Significance
There is increasing interest to understand how proteins fold as they are synthesized on the ribosome. However, we still lack basic knowledge such as how protein size, net charge, and thermodynamic stability impact cotranslational folding. Here, we have studied eight proteins of increasing size and provide a comprehensive picture of how the location in the ribosome exit tunnel where a protein folds correlates with protein size. Moreover, we demonstrate that the force exerted on the nascent chain by protein folding varies linearly with the thermodynamic stability of the folded state, and that the ribosome environment disfavors folding of domains of high net-negative charge. These findings establish important basic facts about cotranslational folding.
Keywords: arrest peptide, ribosome, protein folding, pulse proteolysis
Abstract
During the last five decades, studies of protein folding in dilute buffer solutions have produced a rich picture of this complex process. In the cell, however, proteins can start to fold while still attached to the ribosome (cotranslational folding) and it is not yet clear how the ribosome affects the folding of protein domains of different sizes, thermodynamic stabilities, and net charges. Here, by using arrest peptides as force sensors and on-ribosome pulse proteolysis, we provide a comprehensive picture of how the distance from the peptidyl transferase center in the ribosome at which proteins fold correlates with protein size. Moreover, an analysis of a large collection of mutants of the Escherichia coli ribosomal protein S6 shows that the force exerted on the nascent chain by protein folding varies linearly with the thermodynamic stability of the folded state, and that the ribosome environment disfavors folding of domains of high net-negative charge.
More than 40 y ago Anfinsen postulated that “the native structure is determined only by the protein’s amino acid sequence” (1). His work established the protein folding field, which up until recently has focused almost exclusively on biophysical studies of purified proteins that can be reversibly unfolded/refolded in vitro. However, it is becoming increasingly clear that many proteins start to fold cotranslationally, that is, while still being synthesized on the ribosome (2–6); elements of secondary structure or small protein domains may even fold completely inside the ribosome exit tunnel (7–13). In contrast to in vitro folding, cotranslational folding is still a poorly understood process (14), and we lack basic information such as how protein size and net charge relate to where in the exit tunnel a protein starts to fold, and how protein stability impacts cotranslational folding.
Here, using arrest peptides (APs) as force sensors (15, 16) and on-ribosome pulse proteolysis (17), we have analyzed the cotranslational folding of eight protein domains that display cooperative folding in vitro. The domains are of different size and fold type, of different thermodynamic stability, and of different net charge. We find direct correlations between protein size and the location in the ribosome exit tunnel at which a protein folds, and between thermodynamic stability and the pulling force generated on the nascent chain during folding. Further, it appears that nascent chain segments with high net-negative charge are pushed out of the negatively charged ribosome exit tunnel before they fold. These findings establish important basic facts about cotranslational folding and reinforce the view of the exit tunnel as an environment that can have a strong impact on protein folding.
Results
Folding Assays.
We have used two assays to follow cotranslational folding of protein domains: an AP-based assay that makes it possible to detect the tension generated in the nascent chain when a domain folds (8) and an on-ribosome pulse-proteolysis assay where thermolysin resistance is used as an indicator of folding (17).
Translational APs are short stretches of polypeptide that interact with the upper parts of the ribosome exit tunnel in such a way that they stall translation when the ribosome reaches the last codon in the AP (18). APs are sensitive to external forces pulling on the nascent chain (19), and stalling efficiency is reduced in proportion to the external pulling force (20, 21). Hence, APs can be used as force sensors to report on cotranslational processes such as protein translocation (22), membrane protein biogenesis (21, 23), and protein folding (8, 15, 20, 24).
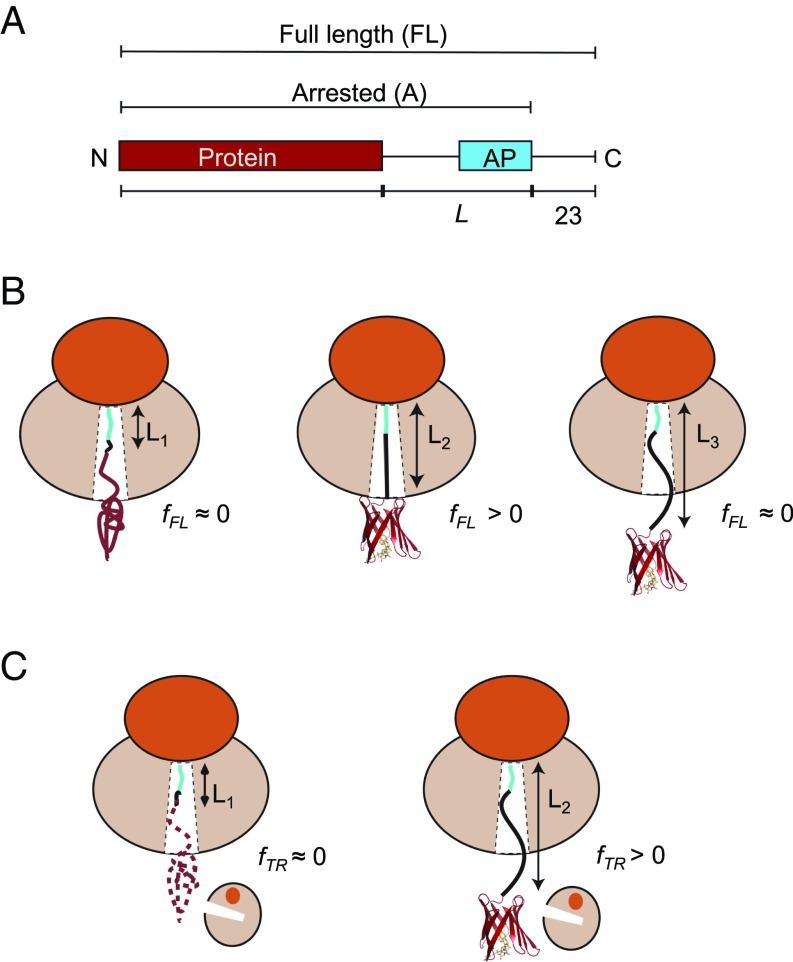
The basic construct used in the AP cotranslational folding assay is composed of the following elements (Fig. 1A): (i) the protein domain to be studied, (ii) a variable-length stretch of alternating glycine and serine (GS) residues, (iii) the AP, and (iv) a 23-residue C-terminal extension that makes it possible to separate arrested and full-length protein products by SDS/PAGE. We define linker length L as the number of residues between the protein domain and the end of the AP. As illustrated in Fig. 1B, for small L (short linkers) there is not enough space in the ribosome exit tunnel to allow folding at the point during translation when the ribosome reaches the end of the AP, and little force is exerted on the AP. At some intermediate value of L, the chain is just long enough to allow folding if stretched, and the force on the AP is increased. At large L, the domain is already folded when the ribosome reaches the codon at end of the AP, and again little force is generated. Since APs stall translation less efficiently when under tension (18, 20, 21), the fraction full-length protein (fFL) produced for constructs with different L reflects the variation in force generated on the AP by the folding reaction, and a plot of fFL vs. L can be used to infer where in the exit tunnel folding occurs during translation (15). Mutagenesis studies (16, 25), as well as visualization of folded protein domains located within the exit tunnel by cryo-EM (8, 15, 16, 24) and molecular dynamics simulations (8, 24), show that the dominant peak in a fFL profile corresponds to folding into the native state (as opposed to, e.g., nonspecific compaction of the nascent chain), at least for small, single-domain proteins; further support for this notion is provided below. fFL profiles were recorded for each protein by in vitro translation in the PURE (Protein synthesis Using Recombinant Elements) system (26), separation of arrested (A) and full-length (FL) forms of the protein by SDS/PAGE, and quantitation of the A and FL bands by phosphoimager analysis.
