Significance
Malonyl-CoA is an important metabolite for the production of many natural products. Here, we repurposed a type III polyketide synthase RppA capable of producing red-colored flaviolin as a malonyl-CoA biosensor in Escherichia coli, Pseudomonas putida, and Corynebacterium glutamicum. Strains with enhanced malonyl-CoA accumulation could easily be identified by colorimetric screening of a library. Gene knockdown targets enabling increased malonyl-CoA accumulation were identified and applied for production of two polyketide (6-methylsalicylic acid and aloesone) and two phenylpropanoid (resveratrol and naringenin) compounds. Without extensive metabolic engineering, 6-methylsalicylic acid could be produced to the highest titer reported for E. coli and also naringenin and resveratrol to high concentrations. Furthermore, microbial production of aloesone was demonstrated.
Keywords: malonyl-CoA, metabolic engineering, natural products, polyketide synthase, biosensor
Abstract
Malonyl-CoA is an important central metabolite for the production of diverse valuable chemicals including natural products, but its intracellular availability is often limited due to the competition with essential cellular metabolism. Several malonyl-CoA biosensors have been developed for high-throughput screening of targets increasing the malonyl-CoA pool. However, they are limited for use only in Escherichia coli and Saccharomyces cerevisiae and require multiple signal transduction steps. Here we report development of a colorimetric malonyl-CoA biosensor applicable in three industrially important bacteria: E. coli, Pseudomonas putida, and Corynebacterium glutamicum. RppA, a type III polyketide synthase producing red-colored flaviolin, was repurposed as a malonyl-CoA biosensor in E. coli. Strains with enhanced malonyl-CoA accumulation were identifiable by the colorimetric screening of cells showing increased red color. Other type III polyketide synthases could also be repurposed as malonyl-CoA biosensors. For target screening, a 1,858 synthetic small regulatory RNA library was constructed and applied to find 14 knockdown gene targets that generally enhanced malonyl-CoA level in E. coli. These knockdown targets were applied to produce two polyketide (6-methylsalicylic acid and aloesone) and two phenylpropanoid (resveratrol and naringenin) compounds. Knocking down these genes alone or in combination, and also in multiple different E. coli strains for two polyketide cases, allowed rapid development of engineered strains capable of enhanced production of 6-methylsalicylic acid, aloesone, resveratrol, and naringenin to 440.3, 30.9, 51.8, and 103.8 mg/L, respectively. The malonyl-CoA biosensor developed here is a simple tool generally applicable to metabolic engineering of microorganisms to achieve enhanced production of malonyl-CoA–derived chemicals.
In the past several decades, metabolic engineering has significantly contributed to the production of various products, including biofuels, drugs, food additives, petro-based chemicals, pharmaceutical proteins, and polymers by rewiring and optimizing the metabolism of microorganisms (1–3). Through integration with systems biology, synthetic biology, and evolutionary engineering, metabolic engineering has become more powerful, allowing accelerated development of high-performance strains (3). One of the bottlenecks in metabolic engineering is the analysis step, such as high-performance liquid chromatography (HPLC), required for monitoring the concentrations of a product or metabolic intermediates during strain development. Thus, much effort has been exerted to develop metabolite sensors that enable rapid monitoring of product and metabolite levels (4–8).
Within a wide portfolio of chemicals that can be produced by microorganisms, malonyl-CoA is of particular interest, as it is a major building block for many value-added chemicals, including polyketides (9, 10), phenylpropanoids (11), and biofuels (12). However, malonyl-CoA availability for the overproduction of target compounds is often limited due to the competition with essential cellular metabolism, such as fatty acid biosynthesis. In addition, quantification of intracellular malonyl-CoA concentration often requires highly accurate but time-consuming analytical methods, such as LC-MS/MS, due to the presence of many different intracellular metabolites (13). Moreover, laborious analytical sample preparation processes coupled with effective and rapid quenching of cellular metabolism is required because malonyl-CoA is a dynamic intermediate with rapid turnover rates and is sensitive to environmental conditions, such as pH and temperature (14, 15). For these reasons, several malonyl-CoA biosensors based on transcription factors have been developed to increase analytical throughput and avoid labor-intensive and time-consuming sample preparation steps (5, 7, 16, 17). However, these transcription factor-based malonyl-CoA sensors, only demonstrated in Escherichia coli (7) and Saccharomyces cerevisiae (5), have limitations, including the requirement of multiple signal transduction steps, which might bias the output signal and impede general application to different microbial strains. In addition, utilization of fluorescence reporter proteins makes it difficult to be employed in microorganisms displaying autofluorescence, such as Pseudomonas species (18).
