Significance
We have charted the evolution of the capsid-binding retroviral restriction factor Fv1 through murid evolution, extending its age to ∼45 million years. Functionality can be found outside of the genus Mus, and shared signatures of positive selection are visible across species. Modeling suggests that maintenance for these extended periods can only be parsimoniously explained by repeated selection events—waves of retroviral infection throughout murid evolution. Our results complement and extend findings with TRIM5α and suggest that conserved features of retroviral capsid lattice assemblies may be common targets in convergent evolution of intrinsic defenses to retroviral infection. Functional constraints on capsid structure may prevent effective escape of host factors and result in cyclical coevolution, which is visible in the evolution of Fv1.
Keywords: restriction factor, evolution, host–virus interactions, retrovirus
Abstract
Both exogenous and endogenous retroviruses have long been studied in mice, and some of the earliest mouse studies focused on the heritability of genetic factors influencing permissivity and resistance to infection. The prototypic retroviral restriction factor, Fv1, is now understood to exhibit a degree of control across multiple retroviral genera and is highly diverse within Mus. To better understand the age and evolutionary history of Fv1, a comprehensive survey of the Muroidea was conducted, allowing the progenitor integration to be dated to ∼45 million years. Intact coding potential is visible beyond Mus, and sequence analysis reveals strong signatures of positive selection also within field mice, Apodemus. Fv1’s survival for such a period implies a recurring and shifting retroviral burden imparting the necessary selective pressures—an influence likely also common to analogous factors. Regions of Fv1 adapt cooperatively, highlighting its preference for repeated structures and suggesting that this functionally constrained aspect of the retroviral capsid lattice presents a common target in the evolution of intrinsic immunity.
While a variety of viruses occasionally integrate as endogenous viral elements (1), the absolute requirement for an integrated proviral stage is the defining feature of retroviral replication. When infection occurs within a germ cell, endogenous retroviruses (ERVs) may be inherited in a Mendelian manner and hence, form a partial “fossil record” of historic viral burdens. Although originally unappreciated, retroviruses, as filterable, transmissible pathogens, have been studied since the late 1800s. The earliest breeding of inbred animals both facilitated and was necessitated by the study of ERVs and exogenous retroviruses as the agents of “heritable cancer” (2). Research developing these themes in mice led to the description of Friend virus susceptibility 1 (Fv1), a dominant locus conferring protection from otherwise lethal challenges with murine leukemia virus (MLV) (3, 4). Within common laboratory lines, two alleles can be observed, Fv1b and Fv1n, that were identified in BALB/c and NIH-Swiss mice, respectively. Each allele confers resistance to virus of the opposing N and B tropism and may be additively combined (5, 6).
The molecular cloning of Fv1 revealed its derivation from a retroviral gag gene (7, 8). While many examples of such co-options for host defense have been reported, these are most frequently products of env operating through receptor blockade (9). Fv1’s presumably more unique mode of restriction, indirectly determined to be through capsid (CA) binding (10), has remained elusive. Similarly, while its domain organization has been characterized, the protein has not proven amenable to crystallization, and all studies to date have had a necessarily genetic basis. Nevertheless, recent work has expanded the scope of restriction beyond the gammaretroviruses to lenti- and spumaviruses (11).
Based on instances of absence within certain Mus species and on its absence in Rattus, previous estimates have placed integration of Fv1’s progenitor virus at 4–7 Mya (12, 13). Despite this apparently recent ancestry, the pol gene of the progenitor virus is lacking, and neither LTR has been discerned (8). Searches for intact representatives of the progenitor revealed no closely related ERVs, and Fv1 shares only 43% amino acid identity with its nearest neighbor in the mouse genome, MuERV-L (ERV with a leucine tRNA primer binding site) (8). This paradox may result from incomplete representation of exogenous viruses among those endogenized and fixed but, equally, may suggest a longer and more complex evolutionary history. Indeed, Southern blotting revealed hybridizing digestion fragments within the genus Mastomys (12), although this was never further studied.
Here, we have sought to more accurately determine the origin of Fv1 and to use a phylogenetic approach to inform on the historical selection pressures that have shaped its restriction specificities and preserved the gene through evolutionary time.
Results
Resolving the History of the Fv1 Locus.
Within Mus, Fv1 (GRCm38 Chr4:147,868,979–147,870,358) is located in an ∼5-kb region between Migration and invasion inhibitory protein (Miip) and Mitofusin 2 (Mfn2) (Fig. 1). The shared direction and relative separation of this pair are common among assembled genomes from humans and mice through to chickens (diverging ∼310 Mya), and they thus present a useful framework within which the presence or absence of Fv1 can be established.
Fig. 1.
Fv1 is found across the Muroidea. Representation of the multiple alignment of 22 species from the 5′ UTR of Miip to the 3′ UTR of Mfn2. Regions masked by RepeatMasker as deriving from repetitive elements (including Fv1) are show in red, with the remaining sequence and genic regions in black and alignment gaps represented as linking lines. The region encompassing Fv1 is shaded in blue and can be seen in genera from Mus to Spalax, where larger regions of the progenitor virus can also be identified (lighter blue shading).
To initially investigate the presence of Fv1 immediately beyond the genus Mus, we analyzed three sequences from the genus Apodemus: Apodemus sylvaticus from an archived genome assembly and Apodemus uralensis and Apodemus semosus from targeted assemblies of the region. Fv1 was present in all instances, with complete ORFs visible in A. sylvaticus and A. uralensis. Both ORFs exhibited activity directed against MLVs when assayed for restriction capacity (Table 1).
Table 1.
Fv1 genes of Apodemus have restriction potential
| Gamma | Lenti | Spuma | |||||
| Species | N-MLV | B-MLV | EIAV | HIV-1 | PFV | SFV | FFV |
| A. sylvaticus | 0.48 | 1.18 | 1.11 | 1.09 | 1.00 | 1.02 | 0.95 |
| A. uralensis | 0.12 | 0.13 | 0.73 | 1.09 | 0.99 | 1.03 | 0.99 |
Bold restriction values denote full activity (0 < 0.3), and italics denote partial activity (0.3 < 0.7). EIAV, equine infectious anemia virus; FFV, feline foamy virus; PFV, prototypic foamy virus; SFV, simian foamy virus.
The divergence of the Apodemini and Murini tribes predates that of the Murini and Praomyini (14), represented by Mastomys, and these data presented likely confirmation of the previously published Southern blotting data (12). Encouraged, we thus sought to extend this analysis to reexamine the point of insertion of Fv1’s progenitor virus. Representative assemblies from rodent genera for which genome sequences have been published to date (SI Appendix, Table S1) were compiled and searched for Fv1. Fv1 was further noted in gerbils (Meriones and Psammomys) within the Muridae, in hamsters (Cricetulus, Mesocricetus, and Phodopus) within the Cricetidae, and in the blind mole rat, Spalax, within the Spalacidae. No sequences identified in this screen contained intact ORFs.
Twenty-two assemblies contained single contigs bridging Miip and Mfn2 and were used to build an alignment of the region (species for which both genes were not assembled together were excluded, as unassembled regions would otherwise be indistinguishable from genuinely absent sequence). Comparisons revealed a high degree of variability due largely to the activity of transposable elements (TEs) (Fig. 1). This variability, combined with the multiple points of erroneous homology presented by TEs, posed a significant challenge and necessitated the use of a repeat-aware alignment program, FSA (15), which can be used in conjunction with RepeatMasker annotations. The region further displayed a propensity for large deletions; one, spanning Fv1, was visible in Rattus norvegicus and explained the absence of hybridization signals within samples from this genus (7, 12). Similar deletions were also visible within other species (Fig. 1).
Thus, although an ORF was absent in many instances, the progenitor integration could be identified throughout the Muridae, Cricetidae, and Spalacidae. Among currently available assemblies, Fv1 was absent in the Dipodidae (Jaculus jaculus) and in all more distantly diverged groups (Fig. 1). Subsequent deletions between Miip and Mfn2 may have occurred since their speciation from the last common ancestor, however, and indeed, otherwise conserved regions of these genes are absent within both J. jaculus and Dipodomys ordii (Fig. 1), highlighting this possibility. Accordingly, these data suggest a minimum insertion time of ∼45 Mya within the common ancestor of the Muroidea (14), which might be further extended to ∼50 Mya if insertion occurred more basally within the Myodonta.
The Hunt for a Candidate Progenitor.
While Fv1’s derivation from an ERV-L is clear, the precise nature of its progenitor remains obscure. Interestingly, our screening revealed regions of an ERV-L–like provirus within Spalax galili that surrounded Fv1, including remnants of the 5′ LTR (Fig. 1). While only this species was found to contain such regions, it nevertheless remained feasible to use consensus data to better represent the ancestral gag gene from which Fv1 derives. We thus conducted a more detailed search of basal genera within the Muroidae.
Fv1 was amplified and sequenced for the murines Phloeomys pallidus and Chiropodomys gliroides and the nesomyine Nesomys audeberti. Separately, the region was assembled from published whole genome sequencing (WGS) data for the cricetoyine Cricetomys gambianus. Again, no intact ORFs were determined, but together, these complete and fragmentary sequences were aligned with those previously identified and used to build a consensus model with a 1,320-bp ORF (Dataset S1). In parallel, we implemented an in silico approach to screen for ERVs more closely related to Fv1’s progenitor virus. Genome assemblies previously obtained (SI Appendix, Table S1) were masked with RepeatMasker, and all ERV-L–derived regions were extracted and clustered for each species in isolation. Consensus sequences were built for each resulting cluster and queried by BLASTn with the ancestral Fv1 model; within those hit, the regions corresponding to Fv1 were aligned and used to form a maximum likelihood (ML) tree (Fig. 2).
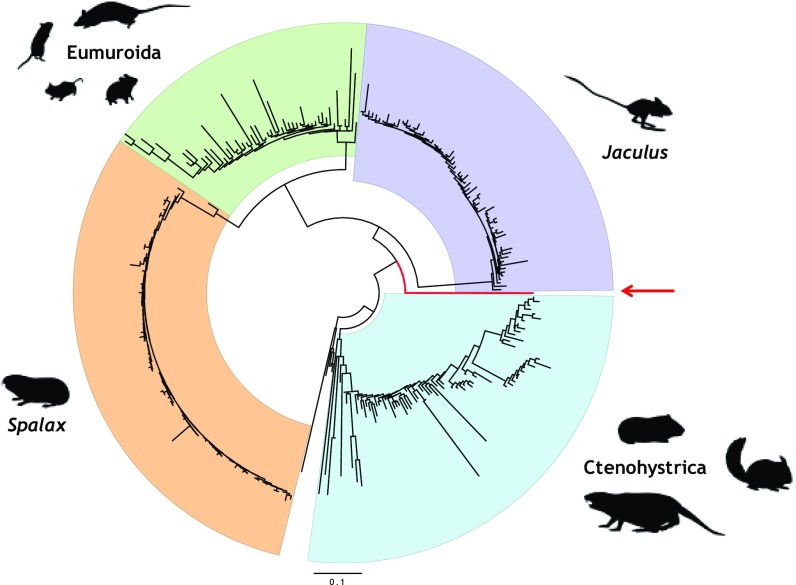
Fig. 2.
An Fv1 consensus sequence is basal to the gag regions of the ERV-L elements of the Muroidea. Circular representation of the ML tree (LogL under a generalized time reversible model (GTR+CAT) = −33,109, scale as substitutions per site) of ERV-L elements obtained from all available rodent assemblies (black branches) alongside an Fv1 consensus (thick red branch). Highlighted are the Ctenohystrica (cyan) and the Myodonta (Fig. 1) represented by Jaculus (purple), Spalax (orange), and the Eumuroida (green). The tree is rooted on the ERV-L elements of the Lagomorpha (unshaded).
Four groups of ERV-L elements radiate from those of the basal Glires: those of the Ctenohystrica, those of the Eumuroida, and two clades representing separate expansions within J. jaculus and S. galili (Fig. 2). Notably, the Fv1 consensus represented a transitionary point at the base of the Myodonta, providing an independent corroboration of Fv1’s age—∼45–50 Mya—and supporting a point of integration toward the far end of this range. In a separate tree, the Fv1 genes of Mus clustered with the Fv1 consensus formed here rather than with the ERV-L elements of the Eumuroida, confirming this unique position and ruling out the potential that restrictive capacity was achieved later through a recombination event. As such, nucleotide homology peaked at 73.9% to a cluster from S. galili and at 73.5% to a cluster from J. jaculus. Similarly, as no clusters grouped closely with Fv1, it is unlikely that alternate integrations of the progenitor virus or multiple copies of Fv1 are present within the species sampled. Indeed, duplication of Fv1 has been described only once in the literature (13).
The 45 My of Fv1.
Overall, comparatively few genera retained Fv1 ORFs, and mutational inactivation or deletion was common. This apparent propensity has previously been noted within Mus, where both gene loss and disruption have occurred (13). Despite this, the observation of any intact coding potentials over such extended periods can likely only be parsimoniously explained by their uninterrupted existence. We thus sought to assess the requirement for and frequency of selection events on Fv1 retention by modeling the likelihood of ORF loss under neutral pressure. Such periods (realized through alterations in the viral burden) may gradually occur as a result of geographic movement, habitat change, or changes in association with other species or more rapidly through the involvement of other cellular factors or through receptor escape (16).
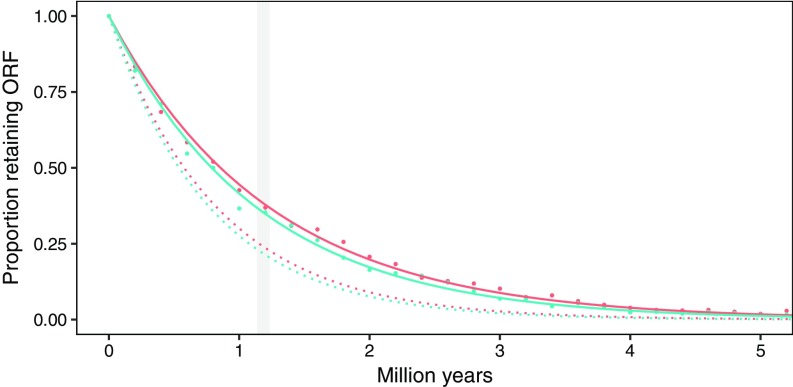
The probability of survival of a monoexonic 1,380-bp ORF (for Fv1b) can be modeled with an exponential decay function. Using empirical estimates of background substitution rates for the mouse [, (17)] and rat [, (17)] as representative examples, half-life values of 0.576 and 0.538, respectively, were calculated (Fig. 3). As these were generic values, we also explicitly simulated the mutation of Fv1b with the above , yielding half-life values of 0.856 and 0.788, respectively (Fig. 3). Using even these higher values, a mean lifetime of 1.14–1.23 My was calculated, and probability of ORF retention fell below 0.05 within 3.5–3.7 My, reaching 1.5e−16 at 45 My.
Fig. 3.
Modeling of ORF loss through time. Modeling (dotted lines) and explicit simulations (solid lines) of the mutation of Fv1b using mutation rate and generation times for Mus (red) and Rattus (green). The range of mean lifetime estimates from the explicit models is shaded gray.
The maintenance of an Fv1 ORF in any lineage over such extended time periods thus necessitates either continuous or intermittent selection reoccurring at a frequency not regularly exceeding ∼1.2 My.
The Fv1s of Apodemus Reveal Signatures of Positive Selection.
To determine the range of genera retaining coding potential, we conducted a larger survey of the Murinae, with a specific focus on the African murines, a frequently recovered evolutionary grouping containing both the Murini and the Apodemyini, which is estimated to have diversified 8.3–10.1 Mya (14, 18) (SI Appendix, Table S2). Positive selection has previously been noted in comparisons of the Fv1 genes of Mus (13), but selective pressures operating over the newly determined timescales will necessarily result in differences visible not only between species but also, between more distantly diverged groups. The Apodemus genus is similarly sized to Mus and represented a useful comparator for determining and comparing signatures of positive selection. We thus sequenced the Fv1 genes for 15 species of Apodemus, obtaining complete ORFs for 11 (SI Appendix, Table S2).
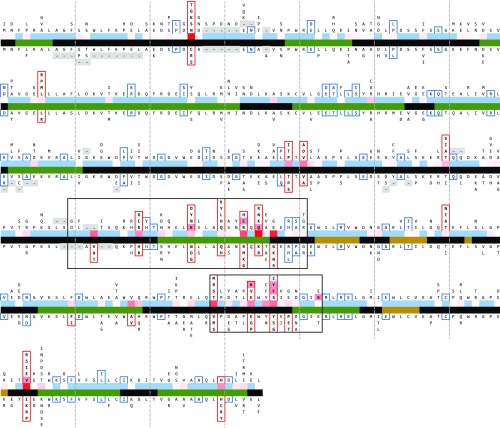
Nucleotide sequence identity was lower within Apodemus (median of 94.2%) than within Mus (96.8%), suggesting a greater sequence diversity. Signatures of positive selection were visible in both genera; overall, 14 sites were subject to pervasive positive selection, with another 7 sites under episodic positive selection within a subset of species of either genus (Fig. 4). Pervasive selection analyses with FEL and FUBAR assume that selection pressures for each site are constant throughout a phylogeny, assessing selection across all branches, whereas episodic selection analysis, with MEME, determines if individual sites have been subject to selection within a subset of branches. Likely due to the increased statistical power afforded by the larger number of sequences, an increased number of sites displayed positive selection within this analysis in comparison with previous assessments (13). Supporting the observation of increased variability within Apodemus, five of seven instances of episodic selection were within this genus.
Fig. 4.
Fv1 variability in Mus and Apodemus. Collapsed representation of the multiple sequence alignment of Mus (extending upward) and Apodemus (extending downward), with the most frequent residue toward the center. Alignment gaps are shaded gray. Sites under pervasive positive selection are boxed red above and below, and those under episodic selection are boxed red on one side only. Sites under pervasive negative selection are boxed blue above and below. For comparison, residues linked to specific restriction activities are shaded red (11). Central coloring represents residue involvement (SI Appendix, Fig. S1B) [above: low (blue) to high (red)] and structural predictions from Jpred4 [below: no prediction/unstructured (black), alpha helix (green), and beta sheet (gold)]. The previously identified variable regions, VA and VB (11), are boxed.
Two known hypervariable areas, VA (Fv1b residues 248–276) and VB (344–358) (11), were again prominent and together, included 11 of 21 positively selected residues. A third region, VC (374–401), while variable within Mus, was relatively invariable within Apodemus and contained only a single residue under positive selection. Continued support for the annotation of VA and VB alone was warranted, therefore, and suggested that these two regions likely form the main contact with CA, with individual downstream residues potentially in structural proximity. To explore this further, we sought to determine a metric for “residue involvement”—the frequency of a particular residue pair changing in combination repeatedly (SI Appendix, Fig. S1A)—and hence, to determine potential linkages. This might occur where alteration in the size or charge of a residue necessitates a corresponding supporting alteration of another residue in close structural proximity. An alignment of Fv1 genes and calculated nodal sequences was walked to determine all pairs of changes at each branch or leaf where one or both residues were under positive selection. This revealed linkages both within and between VA and VB, with residues frequently found to change in combination (SI Appendix, Fig. S1B). Again, however, this did not support the continued annotation of VC.
Analyses of likely secondary structures with this larger dataset confirm previous α-helix predictions within the N-terminal region, thought to form a coiled coil required for dimerization, as well as the presence and length of the unstructured linker region (19) (Fig. 4). Overall, 10 of 14 and 6 of 7 sites under pervasive and episodic positive selection, respectively, were within the C-terminal region, which has been shown to confer restriction specificity (19). Only sparse residue involvement was observed within the N-terminal α-helices, highlighting their likely conserved structural function. Indeed, where nucleotide variation was observed within this region, it revealed many instances of negative (purifying) selection of corresponding residues (Fig. 4), and while length differences were observed upstream of the N-terminal α-helices and downstream within the linker, none were within the region predicted to form a coiled coil. Length adjustment within the linker was otherwise observed within eight species, with an additional three-residue difference separating the genera. Length adjustments within this area were also determined within other genera within the Murinae, which extend up to 15 residues in size in Fv1 sequences described separately (20).
Discussion
Comparisons of viruses with differing sensitivities to restriction revealed Fv1 to be a CA-binding factor (10, 21). More recently, ordered assemblies of CA have been shown to direct Fv1 binding (22). Here, a requirement for multimerized CA strikes parallels to the CA-binding factor TRIM5α, also shown to interact only with regular arrays of CA (23–25). Both factors exhibit similar domain organizations—an N-terminal facilitating multimerization and a C-terminal conferring restriction specificity (22, 24)—and hybrid factors have activity in vitro (26). Now approaching 50 y since its discovery, however, the mechanism of Fv1 restriction has not been elucidated beyond the achievement of a block between reverse transcription and nuclear entry (27). In fact, no definitive modes of action have yet been described for any CA-binding restriction factors, but mechanisms to promote degradation by the proteasome or to sequester, stabilize, or destabilize the viral core after entry are most widely suggested. Indeed, CA mutants with increased or decreased lattice stability display somewhat equivalent infectivity impairments (28).
In the absence of mechanistic or accurate structural detail, which would facilitate understanding of specific interactions and allow prediction or design of restriction capacity, great efforts have been made to better understand genetic variation at the Fv1 locus. This has revealed a scope of restriction extending beyond the gammaretroviruses (11), suggesting that a variety of viruses have historically contributed to positive selection of the gene within Mus (13). Here, we have sought to more accurately detail the evolutionary journey of Fv1 and present a number of sequences, many with intact ORFs, for species outside the genus Mus. An ML tree built with all complete gene sequences determined here and to date within the literature (11, 13, 20) confirms the widespread distribution of the Fv1 gene across the Eumuroida (SI Appendix, Fig. S2). Accurate field identification of wild-caught animals is an issue pervasive across this and other published studies, and we note a number of potential inconsistencies between suggested identities and the position of certain sequences within this tree both for sequences described herein and for those from the literature. Regardless, their presentation with caveats can only benefit Fv1 research, and we include them accordingly (SI Appendix, Table S2).
Previously accepted to have integrated ∼7 Mya (13), we have now dramatically extended this timeframe to ∼45–50 Mya, a conclusion supported by other recent research (20). Assuming Dollo parsimony, where a complex trait arises once but can be lost multiple times, the retention of the Fv1 ORF for such extended periods highlights a role for a continuous or frequently reoccurring selective advantage. In turn, this implies repeated waves of infection by novel viruses, possibly as a result of cross-species transmissions. In the absence of such a pressure, Fv1 would be expected to have a mean lifetime of only ∼1.2 My. In support of this, we have found Fv1 to be frequently lost within the Muroidea, and even within Mus, Fv1 is deleted or the ORF lost in several instances (11). This further suggests that the progenitor viral gag itself conferred a selective advantage at the point of integration or that its co-option was both unconvoluted and rapid, especially given that, in the absence of such a selective advantage, intact proviruses are otherwise expected to be deleterious.
A requirement for swift transference or adaptation of restriction potential in the face of successive waves of retroviral infections necessitates significant plasticity in Fv1’s mechanism of CA recognition, complicating attempts to predict target specificity from primary sequence alone or to link restriction specificities to specific variations in the endogenous retroviral complement of host lineages. Nevertheless, linkage analyses suggest a previously unappreciated role for cooperative change across the protein, especially within and between VA and VB, inside of which positive selection is also largely confined. Together, these data point toward restriction determination by a face composed largely of these two regions, and indeed, many conversions between restriction capacities require only single-residue substitutions within these areas (11). Repeated change at certain sites may derive from functional constraints on the restriction factor but also from multifactorial constraints on the viral target, leading to the resampling of specific residues and cyclical host–virus coevolution (29). Together, this likely suggests that shared properties of CA lattice structures, rather than the specific properties of any individual CA monomer, are the primary means of recognition.
Fv1n, an ∼52-kDa monomer, has previously been shown to be divided into two ∼20-kDa fragments separated by a protease-sensitive linker (19). The N-terminal domain has extensive α-helix predictions; in support of a conserved role in coiled coil formation, N-terminal deletions extending past residue 32 are not tolerated, and at least the first 158 residues are required to confer function in hybrid factors (26, 30). Accordingly, we now reveal frequent negative (purifying) selection within this region. In contrast, extensive deletions can be made within the linker, suggesting that its length is of greater importance than its sequence (30). Correspondingly, we now present evidence of length variations within this region, perhaps suggesting a means of fine-tuning C-terminal domain positioning according to differences in the size or curvature of presented CA lattice structures. This again parallels TRIM5α, where regulated positioning of the SPRY domain is central to attainment of an avid interaction with CA (31, 32).
Positive selection of TRIM5α has previously been shown to have occurred over at least ∼30 My (33, 34) and to have been shaped by lentiviruses over the last 11–16 My (35). Here, we now show that extended coevolution of host factors and their viral partners is also common to murids and undoubtedly necessitates a shifting and reoccurring burden to maintain selection over such timescales. Such fluid interactions likely also present in the TRIM5 gene of the cotton top tamarin, Saguinus oedipus, which has also been shown to restrict multiple genera of retroviruses (36–38). CA-binding restriction factors have evolved multiple times in mammals (39), and it might be hypothesized that analogous factors may be common to any host adaptation to retroviral infection or indeed, to any pathogen presenting regularized structures on infection of a cell. Indeed, it is possible that such historic interactions have also shaped both Fv1 and Trim5α. Drawing a parallel to the recognition of LPS from diverse Gram-negative bacteria (40), CA-binding retroviral restriction factors may represent another class of pattern recognition receptors.
Materials and Methods
Modeling ORF Half-Life.
In standard genetic code, 22 of 549 possible alterations of the 61 amino acid-encoding codons produce stop codons, assuming an equal rate of mutation across the alphabet: . Base transition is, however, twice as frequent as transversion among de novo single-nucleotide variants (41), and only 4 of 183 base transitions produce stop codons. Thus, the probability, , of substitution resulting in stop codon acquisition can be somewhat more accurately represented (42):
Allowing three substitutions per codon, incorporating the background per-base substitution rates, , and the length of the ORF, , allows derivation of the decay constant . Similarly, the decay constant is derived from , , and the probability, , of an indel not being a multiple of the codon length [empirically estimated at 0.83 (43)]:
The half-life, , and mean lifetime, , are thus calculated with standard formulas:
Mutation Simulation.
Two Python programs, mutator (commit c9ae773) and orf_scanner (commit 7d70b14), were written to simulate mutational processes and to assess their impact on ORF length. ORF retention rates for 1,000 replicates were used to fit a standard model for exponential decay within R and to derive mean lifetime, , and half-life,
Software.
The programs developed for this study are available under permissive license at https://github.com/A-N-Other/pedestal. We encourage code reuse and comment.
Other Methods.
Full materials and methods are included in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge the museums that provided samples for this study—without these excellent collections, this work would not have been possible. We thank Milos Macholán (Czech Academy of Sciences), Ondřej Mikula (Czech Academy of Sciences), François Catzeflis (University of Montpellier), Jean-Pierre Quéré (French National Institute for Agricultural Research), and Serge Morand (CIRAD/University of Liège) for samples. We also thank Aris Katzourakis (Oxford University) for his thoughtful comments on the manuscript. G.R.Y., M.W.Y., and J.P.S. were supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001162), the UK Medical Research Council (FC001162), and the Wellcome Trust (FC001162). S.J.S. was supported by the US National Science Foundation (DEB-0841447).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MH001948–MH001969 and MH727610–MH727614).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808516115/-/DCSupplemental.
References
- 1.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crow JF. C. C. Little, cancer and inbred mice. Genetics. 2002;161:1357–1361. doi: 10.1093/genetics/161.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lilly F. Fv-2: Identification and location of a second gene governing the spleen focus response to Friend leukemia virus in mice. J Natl Cancer Inst. 1970;45:163–169. [PubMed] [Google Scholar]
- 4.Pincus T, Hartley JW, Rowe WP. A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J Exp Med. 1971;133:1219–1233. doi: 10.1084/jem.133.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowe WP. Studies of genetic transmission of murine leukemia virus by AKR mice. I. Crosses with Fv-1 n strains of mice. J Exp Med. 1972;136:1272–1285. doi: 10.1084/jem.136.5.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowe WP, Hartley JW. Studies of genetic transmission of murine leukemia virus by AKR mice. II. Crosses with Fv-1 b strains of mice. J Exp Med. 1972;136:1286–1301. doi: 10.1084/jem.136.5.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 8.Bénit L, et al. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol. 1997;71:5652–5657. doi: 10.1128/jvi.71.7.5652-5657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malfavon-Borja R, Feschotte C. Fighting fire with fire: Endogenous retrovirus envelopes as restriction factors. J Virol. 2015;89:4047–4050. doi: 10.1128/JVI.03653-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozak CA, Chakraborti A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 11.Yap MW, Colbeck E, Ellis SA, Stoye JP. Evolution of the retroviral restriction gene Fv1: Inhibition of non-MLV retroviruses. PLoS Pathog. 2014;10:e1003968. doi: 10.1371/journal.ppat.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi CF, et al. Molecular phylogeny of Fv1. Mamm Genome. 1998;9:1049–1055. doi: 10.1007/s003359900923. [DOI] [PubMed] [Google Scholar]
- 13.Yan Y, Buckler-White A, Wollenberg K, Kozak CA. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc Natl Acad Sci USA. 2009;106:3259–3263. doi: 10.1073/pnas.0900181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steppan SJ, Schenk JJ. Muroid rodent phylogenetics: 900-Species tree reveals increasing diversification rates. PLoS One. 2017;12:e0183070. doi: 10.1371/journal.pone.0183070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley RK, et al. Fast statistical alignment. PLOS Comput Biol. 2009;5:e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak CA. The mouse “xenotropic” gammaretroviruses and their XPR1 receptor. Retrovirology. 2010;7:101. doi: 10.1186/1742-4690-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper GM, et al. Characterization of evolutionary rates and constraints in three mammalian genomes. Genome Res. 2004;14:539–548. doi: 10.1101/gr.2034704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lecompte E, et al. Phylogeny and biogeography of African Murinae based on mitochondrial and nuclear gene sequences, with a new tribal classification of the subfamily. BMC Evol Biol. 2008;8:199. doi: 10.1186/1471-2148-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bishop KN, et al. Characterization of an amino-terminal dimerization domain from retroviral restriction factor Fv1. J Virol. 2006;80:8225–8235. doi: 10.1128/JVI.00395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boso G, Buckler-White A, Kozak CA. Ancient evolutionary origin and positive selection of the retroviral restriction factor Fv1 in muroid rodents. J Virol. July 5, 2018 doi: 10.1128/JVI.00850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens A, et al. Retroviral capsid determinants of Fv1 NB and NR tropism. J Virol. 2004;78:9592–9598. doi: 10.1128/JVI.78.18.9592-9598.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilditch L, et al. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci USA. 2011;108:5771–5776. doi: 10.1073/pnas.1100118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forshey BM, Shi J, Aiken C. Structural requirements for recognition of the human immunodeficiency virus type 1 core during host restriction in owl monkey cells. J Virol. 2005;79:869–875. doi: 10.1128/JVI.79.2.869-875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dodding MP, Bock M, Yap MW, Stoye JP. Capsid processing requirements for abrogation of Fv1 and Ref1 restriction. J Virol. 2005;79:10571–10577. doi: 10.1128/JVI.79.16.10571-10577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yap MW, Mortuza GB, Taylor IA, Stoye JP. The design of artificial retroviral restriction factors. Virology. 2007;365:302–314. doi: 10.1016/j.virol.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Jolicoeur P, Rassart E. Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J Virol. 1980;33:183–195. doi: 10.1128/jvi.33.1.183-195.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyerson NR, Sawyer SL. Two-stepping through time: Mammals and viruses. Trends Microbiol. 2011;19:286–294. doi: 10.1016/j.tim.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bishop KN, Bock M, Towers G, Stoye JP. Identification of the regions of Fv1 necessary for murine leukemia virus restriction. J Virol. 2001;75:5182–5188. doi: 10.1128/JVI.75.11.5182-5188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roganowicz MD, et al. TRIM5α SPRY/coiled-coil interactions optimize avid retroviral capsid recognition. PLoS Pathog. 2017;13:e1006686. doi: 10.1371/journal.ppat.1006686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldstone DC, et al. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc Natl Acad Sci USA. 2014;111:9609–9614. doi: 10.1073/pnas.1402448111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawyer SL, Emerman M, Malik HS. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy KR, Kirmaier A, Autissier P, Johnson WE. Evolutionary and functional analysis of old world primate TRIM5 reveals the ancient emergence of primate lentiviruses and convergent evolution targeting a conserved capsid interface. PLoS Pathog. 2015;11:e1005085. doi: 10.1371/journal.ppat.1005085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80:8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diehl WE, Stansell E, Kaiser SM, Emerman M, Hunter E. Identification of postentry restrictions to Mason-Pfizer monkey virus infection in New World monkey cells. J Virol. 2008;82:11140–11151. doi: 10.1128/JVI.00269-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yap MW, et al. Restriction of foamy viruses by primate Trim5alpha. J Virol. 2008;82:5429–5439. doi: 10.1128/JVI.02462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanz-Ramos M, Stoye JP. Capsid-binding retrovirus restriction factors: Discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol. 2013;94:2587–2598. doi: 10.1099/vir.0.058180-0. [DOI] [PubMed] [Google Scholar]
- 40.Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med. 2013;45:e66–e69. doi: 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell CD, Eichler EE. Properties and rates of germline mutations in humans. Trends Genet. 2013;29:575–584. doi: 10.1016/j.tig.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brookfield JFY. Genetic redundancy. In: Hall J, Dunlap J, Friedmann T, Giannelli F, editors. Advances in Genetics. 1st Ed. Vol 36. Elsevier; San Diego: 1997. pp. 143–150. [DOI] [PubMed] [Google Scholar]
- 43.Ratan A, Olson TL, Loughran TP, Jr, Miller W. Identification of indels in next-generation sequencing data. BMC Bioinformatics. 2015;16:42. doi: 10.1186/s12859-015-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.