Fig. 1.
(A) Schematic representation of in vitro translated constructs. The protein under study is placed L residues upstream of the last proline in the AP, and a C-terminal 23-residue extension is added downstream of the AP to ensure that the arrested and full-length forms of the construct can be cleanly separated by SDS/PAGE. (B) The AP-based assay: At linker length L1, the protein is too deep in the exit tunnel to be able to fold, and hence no force is generated on the AP and mostly arrested nascent chains are produced (fFL ≈ 0). At L2, if the linker is stretched beyond its equilibrium length the protein can just reach a location in the exit tunnel where there is sufficient space for it to fold. Some of the folding free energy is therefore stored as elastic energy in the linker, increasing the force on the AP. More full-length protein is produced (fFL > 0). At L3, finally, the protein is already folded when the ribosome reaches the C-terminal end of the AP, and little force is exerted on the AP (fFL ≈ 0). (C) The on-ribosome pulse proteolysis assay: At L1, the protein is located too deep in the exit tunnel to be able to fold and the nascent chain is hence degraded by a brief thermolysin pulse. At L2, the protein is folded and hence resistant to proteolysis. Note that the protein is attached to the ribosome via a (GS)n linker that in itself is insensitive to thermolysin.
The pulse-proteolysis assay (17) is based on the premise that a short exposure of a stalled ribosome-nascent chain complex to thermolysin will lead to degradation of the protein if it is partly or wholly unfolded, but not if it is folded (Fig. 1C). Since thermolysin cleaves only on the N-terminal side of hydrophobic residues (27), the (GS)n linker will not be digested. Like the AP assay, pulse proteolysis reports on folding into the native state rather than nonspecific compaction of the nascent chain (17, 28). Examples of SDS/PAGE gels for both assays are shown in SI Appendix, Fig. S1.
The Onset of Cotranslational Folding Correlates with Domain Size.
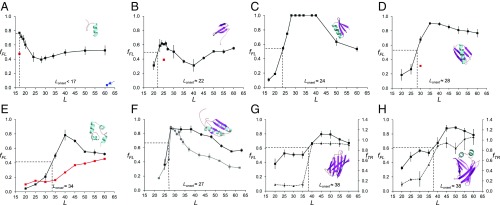
To put the possible relation between protein size and the location in the ribosome exit tunnel where folding commences (4, 7) on a firm quantitative footing, we selected eight protein domains of different folds and with sizes ranging from 28 to 128 residues for study (SI Appendix, Tables S1 and S2): the de novo-designed protein full sequence design 1 (FSD-1) (28 residues) (29), a WW domain (35 residues) (30), the de novo-designed protein EHEE_rd2_0005 (40 residues) (31), protein G (56 residues) (32), a calmodulin fragment (69 residues) (33), ribosomal protein S6 (101 residues) (34), superoxide dismutase 1 (SOD1, 110 residues) (35), and ileal binding protein (ILBP, 128 residues) (36). We used the moderately strong 17-residue Escherichia coli SecM(Ec) AP for all eight proteins (37); in addition we also used the stronger eight-residue SecM(Ms) AP from Mannheimia succiniciproducens (38) for protein S6. All fFL profiles are shown in Fig. 2.
Fig. 2.
Fraction full-length protein (fFL) and thermolysin-resistant protein (fTR) plotted as function of L for the eight proteins discussed in the text. Each data point is an average of three independent experiments (error bars indicate SEM values). Lonset is the L value corresponding to half-maximal peak height (indicated by dotted lines), and Lmax is the L value corresponding to the maximum of the peak. (A) FSD1. The red data point at L = 17 residues is for the destabilizing mutation F21P. The blue data point at L = 61 residues (arrow) is for a construct where the (GS)n linker was replaced by a linker derived from the LepB protein (Materials and Methods). (B) WW domain. The red data point at L = 25 residues is for the destabilizing mutation Y19A. (C) Designed protein EHEE_rd2_0005. (D) Protein G, the red data point at L = 30 residues is for the destabilizing mutation F52L. (E) Calmodulin EF-2–EF-3. The black profile was obtained in 3 mM CaCl2, and the red profile in 0.3 mM EGTA. (F) Ribosomal protein S6. The black profile was obtained with the SecM(Ec) AP, and the gray profile with the SecM(Ms) AP. (G) SOD1 (lacking the metal-loading loops IV and VII). The full curve (squares) is the fFL profile and the dashed curve (triangles) is the thermolysin-resistance profile (fTR). (H) ILBP. The full curve (squares) is the fFL profile and the dashed curve (triangles) is the thermolysin-resistance profile (fTR).
The fFL profile for FSD1 (a designed 28-residue αββ zinc-finger domain that does not require Zn2+ to fold) has a maximum already at Lmax = 17 residues (Fig. 2A), significantly different from the fFL profile previously obtained for the 29-residue zinc-finger protein ADR1a that has Lmax ≈ 25 residues (8). It has been suggested that the α-helix in FSD1 is more rigid than the β-hairpin, and there is low cooperativity between the α-helix and the β-hairpin during folding (39). Circular dichroism and thermal denaturation suggest that the melting temperature of FSD1 mainly reflects the melting of the α-helix (40). As stretches of α-helix can form deep in the exit tunnel (13, 41, 42), it seems likely that the high fFL value at L = 17 residues reflects the formation of the α-helix rather than folding of the whole domain. To confirm that a folding event is responsible for the peak we introduced a point mutation (F21P) that breaks the α-helix in the L = 17 residues construct; this caused fFL to drop (from 0.77 to 0.47, red data point), as expected.
The WW domain is one of the smallest known β-sheet proteins that can fold autonomously (43). The fFL profile for the 35-residue WW domain (an antiparallel three-stranded β-sheet) peaks at Lmax = 23–25 residues and the protein starts to fold at Lonset ≈ 22 residues (Fig. 2B), in line with previous findings (7). Introduction of the mutation Y19A that reduces the thermodynamic stability of the WW domain (30) caused a reduction of fFL at L = 25 residues from 0.60 to 0.39 (red data point), as expected if the peak reflects a folding event.
The 40-residue protein EHEE_rd2_0005 (βαββ fold) is an artificial miniprotein produced by two rounds of design and selection, and claimed to be “the most stable minimal protein ever found (lacking disulfides or metal coordination)” (31). The peak of the force profile has the highest amplitude of all of the tested proteins, with fFL = 1 between L = 28–40 residues (Fig. 2C). Folding starts at Lonset ≈ 24 residues.
The fFL profile for the 56-residue protein G (ββαββ fold) (44) has a pronounced peak that starts at Lonset ≈ 28 residues and reaches a maximum at Lmax ≈ 35 residues (Fig. 2D). To confirm that the peak is due to folding we introduced the destabilizing mutation F52L (45) that decreased fFL from 0.7 to 0.3 at L = 30 residues (red data point).
Wild-type calmodulin contains four Ca2+-binding sites in the so-called EF-hands EF-1 to EF-4. Each EF-hand folds into a helix–loop–helix motif; the 69-residue version of the protein tested here contains only the EF-2 and EF-3 domains (33). This particular version of calmodulin requires Ca2+ to fold and has a well-defined hydrophobic core composed mainly of aromatic residues (33). When translated in the presence of 3 mM calcium chloride, calmodulin EF-2–EF-3 displays a peak in the fFL profile with Lonset ≈ 34 residues and at Lmax ≈ 40 residues (Fig. 2E). No comparable peak is seen when the translation reaction is supplemented with the Ca2+ chelator EGTA (0.3 mM), validating folding as the source of the peak.
Ribosomal protein S6 from Thermus thermophilus is a 101-residue βαββαβ protein with a ferredoxin-like fold (46). It has been subjected to multiple in vitro folding studies (47–52). The fFL profile of S6 has a peak with Lonset ≈ 27 residues and Lmax ≈ 28 residues (Fig. 2F). We also obtained an fFL profile with the stronger SecM(Ms) AP and determined the fFL profile at single-residue resolution around Lmax. With the stronger AP, the peak in the fFL profile is better defined, with Lonset = 28 residues and Lmax = 29 residues, and returns to a low baseline of fFL ≈ 0.3 at long linker lengths.
SOD1 plays an important role in neurodegenerative disease (53), and its misfolding and aggregation due to demetalation has been linked to amyotrophic lateral sclerosis. The version of SOD1 tested here has been through a process of loop removal, leaving a β-barrel that does not bind copper or zinc but still folds cooperatively into a globular domain (35). The SOD1 fFL profile has a major peak at Lmax ≈ 45 residues (Fig. 2G). We further studied the folding of SOD1 using pulse proteolysis (17). The protein acquires full thermolysin resistance (as measured by the fraction protease-resistant arrested form of the protein, fTR) at L ≈ 40 residues, coincident with the onset of folding at Lonset ≈ 38 residues seen in the fFL profile.
The ILBP is the biggest protein analyzed in this study (128 residues). It shows a significant increase in fraction full length at Lonset ≈ 38 residues and a maximum at Lmax = 45–50 residues (Fig. 2H). As was observed with SOD1, thermolysin resistance reaches its maximum coincident with the onset of folding seen in the fFL profile. We also tested a variant of ILBP that is unable to fold without ligand (36), in which the two helices that cap the β-barrel were removed and replaced by a small GSGS linker (depicted in SI Appendix, Fig. S1A). In the absence of ligand, the fFL profile of this variant remains constant throughout the range L = 20–60 residues (SI Appendix, Fig. S1D, red curve). In contrast, upon ligand binding the mutant protein displays an fFL profile similar to that of the wild-type protein, but with somewhat lower amplitude (SI Appendix, Fig. S1D, gray curve). Notably, in the presence of ligand the mutant protein has a lower melting temperature (Tm = 41.2 °C) than the apo wild-type ILBP (Tm = 60.5 °C) (54), supporting the idea that the amplitude of the fFL peak correlates with the thermodynamic stability of the protein.
One notable feature of all of the fFL profiles obtained with the SecM(Ec) AP in Fig. 2 is the rather high fFL values for the longest linker lengths compared with previously reported fFL profiles (8, 15, 24). The most obvious difference between the constructs analyzed here and those analyzed before is that we here use a flexible (GS)n linker (to ensure resistance to thermolysin cleavage) rather than a linker with more variable amino acid composition derived from the E. coli LepB protein. We therefore substituted the (GS)n linker with the previously used LepB-derived linker in the FSD-1 [L = 61] construct. Indeed, fFL was reduced to 0.03, much lower than the value of ∼0.5 seen for large L values with the (GS)n linker (Fig. 2A, blue arrow). The relatively high limiting fFL values at large L values are thus caused by the linker. As seen in the fFL profile for calmodulin EF-2–EF-3 obtained in the absence of Ca2+ (Fig. 2E, red curve), the rise in fFL values caused by the linker becomes noticeable at L ≈ 40 residues [when the (GS)n segment is ∼25 residues long] and hence does not influence the calculated Lonset values (that are all ≤40 residues). An increase in fFL values at a similar linker length is evident also in Fig. 2 A and B.
In summary, we find strong correlations between protein size and the distance from the peptidyl transferase center at which the protein folds and between the onset of folding as determined from fFL and fTR profiles.
fFL Correlates with Thermodynamic Stability.
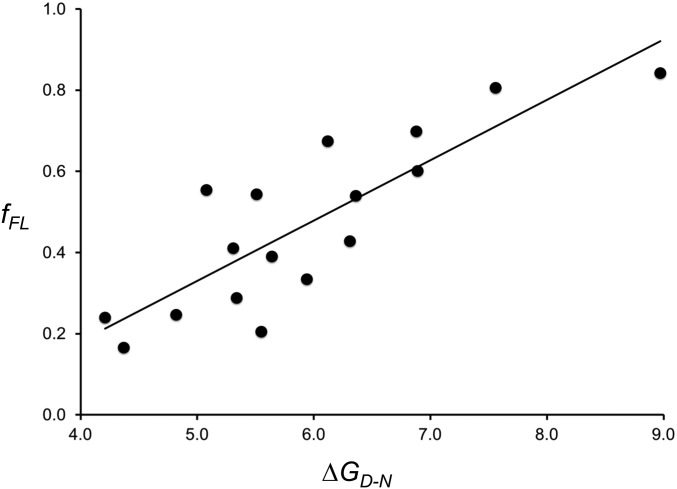
If fFL profiles indeed reflect cotranslational folding to the native state, their amplitude at L = Lmax should reflect the thermodynamic stability of the folded state. The S6 protein provides a good model system to perform this consistency check, since both thermodynamic stability (ΔGD-N) and folding/unfolding rate constants (kf and ku, respectively) have been measured for a large number of point mutants (51). We therefore measured fFL at linker length L = 30 residues, that is, at the middle of the peak of the fFL profile for wild-type S6 obtained with the strong SecM(Ms) AP, for the 16 S6 mutants listed in SI Appendix, Table S3. As seen in Fig. 3, there is a good correlation between fFL and ΔGD-N. A multiple regression analysis including both ΔGD-N and log kf values did not yield a statistically significant improvement in the correlation (P = 0.063, H0 accepted at the 5% level). Folding kinetics thus does not appear to play a significant role in determining fFL values under our experimental conditions. However, since the translation rate is ∼10-fold slower in PURE than in vivo (55), further studies will be required to fully address this last point.
Fig. 3.
Correlation between fFL and ΔGD-N values for wild-type S6 and 16 point mutations listed in SI Appendix, Table S3. The best-fit linear regression line is shown (fFL = 0.15 ΔGD-N − 0.41; R2 = 0.70; P = 2 × 10−5). Statistical analysis was performed using the StatPlus:mac Pro software.
The observations that the appearance of the main peak in the S6 (discussed below), SOD1, and ILBP fFL profiles correlates with the acquisition of thermolysin resistance in the on-ribosome pulse-proteolysis assay, and that the amplitude of the main peak in the S6 fFL profile correlates with the thermodynamic stability of the protein, reinforce the notion that the main peak in an fFL profile represents a bona fide folding reaction rather than, for example, the formation of a nonspecific collapsed state. The sharp onset of the main peak observed for most proteins analyzed in Fig. 2 and in earlier studies (8, 24, 25) provides further support that fFL profiles reflect cooperative folding to the native state.
Net Charge and Cotranslational Folding.
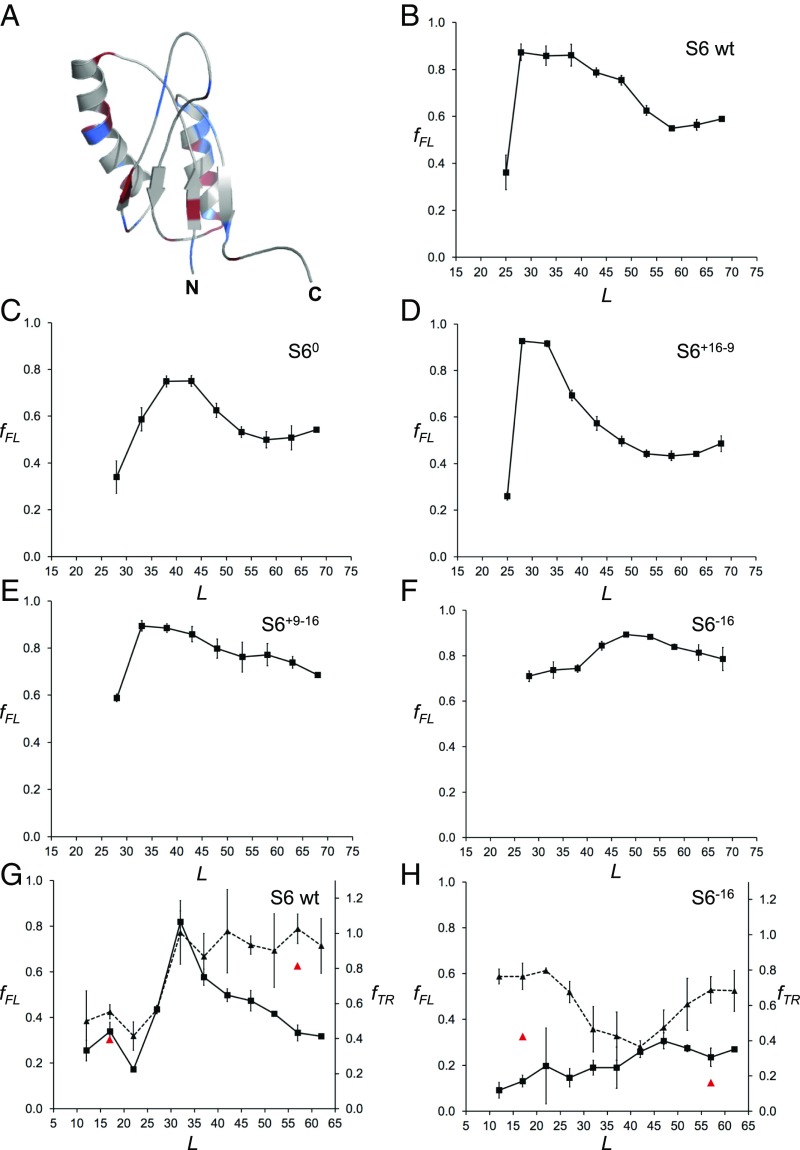
Finally, to determine how the net charge of a folded protein domain affects cotranslational folding on the ribosome, we compared the fFL profiles for wild-type S6 and four mutants of different net charge, three of which have previously been characterized in vitro in terms of ΔGD-N, log kf, and log ku (52) (Fig. 4 and SI Appendix, Table S4). Wild-type S6 contains 16 positively charged and 16 negatively charged residues (zero net charge). The S60 variant also has zero net charge but lacks all charged residues. The fFL profile of S60 is broadly similar to that of wild-type S6, but the peak is shifted to somewhat larger L values (Fig. 4C). The fFL profile of the positively charged variant S6+16–9 is also similar to the profile for wild-type S6 (Fig. 4D), except that the peak is sharper. The in vitro folding studies (52) have shown that a mutant that lacks all negatively charged residues (S6+17) has slightly accelerated folding and unfolding rates and somewhat lower thermodynamic stability compared with wild-type S6 (SI Appendix, Table S4); presumably, the corresponding ΔGD-N, log kf, and log ku values for S6+16–9 are even closer to the wild-type S6 values, explaining the similarity between the two fFL profiles. S60 has a higher unfolding rate than S6+17 and a correspondingly lower thermodynamic stability, consistent with the somewhat higher Lonset and lower amplitude of the fFL peak.
Fig. 4.
(A) NMR structure of wild-type S6 from T. thermophilus (PDB ID code 2kjv). Positively charged residues (Arg+Lys) are colored in blue and negatively charged residues (Asp+Glu) in red. (B) fFL profile of wild-type S6+16–16 (for sequences see SI Appendix, Fig. S2). (C) fFL profile of S60. (D) fFL profile of S6+16–9. (E) fFL profile of S6+9–16. (F) fFL profile of S6−16. (G) fFL (squares, solid curve) and fTR (triangles, dashed curve) profiles for wild-type S6. Red data points at L = 17 and 57 residues are for the destabilizing mutation L10A. (H) fFL (squares, solid curve) and fTR (triangles, dashed curve) profiles for S6−16. Red data points at L = 17 and 57 residues are for the destabilizing mutation L10A. The SecM(Ec) AP was used in A–F and the SecM(Ms) AP in G and H.
In contrast, removal of half of the positive charges from wild-type S6 (variant S6+9–16) causes the peak in the fFL profile to shift from Lmax ≈ 28 to Lmax ≈ 33 residues (Fig. 4E) and leads to a general increase in fFL throughout the profile. Removal of the remaining positively charged residues yields the supercharged but not very stable variant S6−16; in this case, fFL ≥ 0.7 for all L values, and the profile has a broad maximum around Lmax ≈ 50 residues (Fig. 4F).
We also recorded the fFL and fTR profiles for wild-type S6 and the supercharged S6−16 variant using the stronger SecM(Ms) AP (38). The fTR profile for wild-type S6 mirrors the fFL profile, starting to increase at L ≈ 27 residues and plateauing at Lmax ≈ 32 residues (Fig. 4G). In contrast, the S6−16 fTR profile starts out moderately high at small L values, drops to a lower value at L = 30–45 residues, and finally increases again to reach a moderately high plateau at L > 55 residues (Fig. 5H); the fFL profile has no distinct peak, similar to the profile obtained with the SecM(Ec) AP but at lower fFL values (compare Fig. 5F).
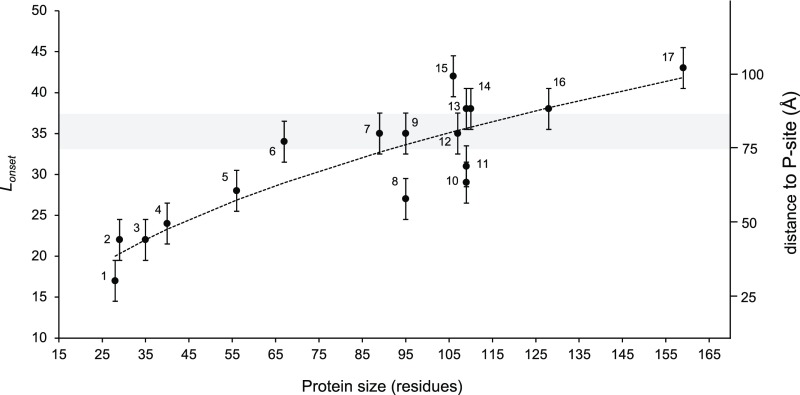
Fig. 5.
Plot of Lonset vs. protein size for proteins included in this and previous studies. The shaded gray area indicates the approximate location of the distal end of the exit tunnel, and numbers on the right indicate the approximate distance from the peptidyl transferase center. Error bars of ±2.5 residues have been added to indicate that the fFL profiles in Fig. 2 were recorded with a five-residue resolution, hence introducing an uncertainty in the Lonset values. The dotted curve shows the linear best-fit obtained from a log-log plot: log10(Lonset) = 0.42 (±0.05) ∙ log10(size) + 0.69 (±0.1) (R2 = 0.80; P = 1.4 × 10−6). Proteins are indicated as follows: 1, FSD1; 2, ADR1a1; 3, WW domain; 4, EHEE; 5, protein G; 6, calmodulin EF-2–EF-3; 7, titin I27; 8, S6; 9, TOP7; 10, spectrin β16; 11, spectrin R16; 12, PENT; 13, spectrin R15; 14, SOD; 15, FLN5; 16, ILBP; and 17, DHFR (see SI Appendix, Table S5 for references).
The relatively high thermolysin resistance of S6−16 at small L values is surprising. To analyze this further, we introduced the destabilizing mutation L10A in both wild-type S6 and S6−16 at linker lengths L = 17 and L = 57 residues. This mutation reduces ΔGD-N for wild-type S6 from 8.97 to 4.62 kcal/mol (51). The destabilization of S6−16 caused by the mutation has not been measured, but the folding free energy of S6−16 itself is only 2.89 kcal/mol (52). As seen in Fig. 4 G and H (red triangles), fTR of wild-type S6 is only marginally affected by the mutation—as expected, since even the mutated protein is relatively stable—but is strongly reduced for S6−16 at both linker lengths.
We conclude that S6 versions that contain many positively charged residues (wild-type and S6+17–9) start to fold well inside the exit tunnel, while S60 and S6+9–16 fold near the distal end of the tunnel. For the negatively charged versions S6+9–16 and S6−16 fFL values are high for all linker lengths despite their low thermodynamic stability, likely reflecting a strong electrostatic repulsion between the negatively charged exit tunnel and the nascent chain (56–58). There is a hint from the fFL and fTR profiles that the supercharged S6−16 version may fold outside the exit tunnel, around L ≈ 50 residues, but the data are not conclusive on this point.
Discussion
In this study, we have focused on understanding how three central protein characteristics—size, thermodynamic stability, and net charge—affect cotranslational protein folding.
Our findings on the relation between domain size and the location in the ribosome exit tunnel where folding commences are summarized in Fig. 5, which includes data not only from the present study but also from previous studies on ADR1a (8), three spectrin domains (15), a penta-repeat β-helix protein (25), DHFR (59), the I27 (24) and FNL5 (60) Ig-domain proteins, and the designed protein Top7 (20) (see SI Appendix, Table S5 for details). From cryo-EM structures of ADR1a (8), spectrin R16 (15), and titin I27 (24) ribosome-nascent chain complexes we know that a ∼35-residue linker is required to span the distance between the P-site and a location at the distal end of the exit tunnel, near a prominent loop in the ribosomal protein uL24 (gray shaded area in Fig. 5).
Small domains (≤70 residues in size) invariably start to fold while still located inside the tunnel (Lonset < 35 residues), and the smaller they are the deeper in the tunnel they fold. Large domains tend to fold at the distal end of, or even outside, the tunnel (Lonset > 35 residues), but, depending on the location of the first accessible folding nucleus in the structure, may also start to fold while still inside the tunnel, as exemplified by the spectrin domains (15). A linear fit of log10(Lonset) vs. log10(size) (SI Appendix, Fig. S3) shows that Lonset scales approximately with the square root of the protein size over the range covered here (dotted line in Fig. 5). Fig. 5 provides the most complete picture to date of how the gradual increase in tunnel dimensions from the so-called constriction site (∼25 Å from the peptidyl transferase site) to the tunnel exit allows protein domains of increasing size to fold.
For the largest proteins analyzed, SOD1 and ILBP, in addition to the main folding peak at L ≈ 45–50 residues, we see an early increase in fFL at L ≈ 25–30 residues that might be indicative of a folding intermediate (Fig. 2 G and H). A similar increase in fFL at small L values was seen previously for the 189-residue protein DHFR (59). In vitro, SOD1 does not seem to fold via an intermediate state but displays two-state behavior (35). Further studies will be necessary to characterize these putative folding intermediates.
Cotranslational folding of the exceptionally stable 40-residue protein EHEE_rd2_0005 generates sufficient force on the AP to yield fFL = 1.0 over the whole range L = 28–40 residues (Fig. 2C), setting it apart from the other proteins studied here. This and previous observations (16, 21) suggest that the amplitude of the peak in the fFL profile reflects the thermodynamic stability of the folded protein. Indeed, there is a good correlation between fFL and ΔGD-N for a large set of S6 mutants (Fig. 3). Since different proteins fold in different locations inside or outside the exit tunnel, and presumably push against the ribosome in different ways during folding, we do not expect the precise quantitative relation between fFL and ΔGD-N found for S6 (discussed above) to hold for other proteins, but only that the general form of the correlation applies.
Our results for protein S6 variants of different charge content show that protein net charge can have a dramatic effect on the fFL profile (Fig. 4). In general, the more positive the net charge, the deeper in the exit tunnel S6 folds. We further note that the fFL profile for the fully uncharged S60 variant is similar to that of wild-type S6, demonstrating that a protein that is totally devoid of charged residues can fold in the exit tunnel. This extends previous results demonstrating efficient folding in vitro of a protein completely devoid of charged residues (52, 61) to cotranslational folding. Strikingly, for the negatively charged variants S6+9–16 and S6−16, high values of fFL persist to much larger L values than for the neutral and positively charged variants, the peaks in the fFL profiles are broad and indistinct, and the maximum amplitudes are high, in contrast to what one would expect from the significantly lower thermodynamic stability of S6−16 (SI Appendix, Table S4). Given the net-negative charge of the exit tunnel (56, 58, 62), we hypothesize that the high fFL values seen for S6+9–16 and S6−16 are caused by negatively charged residues in the nascent chain being pushed out of the exit tunnel by electrostatic forces even before the protein folds, consistent with ribosome-profiling data showing that stretches of nascent chain of high net-negative charge show reduced ribosome densities (63).
More generally, the fact that domains of up to ∼70 residues in size can fold inside the exit tunnel and therefore should be less likely to aggregate or require chaperones for folding might favor the evolution of proteins composed of small (sub)domains that can initiate early folding inside the ribosome. In support of such a scenario, statistical analyses have identified evolutionarily conserved clusters of rare codons located some 20–50 codons downstream of independently folding subdomains that may facilitate their cotranslational folding by slowing down translation (64, 65).
Finally, the observation that cotranslational folding generates force on the nascent chain raises the possibility that new sensor systems composed of a ligand binding domain coupled to an AP may be found in nature, or be constructed for biosensing purposes. In principle, the calmodulin EF-2–EF-3 and ILBP constructs analyzed here could be used as Ca2+ and glycochenodeoxycholic acid sensors, respectively, if fused to, for example, GFP (7).
Materials and Methods
Enzymes and Chemicals.
Enzymes were purchased from Thermo Scientific and New England Biolabs. Thermolysin from Geobacillus stearothermophilus was purchased from Sigma-Aldrich. Oligonucleotides were purchased from Eurofins MWG Operon. DNA/RNA purification kits were obtained from Qiagen. The New England Biolabs PURExpress In Vitro Protein Synthesis Kit was purchased from BioNordika. [35S]methionine was purchased from PerkinElmer. All other reagents were from Sigma-Aldrich.
DNA Manipulations.
The genes of the FSD-1, Pin1 WW domain (Homo sapiens), and EHEE design were assembled from oligonucleotides (66). The gene for protein G (Streptococcus sp.) was purchased from Genewiz. The wild-type calmodulin gene (H. sapiens) was kindly donated by Sara Linse, Lund University, Lund, Sweden and modified by an adaptation of the original QuikChange site-directed mutagenesis protocol (67) to obtain the fragment tested in this study. SOD1 (H. sapiens) gene (35) was kindly donated by Mikael Oliveberg, Stockholm University, Stockholm. The gene for the ILBP from rabbit (Oryctolagus cuniculus) was purchased from Invitrogen and was truncated by an adaptation of the original QuikChange site-directed mutagenesis protocol (67).
The gene for ribosomal protein S6 from T. thermophilus, along with the genes for the five of its mutants (52), were provided by Mikael Oliveberg.
The constructs were cloned in a pET19b vector under the control of a T7 promoter. As shown in SI Appendix, Table S2, the small domain (FSD-1, Pin1 WW domain, and EHEE) constructs contained a sequence corresponding to an N-terminal unstructured fragment of 158 residues from E. coli LepB (21) (to facilitate visualization by SDS/PAGE), a sequence corresponding to a (GS)n flexible linker of 43 or 52 residues (to reach a 60-residues-long tether length together with the AP), the sequence corresponding to the SecM AP from E. coli (FSTPVWISQAQGIRAGP) (68) [referred to here as SecM(Ec)] or M. succiniciproducens (HAPIRGSP) (69) [referred to here as SecM(Ms)] and a sequence corresponding to a 23-residues-long LepB-derived C-terminal fragment (to distinguish between full-length and arrested version of proteins after SDS/PAGE). The genes were inserted into the expression vector by Gibson assembly (70). Constructs with increasingly shorter tether length starting from the N-terminal part of the flexible linker and control constructs for the full-length [SecM(Ec) FSTPVWISQAQGIRAGA, SecM(Ms) HAPIRGSA] and arrested [SecM(Ec): FSTPVWISQAQGIRAG*, SecM(Ms): HAPIRGS*, where * is a stop codon] forms of the proteins were generated by an adaptation of the original QuikChange site-directed mutagenesis protocol (67). PCR amplified products were subjected to DpnI digestion and purified with GeneJET PCR purification Kit or GeneJET Gel Extraction Kit before Gibson assembly. E. coli DH5α competent cells were transformed and plated onto LB agar plate supplemented with carbenicillin.
Single colonies were picked to inoculate LB supplemented with ampicillin and were incubated for 12–18 h at 37 °C with shaking. GeneJET Plasmid Miniprep Kit was used for plasmid isolation and purification. All constructs were verified by sequencing (Eurofins Genomics).
In Vitro Transcription and Translation.
Linear constructs were obtained by PCR amplification using oligonucleotides overlapping the T7 promoter and terminator on purified plasmids. After DpnI digestion and purification (GeneJET PCR Purification Kit; Thermo Scientific), the linear constructs were used as template for protein expression. The NEB PURExpress In Vitro Protein Synthesis Kit was used for in vitro transcription and translation. The reaction mix was supplemented with [32S]methionine and the synthesis of the radiolabeled proteins was performed at 37 °C, for 15 min, under constant shaking at 700 rpm in an Eppendorf 5417R thermomixer. The reaction was stopped by addition of 1:1 volume of 10% ice-cold TCA and the samples were incubated on ice for at least 30 min. After centrifugation at 20817 × g for 5 min at 4 °C, the supernatant was discarded and the pellet was resuspended in sample buffer at 37 °C for 15 min, under constant shaking at 900 rpm. The samples were treated with RNase I (670 μg/mL) at 37 °C for 30 min and were subsequently resolved by SDS/PAGE. All expressions were performed in triplicates.
On-Ribosome Pulse Proteolysis.
After synthesis of radiolabeled proteins as described in the previous section, the reactions were stopped by the addition of chloramphenicol (instead of TCA) at a final concentration of 3.3 mM. Each sample was divided into two aliquots of equal volume, one for pulse proteolysis and the other serving as a nontreated control. Pulse proteolysis was performed by treatment with thermolysin (buffered in 50 mM Tris and 500 μM ZnCl2, pH 7.0) at a final concentration of 0.75 mg/m for 1 min at 37 °C, under constant shaking at 700 rpm. The same conditions were applied for the nontreated controls with the difference that thermolysin was not included in the buffer solution. The reaction was stopped by addition of 3 μL 500 mM EDTA (pH 8.4), and protein precipitation was performed by addition of 1:1 volume of ice-cold 10% TCA and incubation on ice for 30 min. As described in the previous section, after centrifugation at 20817 × g for 5 min, at 4 °C, the supernatant was discarded and the pellet was resuspended in sample buffer at 37 °C for 15 min, under constant shaking at 900 rpm. The samples were supplemented with RNase I (670 μg/mL) at 37 °C, for 30 min, and were subsequently resolved by SDS/PAGE. All pulse-proteolysis assays were performed in triplicate.
Gel Electrophoresis and Quantitation.
Proteins were resolved by SDS/PAGE and visualized on a Fuji FLA-3000 phosphoimager. For fFL measurements, the bands were quantified to estimate the fraction full-length protein fFL = IFL/(IFL + IA), where IFL is the intensity of the band corresponding to the full-length protein and IA is the intensity of the band corresponding to the arrested form of the protein, fFL.
For pulse-proteolysis measurements the thermolysin-resistant fraction was calculated as fTR = IATR/IABuff, where IATR is the intensity of the arrested band after pulse proteolysis and IABuff is the intensity of the nontreated control band. Bands were quantitated using ImageJ (https://imagej.nih.gov/ij) to obtain an intensity cross-section, which was subsequently fit to a Gaussian distribution using in-house software.
Homology Modeling of ILBP Mutant.
The structure [Protein Data Bank (PDB) ID code 2lba] of chicken ILBP protein in complex with glycochenodeoxycholic acid (GDCG) was used to produce a homology model (71) of the truncated mutant (ILBP-tm) protein used in this study (59.6% Seq. Id with 2lba PDB template). We show the estimated position of the ligand (GDCG) in stick representation in SI Appendix, Fig. 1A.
Supplementary Material
Acknowledgments
We thank Mikael Oliveberg, Jens Danielsson, Setareh Arya, and Domhnall Iain Henderson for discussions. This work was supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Cancer Foundation, and the Swedish Research Council (to G.v.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812756115/-/DCSupplemental.
References
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Holtkamp W, et al. Cotranslational protein folding on the ribosome monitored in real time. Science. 2015;350:1104–1107. doi: 10.1126/science.aad0344. [DOI] [PubMed] [Google Scholar]
- 3.Kim SJ, et al. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348:444–448. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- 4.Trovato F, O’Brien EP. Insights into cotranslational nascent protein behavior from computer simulations. Annu Rev Biophys. 2016;45:345–369. doi: 10.1146/annurev-biophys-070915-094153. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien EP, Christodoulou J, Vendruscolo M, Dobson CM. New scenarios of protein folding can occur on the ribosome. J Am Chem Soc. 2011;133:513–526. doi: 10.1021/ja107863z. [DOI] [PubMed] [Google Scholar]
- 6.Nicola AV, Chen W, Helenius A. Co-translational folding of an alphavirus capsid protein in the cytosol of living cells. Nat Cell Biol. 1999;1:341–345. doi: 10.1038/14032. [DOI] [PubMed] [Google Scholar]
- 7.Marino J, von Heijne G, Beckmann R. Small protein domains fold inside the ribosome exit tunnel. FEBS Lett. 2016;590:655–660. doi: 10.1002/1873-3468.12098. [DOI] [PubMed] [Google Scholar]
- 8.Nilsson OB, et al. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 2015;12:1533–1540. doi: 10.1016/j.celrep.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu L, Khanna P, Deutsch C. Transmembrane segments form tertiary hairpins in the folding vestibule of the ribosome. J Mol Biol. 2014;426:185–198. doi: 10.1016/j.jmb.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosolapov A, Deutsch C. Tertiary interactions within the ribosomal exit tunnel. Nat Struct Mol Biol. 2009;16:405–411. doi: 10.1038/nsmb.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti BJ, Elferich J, Yang Z, Shinde U, Skach WR. Cotranslational folding inhibits translocation from within the ribosome-Sec61 translocon complex. Nat Struct Mol Biol. 2014;21:228–235. doi: 10.1038/nsmb.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin PJ, Jongsma CG, Pool MR, Johnson AE. Polytopic membrane protein folding at L17 in the ribosome tunnel initiates cyclical changes at the translocon. J Cell Biol. 2011;195:55–70. doi: 10.1083/jcb.201103118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su T, et al. The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling. eLife. 2017;6:e25642. doi: 10.7554/eLife.25642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hingorani KS, Gierasch LM. Comparing protein folding in vitro and in vivo: Foldability meets the fitness challenge. Curr Opin Struct Biol. 2014;24:81–90. doi: 10.1016/j.sbi.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson OB, et al. Cotranslational folding of spectrin domains via partially structured states. Nat Struct Mol Biol. 2017;24:221–225. doi: 10.1038/nsmb.3355. [DOI] [PubMed] [Google Scholar]
- 16.Farías-Rico JA, Goetz SK, Marino J, von Heijne G. Mutational analysis of protein folding inside the ribosome exit tunnel. FEBS Lett. 2017;591:155–163. doi: 10.1002/1873-3468.12504. [DOI] [PubMed] [Google Scholar]
- 17.Samelson AJ, Jensen MK, Soto RA, Cate JH, Marqusee S. Quantitative determination of ribosome nascent chain stability. Proc Natl Acad Sci USA. 2016;113:13402–13407. doi: 10.1073/pnas.1610272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito K, Chiba S. Arrest peptides: Cis-acting modulators of translation. Annu Rev Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 19.Butkus ME, Prundeanu LB, Oliver DB. Translocon “pulling” of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. J Bacteriol. 2003;185:6719–6722. doi: 10.1128/JB.185.22.6719-6722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman DH, et al. Ribosome. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science. 2015;348:457–460. doi: 10.1126/science.1261909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ismail N, Hedman R, Schiller N, von Heijne G. A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat Struct Mol Biol. 2012;19:1018–1022. doi: 10.1038/nsmb.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ismail N, Hedman R, Lindén M, von Heijne G. Charge-driven dynamics of nascent-chain movement through the SecYEG translocon. Nat Struct Mol Biol. 2015;22:145–149. doi: 10.1038/nsmb.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cymer F, von Heijne G. Cotranslational folding of membrane proteins probed by arrest-peptide-mediated force measurements. Proc Natl Acad Sci USA. 2013;110:14640–14645. doi: 10.1073/pnas.1306787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian P, et al. 2018. The folding pathway of an Ig domain is conserved on and off the ribosome. bioRxiv:10.1101/253013. Preprint, posted June 18, 2018.
- 25.Notari L, Martinez-Carranza M, Farias-Rico JA, Stenmark P, von Heijne G. 2018. Cotranslational folding of a pentarepeat β-helix protein. bioRxiv:10.1101/255810. Preprint, posted July 5, 2018. [DOI] [PubMed]
- 26.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 27.Bogachev MI, Kayumov AR, Markelov OA, Bunde A. Statistical prediction of protein structural, localization and functional properties by the analysis of its fragment mass distributions after proteolytic cleavage. Sci Rep. 2016;6:22286. doi: 10.1038/srep22286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park C, Marqusee S. Pulse proteolysis: A simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 29.Dahiyat BI, Mayo SL. De novo protein design: Fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 30.Jäger M, Nguyen H, Crane JC, Kelly JW, Gruebele M. The folding mechanism of a β-sheet: The WW domain. J Mol Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 31.Rocklin GJ, et al. Global analysis of protein folding using massively parallel design, synthesis, and testing. Science. 2017;357:168–175. doi: 10.1126/science.aan0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gronenborn AM, et al. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science. 1991;253:657–661. doi: 10.1126/science.1871600. [DOI] [PubMed] [Google Scholar]
- 33.Lakowski TM, Lee GM, Okon M, Reid RE, McIntosh LP. Calcium-induced folding of a fragment of calmodulin composed of EF-hands 2 and 3. Protein Sci. 2007;16:1119–1132. doi: 10.1110/ps.072777107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohman A, Oman T, Oliveberg M. Solution structures and backbone dynamics of the ribosomal protein S6 and its permutant P(54-55) Protein Sci. 2010;19:183–189. doi: 10.1002/pro.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danielsson J, Kurnik M, Lang L, Oliveberg M. Cutting off functional loops from homodimeric enzyme superoxide dismutase 1 (SOD1) leaves monomeric β-barrels. J Biol Chem. 2011;286:33070–33083. doi: 10.1074/jbc.M111.251223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rea AM, Thurston V, Searle MS. Mechanism of ligand-induced folding of a natively unfolded helixless variant of rabbit I-BABP. Biochemistry. 2009;48:7556–7564. doi: 10.1021/bi900805s. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, et al. Mechanisms of ribosome stalling by SecM at multiple elongation steps. eLife. 2015;4:e09684. doi: 10.7554/eLife.09684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cymer F, Hedman R, Ismail N, von Heijne G. Exploration of the arrest peptide sequence space reveals arrest-enhanced variants. J Biol Chem. 2015;290:10208–10215. doi: 10.1074/jbc.M115.641555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei H, Duan Y. The role of plastic beta-hairpin and weak hydrophobic core in the stability and unfolding of a full sequence design protein. J Chem Phys. 2004;121:12104–12111. doi: 10.1063/1.1822916. [DOI] [PubMed] [Google Scholar]
- 40.Feng JA, Kao J, Marshall GR. A second look at mini-protein stability: Analysis of FSD-1 using circular dichroism, differential scanning calorimetry, and simulations. Biophys J. 2009;97:2803–2810. doi: 10.1016/j.bpj.2009.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tu LW, Deutsch C. A folding zone in the ribosomal exit tunnel for Kv1.3 helix formation. J Mol Biol. 2010;396:1346–1360. doi: 10.1016/j.jmb.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhushan S, et al. α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol. 2010;17:313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- 43.Pucheta-Martinez E, et al. Changes in the folding landscape of the WW domain provide a molecular mechanism for an inherited genetic syndrome. Sci Rep. 2016;6:30293. doi: 10.1038/srep30293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCallister EL, Alm E, Baker D. Critical role of beta-hairpin formation in protein G folding. Nat Struct Biol. 2000;7:669–673. doi: 10.1038/77971. [DOI] [PubMed] [Google Scholar]
- 45.Gronenborn AM, Frank MK, Clore GM. Core mutants of the immunoglobulin binding domain of streptococcal protein G: Stability and structural integrity. FEBS Lett. 1996;398:312–316. doi: 10.1016/s0014-5793(96)01262-8. [DOI] [PubMed] [Google Scholar]
- 46.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 47.Lindberg M, Tångrot J, Oliveberg M. Complete change of the protein folding transition state upon circular permutation. Nat Struct Biol. 2002;9:818–822. doi: 10.1038/nsb847. [DOI] [PubMed] [Google Scholar]
- 48.Otzen DE, Oliveberg M. Conformational plasticity in folding of the split beta-alpha-beta protein S6: Evidence for burst-phase disruption of the native state. J Mol Biol. 2002;317:613–627. doi: 10.1006/jmbi.2002.5423. [DOI] [PubMed] [Google Scholar]
- 49.Lindberg MO, Haglund E, Hubner IA, Shakhnovich EI, Oliveberg M. Identification of the minimal protein-folding nucleus through loop-entropy perturbations. Proc Natl Acad Sci USA. 2006;103:4083–4088. doi: 10.1073/pnas.0508863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haglund E, et al. Trimming down a protein structure to its bare foldons: Spatial organization of the cooperative unit. J Biol Chem. 2012;287:2731–2738. doi: 10.1074/jbc.M111.312447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haglund E, Lindberg MO, Oliveberg M. Changes of protein folding pathways by circular permutation. Overlapping nuclei promote global cooperativity. J Biol Chem. 2008;283:27904–27915. doi: 10.1074/jbc.M801776200. [DOI] [PubMed] [Google Scholar]
- 52.Kurnik M, Hedberg L, Danielsson J, Oliveberg M. Folding without charges. Proc Natl Acad Sci USA. 2012;109:5705–5710. doi: 10.1073/pnas.1118640109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rakhit R, Chakrabartty A. Structure, folding, and misfolding of Cu,Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:1025–1037. doi: 10.1016/j.bbadis.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 54.Kouvatsos N, et al. Bile acid interactions with rabbit ileal lipid binding protein and an engineered helixless variant reveal novel ligand binding properties of a versatile beta-clam shell protein scaffold. J Mol Biol. 2007;371:1365–1377. doi: 10.1016/j.jmb.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 55.Capece MC, Kornberg GL, Petrov A, Puglisi JD. A simple real-time assay for in vitro translation. RNA. 2015;21:296–305. doi: 10.1261/rna.047159.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petrone PM, Snow CD, Lucent D, Pande VS. Side-chain recognition and gating in the ribosome exit tunnel. Proc Natl Acad Sci USA. 2008;105:16549–16554. doi: 10.1073/pnas.0801795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knight AM, et al. Electrostatic effect of the ribosomal surface on nascent polypeptide dynamics. ACS Chem Biol. 2013;8:1195–1204. doi: 10.1021/cb400030n. [DOI] [PubMed] [Google Scholar]
- 58.Lu J, Kobertz WR, Deutsch C. Mapping the electrostatic potential within the ribosomal exit tunnel. J Mol Biol. 2007;371:1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 59.Nilsson OB, Müller-Lucks A, Kramer G, Bukau B, von Heijne G. Trigger factor reduces the force exerted on the nascent chain by a cotranslationally folding protein. J Mol Biol. 2016;428:1356–1364. doi: 10.1016/j.jmb.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 60.Cabrita LD, et al. A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding. Nat Struct Mol Biol. 2016;23:278–285. doi: 10.1038/nsmb.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Højgaard C, et al. A soluble, folded protein without charged amino acid residues. Biochemistry. 2016;55:3949–3956. doi: 10.1021/acs.biochem.6b00269. [DOI] [PubMed] [Google Scholar]
- 62.Fedyukina DV, Jennaro TS, Cavagnero S. Charge segregation and low hydrophobicity are key features of ribosomal proteins from different organisms. J Biol Chem. 2014;289:6740–6750. doi: 10.1074/jbc.M113.507707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Requião RD, et al. Protein charge distribution in proteomes and its impact on translation. PLOS Comput Biol. 2017;13:e1005549. doi: 10.1371/journal.pcbi.1005549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacobs WM, Shakhnovich EI. Evidence of evolutionary selection for cotranslational folding. Proc Natl Acad Sci USA. 2017;114:11434–11439. doi: 10.1073/pnas.1705772114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaney JL, et al. Widespread position-specific conservation of synonymous rare codons within coding sequences. PLOS Comput Biol. 2017;13:e1005531. doi: 10.1371/journal.pcbi.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rouillard JM, et al. Gene2Oligo: Oligonucleotide design for in vitro gene synthesis. Nucleic Acids Res. 2004;32:W176–W180. doi: 10.1093/nar/gkh401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 69.Yap MN, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 71.Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci Rep. 2017;7:10480. doi: 10.1038/s41598-017-09654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.