To overcome the above limitations, herein we report development of a simple and robust enzyme-coupled malonyl-CoA biosensor by repurposing 1,3,6,8-tetrahydroxynaphthalene synthase (THNS; also referred to as RppA), a type III polyketide synthase (PKS); here, an enzyme-coupled biosensor refers to a biosensor comprising an enzyme repurposed to convert a given target metabolite into a detectable metabolite (8). RppA is responsible for the conversion of malonyl-CoA to 1,3,6,8-tetrahydroxynaphthalene (THN), which is then spontaneously converted to flaviolin (19). Because flaviolin displays a red color, it can be utilized as a direct colorimetric indicator of intracellular malonyl-CoA level. RppA was used to develop an effective enzyme-coupled malonyl-CoA biosensor in three industrially important bacteria including E. coli, Pseudomonas putida, and Corynebacterium glutamicum. This malonyl-CoA biosensor is advantageous as it enables one-step signal conversion from input malonyl-CoA to output color signal, allowing simple and rapid colorimetric screening of malonyl-CoA overproducing bacterial strains. We also show that not only RppA but also other type III PKSs capable of producing colored metabolites from malonyl-CoA can be utilized as malonyl-CoA biosensors in a similar manner. To apply the RppA biosensor for screening E. coli strains with enhanced malonyl-CoA production, we constructed and introduced an E. coli genome-scale synthetic small regulatory RNA (sRNA) library into the sensor strain harboring RppA for screening knockdown gene targets. The screened positive sRNA targets were used to engineer E. coli strains capable of accumulating malonyl-CoA to higher levels, and consequently to develop four different E. coli strains producing two polyketide and two phenylpropanoid products.
Results
Type III PKS RppA Enables the Production of Flaviolin.
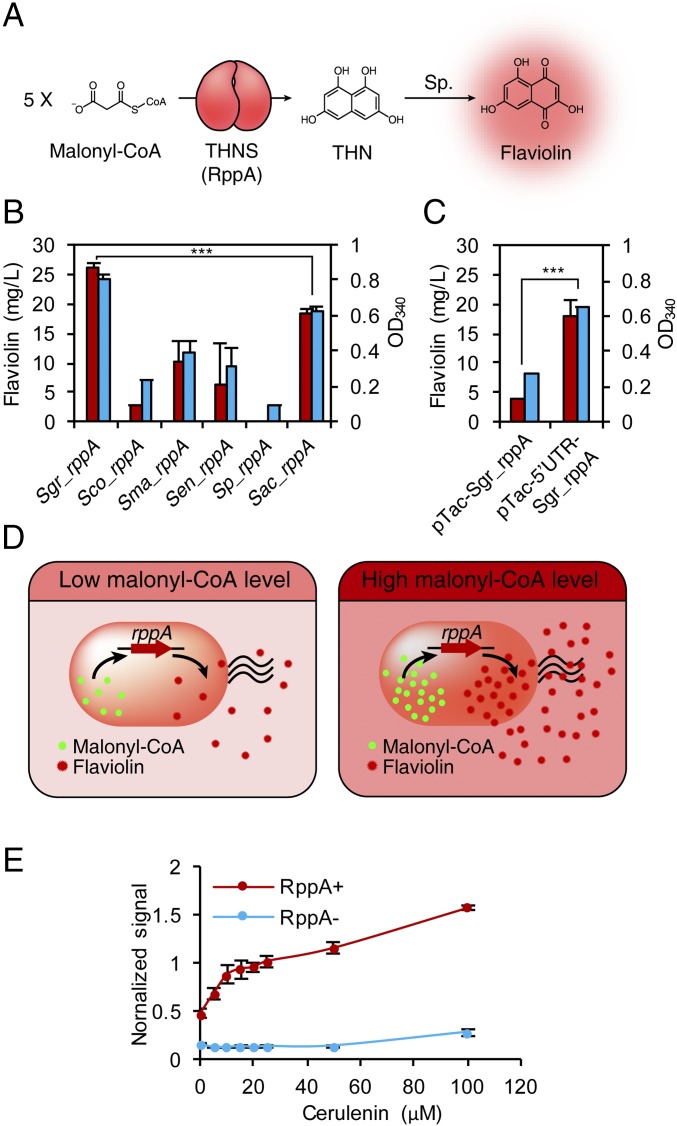
As the first step of flaviolin biosynthesis, five molecules of malonyl-CoA are converted to THN by RppA (Fig. 1A) (19). Then, THN can be spontaneously converted to flaviolin by a nonenzymatic oxidation reaction (Fig. 1A and SI Appendix, Text S1 and Fig. S1A). Because several actinomycetes are reported to harbor rppA (the gene encoding RppA), different rppA genes were tested for the production of flaviolin in E. coli (SI Appendix, Table S1). Plasmid pET-30a(+) was used to express the genes under the strong T7 promoter. Among the six different rppA genes from Streptomyces griseus (19, 20), Streptomyces coelicolor (21), Streptomyces avermitilis (22), Saccharopolyspora erythraea (23), Streptomyces peucetius (24), and Streptomyces aculeolatus (25) tested, the one from S. griseus (Sgr_RppA) allowed production of flaviolin to the highest titer (26.0 mg/L) when cultured in shake flask containing 50 mL of M9 minimal medium supplemented with 10 g/L of glucose (Fig. 1B). It is also reported that the Km value of Sgr_RppA is ∼1 μM, showing a high affinity toward malonyl-CoA (20, 26). The authenticity of flaviolin produced from the engineered E. coli strains was confirmed by LC-MS and MS/MS analysis (SI Appendix, Fig. S1B). The produced flaviolin is readily secreted from the cell (SI Appendix, Fig. S1C). Having confirmed that E. coli expressing the S. griseus rppA gene produced flaviolin, the rppA expression platform was established in a tac promoter-based expression cassette vector for its use in various E. coli strains beyond the DE3 strains; for this, the rppA gene was transferred to the pTacCDFS vector, a pCDFDuet-1 (Novagen) derivative with a tac promoter-based expression cassette. To optimize rppA expression, its 5′ untranslated region (5′UTR) was altered using the previously reported UTR designer (27). The 5′UTR-optimized rppA construct (pTac-5′UTR-Sgr_rppA) resulted in less production of flaviolin (18.1 mg/L) (Fig. 1C) compared with that obtained by expressing the rppA gene under the T7 promoter. However, flaviolin produced by employing the 5′UTR-optimized rppA construct gave enough red color to develop a colorimetric malonyl-CoA biosensor (Fig. 1D).
Fig. 1.
A type III PKS RppA can be repurposed as a malonyl-CoA biosensor. (A) RppA converts five molecules of malonyl-CoA into one molecule of red-colored flaviolin. Malonyl-CoA is first converted to THN, which is catalyzed by RppA (THNS). Then, THN is nonenzymatically oxidized to flaviolin. Sp. denotes spontaneous nonenzymatic oxidation reaction. (B) Flaviolin production from the E. coli BL21(DE3) strains introduced with the given rppA genes isolated from different bacteria. (C) Optimization of flaviolin production. Flaviolin production from E. coli BL21(DE3) harboring the given plasmids are shown. In B and C, flaviolin titers are represented as red bars; absorbances of culture supernatants at 340 nm are represented as blue bars. ***P < 0.001, determined by two-tailed Student’s t test. (D) Schematic description of the repurposed type III PKS RppA as a malonyl-CoA biosensor. Because RppA actively converts malonyl-CoA into red-colored flaviolin, higher intracellular level of malonyl-CoA leads to higher production and secretion of flaviolin into the medium. Thus, superb malonyl-CoA producers can be identified as cultures with deeper red colors. (E) Characterization of the RppA biosensor. Normalized signals generated from the sensor strain showed dose-dependent responses to intracellular malonyl-CoA abundance. Intracellular malonyl-CoA abundance was titrated using different concentrations of cerulenin added to the medium. Signals normalized with cell growth are plotted either with (RppA+) or without (RppA−) RppA expression. Error bars, mean ± SD (SD; n = 3).
RppA Can Be Used as a Malonyl-CoA Biosensor in E. coli.
After the construction of the rppA expression platform, we examined whether RppA can be used as a malonyl-CoA biosensor. First, to define the signal of RppA biosensor, absorbance of flaviolin at different wavelengths was tested. The λmax of flaviolin is reported to be ∼300 nm (28). However, when the culture supernatant of the rppA-expressing strain and that of the rppA− control strain were compared, it was found that absorbance at 340 nm gave the least background noise from the control strain while displaying sufficient value when flaviolin was present (SI Appendix, Fig. S1D). Thus, the signal of the RppA biosensor was defined as absorbance of culture supernatant at 340 nm (OD340). The signal of the RppA-harboring strain showed a strong correlation with the actual flaviolin concentration (Fig. 1 B and C). Generally, for characterizing a biosensor and determining its dose-dependent dynamic response range, different concentrations of a target compound has to be added to measure the change in output signal intensity. However, because malonyl-CoA is an intracellular intermediate not transported through cell membrane, exogenous feeding of malonyl-CoA is not possible. Instead, we employed an indirect method using cerulenin, which is a well-characterized chemical inhibitor for fatty acid elongation, specifically blocking the activity of β-ketoacyl-acyl carrier protein (ACP) synthase (29). It was previously observed that intracellular malonyl-CoA concentration in E. coli increased as the added cerulenin concentration increased (7), which is consistent with the observations in this study (SI Appendix, Fig. S2A). Because cerulenin alters intracellular malonyl-CoA levels in a single-cell basis, a single-cell–based sensor signal should be measured. The signal (OD340 of culture supernatant) normalized with cell growth (OD600) was monitored with different concentrations of added cerulenin. As a result, it was observed that the normalized signal from the strain expressing the rppA gene [denoted as RppA+; E. coli BL21(DE3) harboring pTac-5′UTR-Sgr_rppA; hereafter referred to as the sensor strain] increased as cerulenin concentration increased, while that of the control strain [denoted as RppA−; E. coli BL21(DE3) without RppA] remained constant (Fig. 1E). The relative concentration of flaviolin produced from the sensor strain normalized with cell growth also increased as cerulenin concentration increased (SI Appendix, Fig. S2B). This could also be clearly seen to the naked eyes (SI Appendix, Fig. S2C). Thus, the RppA malonyl-CoA biosensor is expected to enable simple and rapid screening of malonyl-CoA overproducing strains. To confirm the applicability of the RppA biosensor in different E. coli strains, plasmid pTac-5′UTR-Sgr_rppA was introduced into 16 different E. coli strains (SI Appendix, Table S2); all of the resultant strains successfully produced flaviolin and displayed red color (SI Appendix, Fig. S2D).
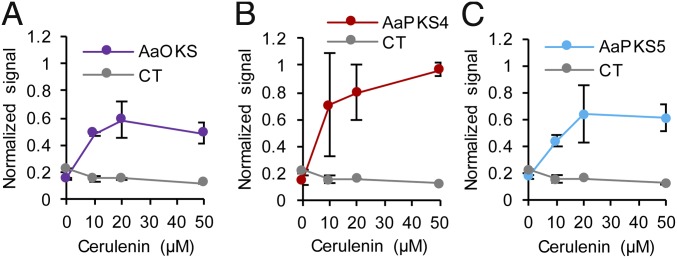
To see whether other type III PKSs can also be used as malonyl-CoA biosensors, five enzymes from Aloe arborescens that use malonyl-CoA for producing polyketides were selected and tested: pentaketide chromone synthase mutant AaPCSm (M207G) (30), octaketide synthase AaOKS (31), aloesone synthase mutant AaPKS3m (A207G) (32), octaketide synthase AaPKS4 (32), and octaketide synthase AaPKS5 (32) (SI Appendix, Table S1). When E. coli BL21(DE3) strains harboring these enzymes were cultivated in test tubes containing modified R/2 medium, those strains harboring AaOKS, AaPKS4, and AaPKS5 showed a slight color change. LC-MS analysis was performed to deduce the identity of the produced polyketides that possibly contributed to the color change (SI Appendix, Fig. S3A). Then, cerulenin titration experiments with the strains expressing these three PKSs suggested that the color signal was dependent on intracellular malonyl-CoA abundance (Fig. 2 and SI Appendix, Fig. S3B); here, the signal of the biosensors was defined as absorbance of culture supernatant at 300 nm (OD300) (SI Appendix, Fig. S3C). Thus, it was concluded that not only RppA but also any other type III PKSs capable of producing colored metabolites from malonyl-CoA can be utilized as malonyl-CoA biosensors. Because RppA displayed wider dynamic response range and more vivid color signal, it was further utilized for the later sections of this study.
Fig. 2.
Characterization of type III PKSs as malonyl-CoA biosensors. Normalized signals generated from the sensor strains harboring (A) AaOKS, (B) AaPKS4, and (C) AaPKS5 showed dose-dependent responses to intracellular malonyl-CoA abundance. Intracellular malonyl-CoA abundance was titrated using different concentrations of cerulenin added to the medium. Signals normalized with cell growth are plotted either with or without PKS expression. AaOKS, octaketide synthase from A. arborescens; AaPKS4, octaketide synthase from A. arborescens; AaPKS5, octaketide synthase from A. arborescens; CT, control. Error bars, mean ± SD (n = 3).
Construction and Application of an E. coli Genome-Scale Synthetic sRNA Library.
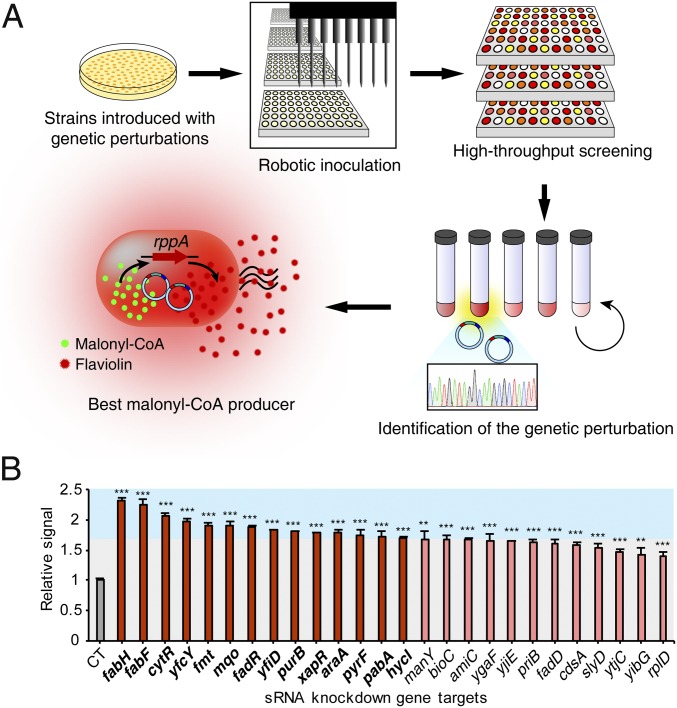
The RppA malonyl-CoA biosensor system developed above was applied for identifying gene knockdown targets increasing the malonyl-CoA level. For the systematic gene knockdown studies in E. coli, the synthetic sRNA technology (33, 34) was employed. To include all major metabolic and regulatory genes in E. coli, a genome-scale synthetic sRNA library targeting 1,858 genes within the E. coli K-12 W3110 strain was constructed in this study (SI Appendix, Fig. S4 and Dataset S1). The new synthetic sRNA library covers 45% of all genes in E. coli and includes all genes that encode proteins with known functions (35). The gene targets were selected based on an in silico E. coli metabolic model, a database, and literatures (SI Appendix, Text S2). This synthetic sRNA library was transformed into E. coli strains and the RppA biosensor was used for high-throughput screening of malonyl-CoA overproducing strains (Fig. 3A).
Fig. 3.
High-throughput screening of E. coli strains with enhanced malonyl-CoA accumulation. (A) Overall procedures of high-throughput screening for the identification of the best malonyl-CoA producer. Colonies composed of the sensor strains introduced with given genetic perturbations (e.g., synthetic small regulatory RNA library) are inoculated into microplates by using a robotic high-throughput screening system. After cultivation, strains showing enhanced signal are isolated and cultivated again in test tubes. Plasmid DNAs or genomic DNAs from the isolated strains can be extracted and sequenced for identifying the beneficial genetic perturbations. The screened genetic perturbations (either a library component or mutation) can be back-transformed into the initial sensor strain to confirm the sole contribution of the introduced genetic perturbations to malonyl-CoA production. Thus, the best malonyl-CoA producer can be isolated. (B) Relative signals from sensor strains harboring the 26 initially screened sRNAs. The final 14 knockdown gene targets, which are displayed as deeper red bars and bold letters, were screened according to the set threshold (70% increase in signal compared with that of the control sensor strain without sRNA). The graph is divided into two sections—blue and gray—according to the set threshold. Error bars, mean ± SD (n = 3). **P < 0.01, ***P < 0.001, determined by two-tailed Student’s t test.
To examine the efficacy of RppA biosensor-based screening, E. coli BL21(DE3) was initially employed. The pooled E. coli genome-scale synthetic sRNA library was introduced into E. coli BL21(DE3) harboring pTac-5′UTR-Sgr_rppA. To comprehensively select the strains introduced with all of the genome-scale sRNA library components, 11,488 colonies covering more than sixfold of the library size were selected and screened (SI Appendix, Fig. S5A). For colorimetric screening, a robotic high-throughput screening system was employed (Fig. 3A). Previous studies involving the transcription factor-based malonyl-CoA biosensor utilized fluorescence-activated cell sorting (FACS) for screening malonyl-CoA overproducers (5). FACS-based screening basically allows selection of a single cell having high specific productivity (e.g., gram product per gram dry cell weight per hour) for a target compound. On the other hand, colorimetric screening allows selection of a strain that gives high volumetric productivity (e.g., gram product per liter per hour) of a target compound because the color signal observed represents combination of cell growth and specific productivity (i.e., the color observed is contributed by color intensity of individual cell and also by cell density in the screening well). Thus, we did not normalize signal with cell growth.
As a result of this robotic high-throughput screening (SI Appendix, Materials and Methods), 231 strains harboring synthetic sRNAs showing either stronger signal or color (to the naked eyes) than the control were initially selected (SI Appendix, Fig. S5B); it should be noted that these initial strains were selected under the least-stringent threshold to obtain all possible candidate strains because cell growth is not consistent in the small wells for high-throughput screening. Thus, the screened strains were cultivated again in test tubes, and 70 strains displaying highly increased signal compared with the control strain without sRNA were chosen. The plasmids in these 70 strains were sequenced to identify the introduced synthetic sRNAs (SI Appendix, Fig. S6); three synthetic sRNAs targeting argB, fabF, and nudD were observed twice. Of these 67 initially screened synthetic sRNAs, 26 exhibited more than a 45% increase in the signal. To eliminate potential false-positives, these 26 sRNA vectors were transformed back to the original sensor strain to confirm the results; all of them showed signal increase, suggesting that there was no false-positive. Among them, 14 sRNAs that resulted in more than 70% increase in signal were selected as the final knockdown gene targets (Fig. 3B and SI Appendix, Table S3): fabH, fabF, cytR, ytcY, fmt, mqo, fadR, yfiD, purB, xapR, araA, pyrF, pabA, and hycI. In addition to screening knockdown gene targets, amplification gene targets were also assessed by testing gene-amplification targets obtained from the in silico FVSEOF (flux variability scanning based on enforced objective flux) algorithm (36) (SI Appendix, Text S3 and Fig. S7 A and C). Although eight of nine overexpressed gene targets showed increased signal, none of them endowed the sensor strain with higher signal than the set threshold (more than 70% increase in signal) (SI Appendix, Fig. S7B).
The ultimate purpose of employing the malonyl-CoA biosensor is to overproduce chemicals of interest derived from malonyl-CoA by applying the screened gene-manipulation targets. Therefore, the effects of knocking down the screened 14 gene targets were assessed by introducing the corresponding synthetic sRNAs into E. coli strains capable of producing representative natural compounds derived from malonyl-CoA. We chose two polyketide (6-methylsalicylic acid and aloesone) and two phenylpropanoid (resveratrol and naringenin) compounds as the target products.
Enhanced Production of Polyketides by Knocking Down the Screened Gene Targets.
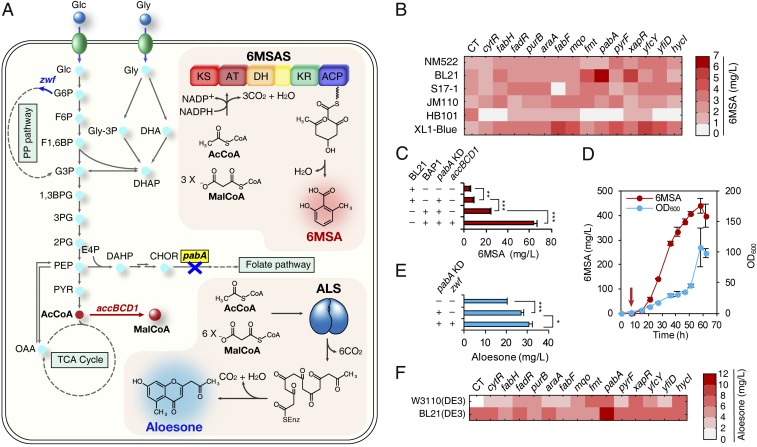
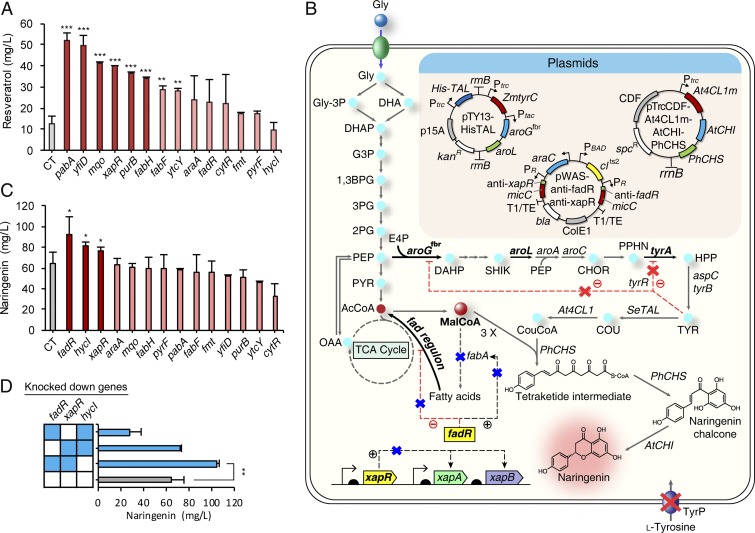
Polyketides, produced from multiple claisen condensation reactions, are compounds having multiple β-keto groups with various degrees of modifications, such as reduction, cyclization, and glycosylation (9, 10). Naturally produced from Penicilium griseofulvum (also referred to as Penicilium patulum), 6-methylsalicylic acid (6MSA) is a representative fungal polyketide reported to possess antibacterial and antifungal activities (Fig. 4A) (37). A type I iterative PKS, 6MSA synthase (6MSAS), synthesizes 6MSA by condensation of one molecule of acetyl-CoA and three molecules of malonyl-CoA. Heterologous 6MSA production was reported both in E. coli (38) and in S. cerevisiae (38, 39); in E. coli, 75 mg/L of 6MSA was produced. For the construction of an E. coli strain capable of producing 6MSA, P. griseofulvum Pg6MSAS was cloned into pTac15K, followed by the introduction of Bacillus subtilis sfp (encoding 4′-phosphopantetheinyl transferase) as an operon (SI Appendix, Fig. S8A). The constructed plasmid pTac-Pg6MSAS-sfp was introduced into E. coli BL21(DE3), and expression of the enzymes was confirmed by SDS/PAGE (SI Appendix, Fig. S8C). Different carbon sources (glucose and glycerol) were compared for the production of 6MSA; 4.7 mg/L of 6MSA was produced from glycerol (SI Appendix, Fig. S9A), while a negligible amount of 6MSA was produced from glucose. The authenticity of the produced 6MSA was confirmed by LC-MS (SI Appendix, Fig. S9 B and C). Because production capacities and metabolic network configurations differ among different E. coli strains, we screened 16 E. coli strains for 6MSA production by test tube scale cultivation (SI Appendix, Fig. S9D and Table S2). Six strains showing higher than 1 mg/L of 6MSA production were selected, and were introduced with each of the previously screened 14 sRNAs for increased malonyl-CoA accumulation, and consequently 6MSA. Test tube cultures were performed for 84 engineered E. coli strains (Fig. 4B and Dataset S2). Among them, E. coli BL21(DE3) harboring pTac-Pg6MSAS-sfp and pWAS–anti-pabA (for the expression of anti-pabA sRNA) showed the highest 6MSA production (6.1 mg/L). Shake flask cultivation of this strain produced 8.1 mg/L of 6MSA (Fig. 4C and SI Appendix, