °C, followed by incubation ±10 nM ETVSK*QSNV (ETV*), YVVPFVAK*V (YVV*), or EGVSK*QSNG (ETVΔ2/9) for 15 min at 37 °C. In D, histograms show the typical binding observed for ETVSK*QSNV and YVVPFVAK*V and E shows the MFI of fluorescent peptide binding ±SD from three independent experiments. (F) Dose–response curves ±SD from three independent experiments of ETVSK*QSNV binding to IFN-γ–treated HeLaM and HeLaM-HLA-ABCKO cells treated ±100 nM TAPBPR with increasing concentrations of peptide for 15 min at 37 °C. (G) Bar graph show the MFI of fluorescent peptide binding to IFN-γ–treated HeLaM-HLA-ABCKO cells reconstituted with HLA-A*02:01 ± SD from two independent experiments with duplicates. Cells were incubated in the absence or presence of 1 µM soluble TAPBPRWT or TAPBPRTN5 for 15 min, followed by incubation with 10 nM NLVPK*VATV (NLV*), YVVPFVAK*V (YVV*), YLLEK*LWRL (YLL*), CLGGK*LTMV (CLG*), ETVSK*QSNV (ETV*), or SRYWK*IRTR (SRY*) for 60 min. ***P ≤ 0.001, ****P ≤ 0.0001, n/s not significant, using unpaired two-tailed t test.
Next, we explored the capability of soluble TAPBPR to promote peptide exchange on surface MHC I molecules by testing its ability to replace naturally presented peptides, with an exogenously added fluorescent peptide. Cells were pretreated ± soluble TAPBPR for 15 min, followed by incubation ± fluorescent peptide with varying affinity for HLA-A*68:02 for an additional 15 min (Fig. 3C). Subsequently, the amount of fluorescent peptide bound to cells was determined using flow cytometry. We found soluble TAPBPRWT significantly enhanced the association of fluorescent peptides specific for HLA-A*68:02, ETVSK*QSNV, and YVVPKVAK*V, onto HeLaM cells (Fig. 3 D and E). Negligible peptide binding was observed either in the absence of soluble TAPBPRWT or in the presence of soluble TAPBPRTN5 (Fig. 3 D and E). No association was observed for the nonbinding peptide, EGVSK*QSNG, under any of the conditions tested (Fig. 3E and SI Appendix, Fig. S4 B and C). Strikingly, soluble TAPBPRWT promoted peptide association onto cells at extremely low levels of exogenous peptide, requiring ∼1,000-fold less peptide to obtain the equivalent peptide binding observed in the absence of TAPBPR (Fig. 3F). Exogenous peptide binding to cells via soluble TAPBPRWT occurred directly onto MHC I since peptide association was only observed on MHC I competent cells and not on MHC I-deficient counterparts (Fig. 3F and SI Appendix, Fig. S4B) and binding was restored upon HLA-A*68:02 reconstitution (SI Appendix, Fig. S4 C and D). Furthermore, soluble TAPBPRTN5, which cannot bind to MHC I, was incapable of promoting peptide association (Fig. 3F). These results clearly demonstrate that the luminal domain of TAPBPR alone is sufficient to promote peptide exchange on surface HLA-A*68:02 molecules. Thus, by incubating intact cells with soluble TAPBPR, we now have a second assay on a cellular membrane to explore TAPBPR-mediated peptide editing on MHC I.
Soluble TAPBPR Facilitates Peptide Exchange on Surface HLA-A*02:01 Molecules.
We extended our analysis to test the ability of soluble TAPBPR to load a range of exogenous peptides onto another human MHC I molecule, HLA-A*02:01, expressed on HeLaM-HLA-ABCK0-A2+cells. In TAPBPR pulldown experiments, we observed an association between soluble TAPBPRWT, but not TAPBPRTN5, with HLA-A2 expressed in HeLaM-HLA-ABCKO-A2+ cells (SI Appendix, Fig. S5A). Soluble TAPBPRWT significantly promoted the binding of fluorescently labeled variants of: NLVPMVATV [an immunogenic peptide derived from the CMV protein pp65 (16)], YVVPFVAKV [derived from human CCR4-NOT transcription complex subunit 1 (6)], and YLLEMLWRL [an immunogenic peptide derived from the EBV protein Latent membrane protein 1 (LMP1) (17)] (Fig. 3G and SI Appendix, Fig. S5B). The TAPBPR-promoted loading of these peptides was dependent on HLA-A2, as fluorescent peptide binding was not detectable on HLA-A2− cells (SI Appendix, Fig. S5 C and D). Soluble TAPBPRWT did not promote binding of peptides specific for other MHC I molecules onto HLA-A2 and also did not significantly enhance the association of the HLA-A2 binding peptide CLGGLLTMV, an immunogenic peptide derived from the Epstein Barr virus (EBV) protein LMP2 (Fig. 3G and SI Appendix, Fig. S5B). Taken together, these data strongly suggest that soluble TAPBPR can promote the loading of exogenous peptide onto surface MHC I in an affinity-based manner.
TAPBPR Loads Immunogenic Peptides onto Human Tumor Cells, Thereby Inducing Their Recognition by T Cells.
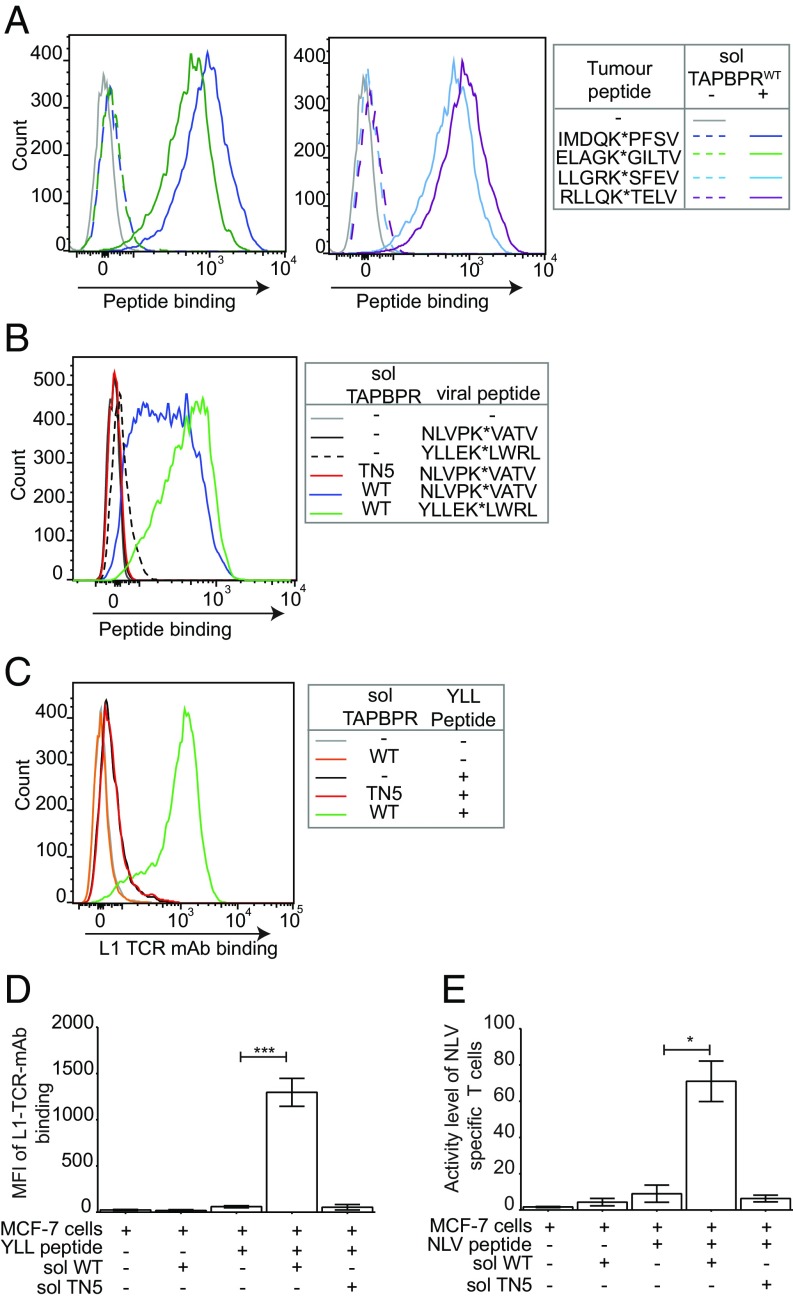
Having identified that adding soluble TAPBPR to intact cells is an efficient and extremely fast way of overriding the endogenous antigen processing and presentation pathway of cells, we were interested in testing whether this may have any translational potential. As the ability to increase neoantigen or indeed foreign antigen presentation on tumors would prove extremely useful in overcoming low immunogenicity often observed in tumors (18), we next tested whether soluble TAPBPR could enhance the binding of both tumor-derived and viral peptides onto tumor cells. We found that soluble TAPBPRWT significantly enhanced the loading of fluorescent derivatives of the tumor antigens IMDQVPFSV (derived from gp100) (19), ELAGIGILTV (from Melan-A/MART-1) (20), LLGRNSFEV (derived from p53) (21), and RLLQETELV (from HER-2/neu) (22) (Fig. 4A) onto HLA-A*02:01 naturally expressed on MCF-7, a breast cancer cell line. Soluble TAPBPRWT also promoted the association of fluorescently labeled derivatives of the viral peptides YLLEMLWRL (from EBV LMP1) and NLVPMVATV (from CMV) onto MCF-7 (Fig. 4B).
Fig. 4.
Antigenic peptides loaded onto MHC I via TAPBPR are available to the T cell receptor. MCF-7 cells were treated ±1 µM soluble TAPBPRWT or TAPBPRTN5 for 15 min at 37 °C followed by 60-min incubation ±10 nM (A) IMDQK*PFSV, ELAGK*GILTV, LLGRK*SFEV, or RLLQK*TELV; (B) NLVPK*VATV or YLLEK*LWRL; or (C and D) YLLEMLWRL (YLL) followed by staining with the TCR-like mAb L1 specific for YLLEMLWRL/HLA-A2 complexes. (D) The MFI of L1 binding to MCF-7 cells ±SD from three independent experiments. (E) Bar graphs show T cell activity measured by IFN-γ secretion in fluorospot assays of a HLA-A2 restricted NLVPMVATV specific CD8+ T cell line when incubated with MCF-7 target cells as treated in B, with the exception that nonfluorescent NLVPMVATV peptide at 100 pM was used. Results are from triplicate wells representative of two independent experiments. Error bars ± SD. Note: In A, B, and E IFN-γ–treated cells were used. Equivalent experiments of B–E were performed using HeLaM-HLA-ABCKO expressing HLA-A*02:01 and can be found in SI Appendix, Fig. S7. *P ≤ 0.05, ***P ≤ 0.001 using unpaired two-tailed t test.
We subsequently determined whether the peptides loaded via TAPBPR were available for T cell receptor (TCR) detection. Encouragingly, soluble TAPBPR dissociates from cells upon high-affinity peptide binding onto surface MHC I molecules (SI Appendix, Fig. S6), raising the possibility that TAPBPR-loaded peptide:MHC complexes might be fully accessible for TCR detection. We found that YLLEMLWRL loaded onto MCF-7 cells by TAPBPR was strongly detected by the anti-EBV TCR-like mAb L1, specific for LMP1125–133 presented on HLA-A*02:01 (17) (Fig. 4 C and D). Furthermore, NLVPMVATV loaded onto MCF-7 cells by soluble TAPBPR significantly increased the stimulation, measured by IFN-γ secretion, of human CD8+ T cells specific for pp65495–503 presented on HLA-A2 (16), compared with cells incubated with peptide alone or in the presence of soluble TAPBPRTN5 (Fig. 4E). We have further verified these findings using HeLaM-HLA-ABCKO-A2+ (SI Appendix, Fig. S7). These results demonstrate that soluble TAPBPR can efficiently load antigenic peptides onto tumor cell lines for recognition by CD8+ T cells.
Soluble TAPBPR Induces Tumor Cell Killing by CD8+ T Lymphocytes.
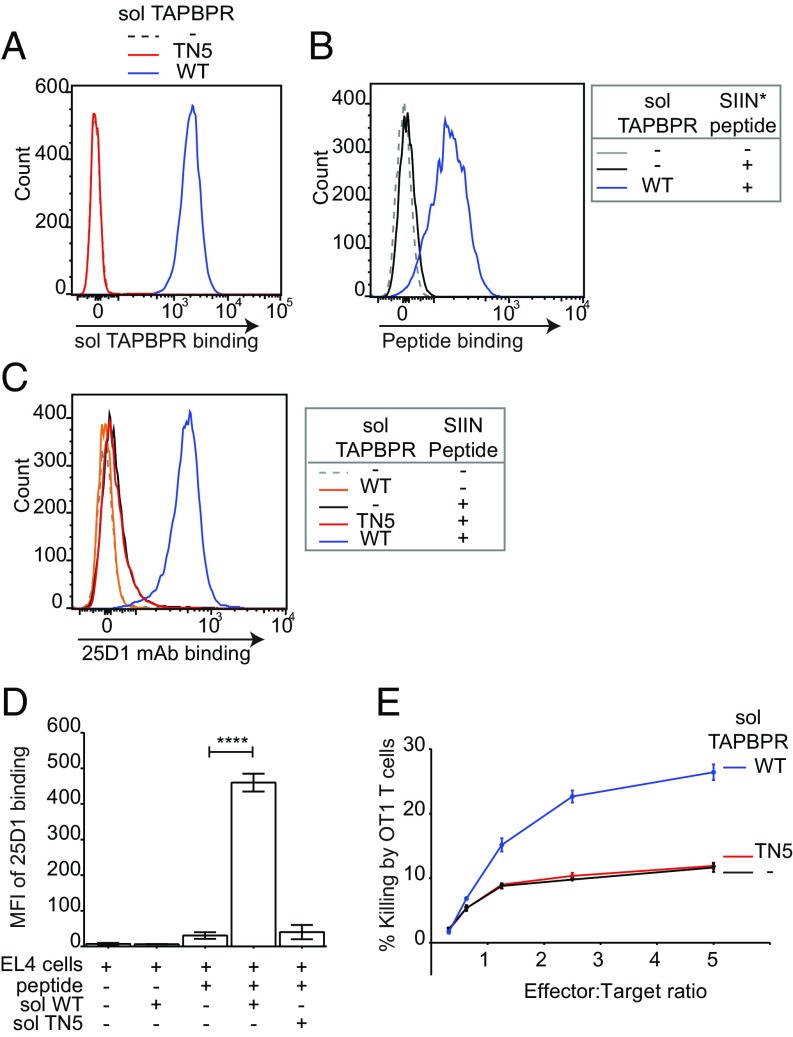
Although the results above suggest that soluble TAPBPR could potentially be utilized to decorate target cells with immunogenic peptides and enhance T cell responses against tumors, it was important to determine whether this could result in enhanced killing of tumor cells. To investigate this possibility, we assessed killing of murine EL4 tumor cells by OT1 T cells in the presence of human TAPBPR and very low concentrations of SIINFEKL peptide. Soluble human TAPBPRWT bound to EL4 cells (Fig. 5A) and significantly enhanced the loading of SIINFEKL onto H-2Kb expressed on EL4 (Fig. 5 B–D). When we tested the ability of OT1 cytotoxic T cells, which recognize SIINFEKL in the context of H-2Kb, to lyse peptide-pulsed EL4 target cells, we observed a significant enhancement in killing in the presence of soluble human TAPBPRWT, but not in the presence of TAPBPRTN5 (Fig. 5E). These results demonstrate that TAPBPR can be utilized to enhance the killing of tumors by peptide-specific CD8+ T lymphocytes.
Fig. 5.
Soluble TAPBPR enhances T cell killing of tumor cells. EL4 cells were incubated ±1 µM soluble TAPBPRWT or TAPBPRTN5 for 15 min at 37 °C, followed by (A) detection of surface bound TAPBPR using PeTe-4, (B) incubation ±1 nM SIINFEK*L for 30 min at 37 °C, or (C) incubation ±1 nM nonlabeled SIINFEKL peptide for 30 min, followed by staining with the 25-D1.16 mAb (recognizes SIINFEKL/H-2Kb complexes). Histograms are representative of three independent experiments. (D) Bar graphs show the MFI of 25-D1.16 ± SD from three independent experiments. (E) OT1 killing of EL4 cells treated ±1 µM soluble TAPBPRWT or TAPBPRTN5, followed by incubation with 1 nM SIINFEKL peptide. Error bars ± SEM from triplicate wells. Data are representative of three independent experiments. Note: surface expressed H-2Kb are relatively more peptide receptive compared with human MHC I molecules. At 10 nM SIINFEKL, some exogenous peptide binding was observed in the absence of soluble TAPBPRWT. Because OT1 T cells are highly efficient cytotoxic cells, killing 80–100% of targets after 1–4 h, we decreased the concentration of SIINFEKL used in these experiments to 1 nM to differentiate between TAPBPR-mediated and background peptide binding; otherwise, we would not observe an additive effect of soluble TAPBPR on target cell killing. ****P ≤ 0.0001 using unpaired two-tailed t test.
Discussion
Although TAPBPR usually functions as an intracellular peptide editor on MHC I molecules, we demonstrate that when given access to the surface pool of MHC I molecules, either through targeting full-length TAPBPR to the PM or by adding soluble TAPBPR to cells, TAPBPR retains its function as a peptide-exchange catalyst. Thus, we have developed two cell-based peptide-exchange systems for MHC I, which complement those already established (11, 12). Here, we have shown that TAPBPR can mediate peptide editing on three distinct MHC I molecules (HLA-A*68:02, HLA-A*02:01, and H-2Kb) expressed on the surface of cells. As expected, the efficiency of TAPBPR-mediated peptide exchange is dependent on affinity of the incoming peptide for a particular MHC I.
Intriguingly, our work, particularly when using soluble TAPBPR, demonstrates that TAPBPR can dissociate peptides that apparently have relatively high affinity for MHC I, given that it works on MHC complexes expressed on the surface of cells with an intact antigen-presentation pathway and thus on molecules that have already undergone the process of chaperone-mediated quality control. This raises interesting questions regarding the precise criteria by which TAPBPR selects peptides. This ability of TAPBPR to outcompete apparently good peptides from MHC I relatively quickly may explain why TAPBPR levels in cells are quite low.
Our cell-based assays for determining the ability of TAPBPR to catalyze peptide exchange on MHC I molecules offer a number of advantages over the already-established cell-free assays, representing a more physiological system for exploring this concept. First, in contrast to the cell-free systems (6, 7, 11, 12), our assays here assess the interaction between TAPBPR and MHC I molecules in their naturally occurring membrane-bound conformations, taking into account the restrictions imposed by a cellular membrane, either on both the MHC I molecules and on TAPBPR, or on MHC I alone. Second, as opposed to the bacterial refolds used in the Chen and Bouvier assay (11), the MHC I molecules present in our system are subjected to the naturally occurring posttranslational modifications within the cell, as is also the case in Wearsch and Cresswell’s (12) assay; moreover, the MHC I molecules here are loaded with a broad spectrum of peptides instead of being refolded around single individual ones, creating a less-biased and broader range of ligands for TAPBPR. In addition, the cellular assays offer the possibility to screen the ability of TAPBPR to function as a peptide-exchange catalyst on a broad range of MHC molecules in a highly efficient manner, simply by using the MHC I molecules expressed on cells, and without the need to make bacterial refolds of individual MHC I.
In contrast to TAPBPR, we found that tapasin was not able to perform its peptide-editing function on surface-expressed MHC I molecules. There are a number of potential reasons to explain the difference in the ability of the two peptide editors to function on surface MHC I molecules. First, because surface-expressed MHC I complexes are loaded with good peptides, they may no longer be accessible to tapasin-mediated peptide editing. Second, TAPBPR appears to have higher affinity for MHC I than tapasin (7), a property that contributed to the recent success of crystallizing the TAPBPR:MHC I complex (8, 9). Third, the luminal domain of TAPBPR alone is known to efficiently mediate peptide exchange (6, 7), while tapasin either needs other cofactors or artificial zippering to MHC I (11, 12). Thus, when tapasin is targeted to the PM, it will lack the other cofactors that it requires to work efficiently as a peptide editor. Finally, we have recently shown that TAPBPR interacts with MHC I in a glycan-independent manner and appears to have no particular preference for the glycan attached (23). This is in contrast to tapasin, which can only associate efficiently with monoglucosylated MHC I via its interactions with calreticulin. Thus, TAPBPR appears to have the ability to function on a wider pool of MHC I molecules with the broader range of N-linked glycan attachments than tapasin. This now appears to include MHC I with mature sugar attachments.
Strikingly, TAPBPR mediates exogenous peptide binding to surface expressed MHC I molecules at an extremely high rate and at very low peptide concentrations. Thus, we have identified a highly efficient way of overriding the endogenous antigen-processing and -presentation pathway of cells.
This discovery has a number of potential future applications. To begin with, soluble TAPBPR may prove an extremely useful tool for researchers studying immune responses to viruses and tumors. By utilizing soluble TAPBPR on cells, investigators will be able to manipulate the peptides presented directly onto surface-expressed MHC I molecules, replacing endogenous cargo with specific peptides of choice, such as those derived from viral proteins or tumor antigens. Currently, peptide-pulsing alone (i.e., without a catalyst) is commonly used to load exogenous peptides onto MHC I. However, this requires high concentrations of peptide, particularly for human MHC I, over a long time period and is often performed on antigen-processing–deficient cells or on cell incubated at 26 °C to increase its efficiency. The level of peptide loading achieved using soluble TAPBPR on intact cells at 37 °C for 15 min is vastly superior to that observed when cells are incubated at 26 °C (>eightfold higher) or to that observed on TAP− cells (SI Appendix, Fig. S8). Thus, the addition of soluble TAPBPR will permit efficient peptide loading at low concentrations of peptide on any desired cell line at 37 °C almost instantaneously, which may have additional benefits when moving from in vitro to in vivo experimentation, given that the half-life of 8–10mer peptides is likely to be extremely short. Furthermore, by creating peptide-receptive MHC I molecules, TAPBPR may permit the study of immune responses to exogenous peptides with lower affinity for MHC, which is more difficult when using peptide alone.
Perhaps the most exciting implication of our findings is the translational potential of utilizing TAPBPR to load immunogenic peptides onto tumor cells to target them for recognition by CTLs. With the recent advances in cancer immunotherapy, powerful antitumor responses of CTLs can be exploited to eliminate cancer (24, 25). Central to CTL-mediated tumor cell elimination is the recognition of immunogenic peptides presented on MHC I molecules. Neoantigens, which arise de novo from tumor-specific mutations, are considered ideal targets, as they are only expressed on cancer cells and thereby avoid central tolerance. However, their presentation on MHC I is likely to be low (26, 27). Therefore, the ability to increase the expression of such neoantigens, or indeed to induce foreign antigen presentation on tumors, would be a fundamental step forward in overcoming low immunogenicity often observed in tumors. Thus, our ability to use TAPBPR to increase the immunogenicity of cells may represent a major advance for the future of immunotherapy to improve treatment outcomes in patients with tumors resistant to current therapies.
Materials and Methods
Constructs.
The expression of full-length TAPBPRWT in the lentiviral vector pHRSIN-C56W-UbEM, which produces the protein of interest under the control of the spleen focus-forming virus promoter and the GFP derivative emerald under the control of an ubiquitin promoter, has been as previously described (5, 15). Tapasin was amplified from cDNA isolated from human foreskin fibroblasts using primers tapasinWT-BamHI-for and tapasinWT-NotI-rev (See SI Appendix, Table S1 for primer sequences), followed by cloning into pHRSIN-C56W-UbEM. The chimeric constructs TAPBPRPM and tapasinPM were generated using a two-step PCR procedure, where the ectodomain and transmembrane domain of either TAPBPR (amplified using primers TAPBPRWT-BamHI-for and TAPBPRPM-rev) or tapasin (amplified using primers tapasinWT-BamHI-for and tapasinPM-rev) were fused to the cytoplasmic tail of CD8 (amplified with primers TAPBPRPM-for and CD8 tail-NotI-rev, or tapasinPM-for and CD8 tail-NotI-rev, respectively). TAPBPRER was produced using a similar procedure, in which the ectodomain of TAPBPR (amplified with primers TAPBPRWT-BamHI-for and TAPBPRER-rev) was fused to the transmembrane and cytoplasmic domains of tapasin (amplified using primers TAPBPRER-for and tapasinWT-NotI-rev). Subsequently, these three chimeric inserts were cloned into pHRSIN-C56W-UbEM. The luminal domains of either TAPBPRWT or TAPBPRTN5 (6) were also cloned in a PiggyBac transposon vector (using primer TAPBPR-soluble-for and TAPBPR-soluble-rev) to produce secreted versions of these proteins, containing a His tag at the C terminus in a mammalian expression system. The full-length HLA-A*02:01 and HLA-A*68:02 constructs were cloned into the lentiviral vector pHRSINcPPT-SGW (15). HCMV pp65 was cloned into the lentiviral vector pHRSIN-C56W-UbEM.
Cell Culture.
HeLaM cells, a variant HeLa cell line that is more responsive to IFN (28) (a gift from Paul Lehner, University of Cambridge, UK), their modified variants, HEK-293T cells (a gift from Paul Lehner, University of Cambridge, UK), MCF-7, and H-2b EL4 cells were maintained in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Gibco, Thermo Fisher Scientific), 100 U/mL penicillin, and 100 µg/mL streptomycin (Gibco, Thermo Fisher Scientific) at 37 °C with 5% CO2. To induce expression of endogenously expressed TAPBPR and up-regulate other components of the MHC I antigen-processing and -presentation pathway, HeLaM and MCF-7 cells were treated with 200 U/mL IFN-γ (Peprotech) for 48–72 h, where indicated.
Lentiviral Transduction and Transfections.
Lentivirus was produced by transfecting HEK-293T cells with lentiviral vectors along with the packaging vector pCMVΔR8.91 and the envelope vector pMD.G using Fugene (Promega). Viral supernatant was collected at 48 h and used to transduce different cell lines, as follows: TAPBPRWT, TAPBPRTN5, TAPBPRPM, TAPBPRER, tapasinWT, and tapasinPM were reconstituted in a TAPBPR-deficient HeLaM cell line (HeLaM-TAPBPRKO) (10); HLA-A*02:01 and HLA-A*68:02 were reconstituted in a HeLaM cell line deficient of the HLA-A, -B, and -C (HeLaM-HLA-ABCKO) (23). HeLa HLA-A2 pp65 cells were generated by transducing HeLaM cells first with HLA-A2-pHRSINcPPT-SGW, followed by pp65-pHRSIN-C56W-UbEM.
Antibodies.
TAPBPR was detected using either PeTe4, a mouse monoclonal antibody (mAb) specific for the native conformation of TAPBPR, raised against amino acids 22–406 of human TAPBPR (5) that does not cross-react with tapasin (15), or ab57411, a mouse mAb raised against amino acids 23–122 of TAPBPR that is reactive to denatured TAPBPR (Abcam). Tapasin was detected using Pasta-1 (29) (a kind gift from Peter Cresswell, Yale University, New Haven, CT). UGT1 was detected using the rabbit mAb ab124879 (Abcam). MHC I heavy chains were detected using mAb HC10 (30). OVA257–264 (SIINFEKL) peptide on H-2Kb was detected using the mAb 25D-1.16 (Thermofisher). The EBV-derived peptide LMP1125–133 (YLLEMLWRL) in association with HLA-A*02:01 was detected using the TCR-like mAb L1 (17) (a kind gift from Paul MacAry, National University of Singapore, Singapore). A mouse IgG2a isotype control was also used as a control (Sigma-Aldrich).
MHC Class I Binding Peptides.
The following MHC I-specific peptides were used: HLA-A*68:02-binding peptide ETVSEQSNV, its derivative EGVSEQSNG, obtained by replacing its anchor residues (amino acids on positions 2 and 9) with glycine, as well as their fluorescently labeled versions ETVSK*QSNV and EGVSK*QSNG, respectively [K* represents a lysine labeled with 5-carboxytetramethylrhodaime (TAMRA)]; HLA-A*02:01 binding peptides NLVPMVATV, YLLEMLWRL, CLGGLLTMV, YVVPFVAKV, IMDQVPFSV, LLGRNSFEV, ELAGIGILTV and RLLQETELV, together with their fluorescently labeled variants NLVPK*VATV, YLLEK*LWRL, CLGGK*LTMV, YVVPFVAK*V, IMDQK*PFSV, LLGRK*SFEV, ELAGK*GILTV, and RLLQK*TELV, respectively; HLA-B*27:05 specific peptide SRYWAIRTR and its fluorescently labeled variant SRYWK*IRTR; H-2Kb specific peptide SIINFEKL and its fluorescently-labeled variant SIINFEK*L. All peptides were purchased from Peptide Synthetics.
Expression and Purification of TAPBPR Protein.
Secreted forms of either TAPBPRWT or TAPBPRTN5 were expressed in HEK 293T cells, using the PiggyBac expression system. The C-terminally His-tagged ectodomain of either protein was cloned into the PB-T-PAF vector. 293T cells were cotransfected in six-well plates with 0.9 µg PB vector and 0.15 µg of both PB-RN and PBase (at a ratio of 6:1:1). Forty-eight hours after transfection, cells were transferred for at least 5 d into selection media: DMEM supplemented with 10% FBS, 1% pen/strep, 3 µg/mL puromycin (Invivogen), and 700 µg/mL geneticin (Thermo Fisher Scientific). To induce protein expression, cells were harvested and transferred into DMEM supplemented with 5% FBS, 1% pen/strep, and 2 µg/mL doxycycline (Sigma-Aldrich). After 7 d, the media was collected and TAPBPR was purified using Ni-NTA affinity chromatography. For purity assessment, elution fractions were analyzed by SDS/PAGE, followed by Coomassie staining.
Flow Cytometry.
Following trypsinization, cells were washed in 1% BSA, dissolved in 1× PBS at 4 °C, and then stained for 30 min at 4 °C in 1% BSA containing one of the following antibodies: PeTe4, Pasta-1, TCR-like mAb L1, 25-D1.16, or with an isotype control antibody. After washing the cells to remove excess unbound antibody, the primary antibodies bound to the cells were detected by incubation at 4 °C for 25 min with goat anti-mouse Alexa-Fluor 647 IgG (Invitrogen Molecular Probes; Thermo Fisher Scientific). After a subsequent three rounds of washing, the fluorescence levels were detected using a BD FACScan analyzer with Cytek modifications and analyzed using FlowJo (FlowJo).
Immunoprecipitation, Gel Electrophoresis, and Western Blotting.
Cells were harvested then washed in PBS. For immunoprecipitation of the TAPBPR fraction present at the PM, cells were incubated with 2 µg PeTe4 antibody in 1% BSA in 1× PBS for 1 h with rotation at 4 °C. Excess antibody was removed by washing the cells five times in 1× PBS at 4 °C. Cells were then lysed in 1% Triton X-100 (VWR), Tris-buffered saline (TBS) (20 mM Tris⋅HCl, 150 mM NaCl, 2.5 mM CaCl2), supplemented with 10 mM NEM, 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich), and protease inhibitor mixture (Roche) for 30 min at 4 °C. Nuclei and cell debris were pelleted by centrifugation at 13,000 × g for 15 min and supernatants were collected. The TAPBPR fraction originally present the PM, bound to the PeTe4 antibody, was then precipitated using protein A Sepharose beads alone (GE Healthcare) for 2 h at 4 °C with rotation. Following the immunoprecipitation of the PM TAPBPR fraction, the flow-through was collected and subjected to a subsequent round of immunoprecipitation, this time using protein A Sepharose beads conjugated to PeTe4 antibody, to pull down the intracellular TAPBPR fraction. Following immunoprecipitation, beads were washed thoroughly in 0.1% detergent-TBS to remove unbound protein. For separation by gel electrophoresis, the samples were heated at 94 °C for 10 min in sample buffer (125 mM Tris⋅HCl pH 6.8, 4% SDS, 20% glycerol, 0.04% Bromophenol blue), supplemented with 100 mM β-mercaptoethanol. To detect the samples by Western blotting, proteins were transferred onto an Immobilon transfer membrane (Merck Millipore). Membranes were blocked using 5% (wt/vol) dried milk and 0.05% (vol/vol) Tween 20 in PBS for 30 min and subsequently incubated with the indicated primary antibody for 1–16 h. After washing, membranes were incubated with species-specific HRP-conjugated secondary antibodies, washed, and detected by enhanced chemiluminescence using Western Lightning (Perkin-Elmer) and Super RX film (Fujifilm). Films were scanned on a CanoScan8800F using MX Navigator Software (Canon).
For pulldown experiments using soluble TAPBPR proteins, protein A Sepharose precleared lysates from IFN-γ–stimulated HeLaM-TAPBPRKO cells were incubated with 5 µg of the soluble TAPBPR variant for 90 min at 4 °C. Immunoprecipitation of soluble TAPBPR was performed using PeTe4 as above. Soluble TAPBPR was detected on Western blots with the antipoly His primary antibody.
Peptide Binding.
Target cell lines were seeded at 25,000–30,000 cells per well in 12-well plates and stimulated with IFN-γ. Following the stimulation period, the cells were washed three times with 1× PBS and incubated with 300 μL prewarmed opti-MEM (Thermo Fisher Scientific). In case the peptide binding was done in the presence of recombinant TAPBPR, the cells were then treated with or without recombinant TAPBPR (100 nM for HLA-A*68:02 or 1 µM for HLA-A*02:01 and H-2Kb). After 15 min, the desired TAMRA-labeled peptide was added to the cells and incubated at 37 °C (15 min for HLA-A*68:02, 60 min for HLA-A*02:01 or 30 min for H-2Kb). In case the peptide binding was facilitated by overexpressed TAPBPR, the labeled peptide was directly added to the cells, without using recombinant TAPBPR. Following the peptide treatment, the cells were washed three times in 1× PBS and harvested. The level of bound peptide/cell was determined by flow cytometry, using the YelFL1 channel (Cytek).
Peptide Exchange.
HeLaM-TAPBPRKO cells, reconstituted with TAPBPR were seeded at 25,000 cells per well and stimulated with IFN-γ. The cells were then washed and treated with 10 nM TAMRA-labeled peptide of interest diluted in opti-MEM for 15 min at 37 °C, as described above. Following the binding step, the peptide-containing medium was removed, the cells were washed, and then treated with media alone or with different concentrations of nonlabeled peptide for another 15 min at 37 °C. The cells were then washed and harvested and the level of bound peptide per cell was determined by flow cytometry, using the YelFL1 channel (Cytek).
FluoroSpot T Cell Assay.
Expansion of HCMV specific CD8+ T cells.
CD8+ T cells were isolated from peripheral blood mononuclear cells using MACS anti-CD8 direct beads (Miltenyi Biotec) magnetic separation and then resuspended in supplemented RPMI + 10% FBS (Invitrogen) + 10% heat inactivated autologous donor serum. Cells were stimulated with peptide pulsed irradiated autologous peripheral blood mononuclear cells in the presence of 2.5 IU/mL human recombinant IL-2 (National Institute for Biological Standards and Control, Potters Bar, United Kingdom) in round-bottom 96-well plates at 37 °C +5% CO2 for 10–14 d, fresh media was replenished every 5 d. Specificity of expanded CD8+ T cell cultures were tested for specificity using IFN-γ FLUOROSPOT assays stimulated with HeLa HLA-A2 pp65 cells. Individual HLA-restricted peptides from HCMV pp65 used in this study were HLA-A2 NLVPMVATV (pp65 495–504 aa).
Ethical approval was obtained from the Addenbrookes National Health Service Hospital Trust Institutional Review Board (Cambridge Research Ethics Committee) for this study. Informed written consent was obtained from all recipients in accordance with the Declaration of Helsinki (LREC 97/092).
Experimental set-up.
Target cells (MCF-7 or HeLaM-HLA-ABCKO reconstituted with HLA-A*02:01 heavy chain) were seeded at 80,000 cells per well of a six-well plate and stimulated with IFN-γ for 72 h. Cells were then washed three times with 1× PBS and incubated with 600 μL prewarmed opti-MEM, containing either soluble TAPBPRWT, TAPBPRTN5, or without TAPBPR. After 15 min, 100 pM NLVPMVATV peptide was added to the desired samples and incubated for another 60 min. Following peptide treatment, cells were washed three times in 1× PBS and harvested. Each sample was then washed again twice in 1× PBS and resuspended in X-VIVO 15 media (Lonza) at 1 × 106 cells/mL. Target cells were then irradiated for 20 min, to cease proliferation throughout the experiment. Triplicate wells of NLVPMVATV specific CD8+ T cells in X-VIVO 15 media were incubated in coated Fluorospot plates [human IFN-γ FLUOROSPOT (Mabtech AB)], at 8,000 cells per well, with target cells, at 50,000 cells per well, at 37 °C in a humidified CO2 atmosphere for 20 h. The cells and medium were decanted from the plate and the assay developed following the manufacturer’s instructions. Developed plates were read using an AID iSpot reader (Autoimmun Diagnostika) and counted using EliSpot v7 software (Autoimmun Diagnostika).
Mice.
OT-I RAG2−/− mice were a generous gift from Suzanne Turner (Department of Pathology, University of Cambridge, Cambridge, UK) and were bred and housed in accordance with United Kingdom Home Office regulations.
Cytotoxicity Assay.
To generate OT-I CTLs, spleens were extracted from OT-I RAG2−/− mice and single-cell suspensions of splenocytes were obtained using a 70-μm cell strainer (Greiner Bio-one). Splenocytes were stimulated with 10 nM OVA257–264 (SIINFEKL) peptide (Peprotech). After 3 d of culture cells were washed, seeded into fresh T cell media daily, and used 3–4 d later. T cells were cultured in RPMI medium 1640 with l-glutamine (Gibco, Thermo Fisher Scientific), 10% heat-inactivated FCS (Biosera), 50 μM of β-mercaptoethanol, 1 mM sodium pyruvate (Gibco, Thermo Fisher Scientific), 10 mM Hepes (Sigma-Aldrich), 50 IU/mL recombinant murine IL-2 (Peprotech), and 50 U/mL penicillin and streptomycin (Gibco, Thermo Fisher Scientific) (T cell media).
The CytoTox96 Non-Radioactive Cytotoxicity Assay (Promega) was used to measure EL4 target cell death. Target H-2b EL4 cells were washed the day before the experiment and resuspended in fresh DMEM at 3 × 105 to 4 × 105 cells/mL. The following morning, the EL4 cells were washed once and resuspended in warm opti-MEM at 5 × 105 cells/mL. The cells were treated initially with 1 μM soluble TAPBPR alone or with carrier alone for 10 min, after which either 1 nM OVA257–264 (SIINFEKL) peptide or carrier alone was added to the cells for another 30 min. Cells were then washed 1× in Optimem and 2× in killing assay media (RPMI medium minus phenol red, 2% FCS), and resuspended in killing assay media at 1 × 105 cells/mL in a round-bottom 96-well plate. Effector OT-I CTLs were washed in killing assay media once and then added to the plate at titrated effector to target cell (E:T) ratios. Plates were incubated at 37 °C and after 6–7 h EL4 killing was assessed by release of lactate dehydrogenase in the supernatant.
Supplementary Material
Acknowledgments
We thank Paul MacAry (University of Singapore) for the generous provision of T cell receptor-like mAbs that recognize Epstein Barr virus-derived peptides in the context of HLA-A*02:01; and Klaus Okkenhaug, John Trowsdale, and Ben Challis, all from the University of Cambridge, for proofreading our manuscript. L.H.B. and A.N. were funded by Wellcome Senior Research Fellowship 104647/Z/14/Z (to L.H.B.). F.T.I. was funded by Wellcome Doctoral Studentship 109076/Z/15/A. M.d.l.R. is supported by Cancer Research UK Cambridge Institute Core Grant C14303/A17197 and Sir Henry Dale Fellowship 107609/Z/15/Z jointly funded by the Wellcome and the Royal Society. M.R.W. is supported by Medical Research Council Grant MR/K021087/1 (Great Britain).
Footnotes
Conflict of interest statement: L.H.B., A.N., and F.T.I. have filed a patent based on this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809465115/-/DCSupplemental.
References
- 1.Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 2.Ortmann B, et al. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997;277:1306–1309. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Sjögren HO, Hellman U, Pettersson RF, Wang P. Cloning and functional characterization of a subunit of the transporter associated with antigen processing. Proc Natl Acad Sci USA. 1997;94:8708–8713. doi: 10.1073/pnas.94.16.8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–520. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 5.Boyle LH, et al. Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc Natl Acad Sci USA. 2013;110:3465–3470. doi: 10.1073/pnas.1222342110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermann C, et al. TAPBPR alters MHC class I peptide presentation by functioning as a peptide exchange catalyst. eLife. 2015;4:e09617. doi: 10.7554/eLife.09617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morozov GI, et al. Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc Natl Acad Sci USA. 2016;113:E1006–E1015. doi: 10.1073/pnas.1519894113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas C, Tampé R. Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing. Science. 2017;358:1060–1064. doi: 10.1126/science.aao6001. [DOI] [PubMed] [Google Scholar]
- 9.Jiang J, et al. Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation. Science. 2017;358:1064–1068. doi: 10.1126/science.aao5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neerincx A, et al. TAPBPR bridges UDP-glucose:glycoprotein glucosyltransferase 1 onto MHC class I to provide quality control in the antigen presentation pathway. eLife. 2017;6:e23049. doi: 10.7554/eLife.23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M, Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 2007;26:1681–1690. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wearsch PA, Cresswell P. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat Immunol. 2007;8:873–881. doi: 10.1038/ni1485. [DOI] [PubMed] [Google Scholar]
- 13.Hogan KT, et al. The peptide recognized by HLA-A68.2-restricted, squamous cell carcinoma of the lung-specific cytotoxic T lymphocytes is derived from a mutated elongation factor 2 gene. Cancer Res. 1998;58:5144–5150. [PubMed] [Google Scholar]
- 14.Nilsson T, Jackson M, Peterson PA. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 15.Hermann C, Strittmatter LM, Deane JE, Boyle LH. The binding of TAPBPR and tapasin to MHC class I is mutually exclusive. J Immunol. 2013;191:5743–5750. doi: 10.4049/jimmunol.1300929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wills MR, et al. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: Frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sim AC, et al. Defining the expression hierarchy of latent T-cell epitopes in Epstein-Barr virus infection with TCR-like antibodies. Sci Rep. 2013;3:3232. doi: 10.1038/srep03232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blankenstein T, Coulie PG, Gilboa E, Jaffee EM. The determinants of tumour immunogenicity. Nat Rev Cancer. 2012;12:307–313. doi: 10.1038/nrc3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parkhurst MR, et al. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J Immunol. 1996;157:2539–2548. [PubMed] [Google Scholar]
- 20.Valmori D, et al. Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J Immunol. 1998;160:1750–1758. [PubMed] [Google Scholar]
- 21.Theobald M, Biggs J, Dittmer D, Levine AJ, Sherman LA. Targeting p53 as a general tumor antigen. Proc Natl Acad Sci USA. 1995;92:11993–11997. doi: 10.1073/pnas.92.26.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawashima I, et al. The multi-epitope approach for immunotherapy for cancer: Identification of several CTL epitopes from various tumor-associated antigens expressed on solid epithelial tumors. Hum Immunol. 1998;59:1–14. doi: 10.1016/s0198-8859(97)00255-3. [DOI] [PubMed] [Google Scholar]
- 23.Neerincx A, Boyle LH. Preferential interaction of MHC class I with TAPBPR in the absence of glycosylation. Mol Immunol. August 1, 2018 doi: 10.1016/j.molimm.2018.06.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 27.Kalaora S, et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget. 2016;7:5110–5117. doi: 10.18632/oncotarget.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiwari RK, Kusari J, Sen GC. Functional equivalents of interferon-mediated signals needed for induction of an mRNA can be generated by double-stranded RNA and growth factors. EMBO J. 1987;6:3373–3378. doi: 10.1002/j.1460-2075.1987.tb02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dick TP, Bangia N, Peaper DR, Cresswell P. Disulfide bond isomerization and the assembly of MHC class I-peptide complexes. Immunity. 2002;16:87–98. doi: 10.1016/s1074-7613(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 30.Stam NJ, Spits H, Ploegh HL. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J Immunol. 1986;137:2299–2306. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.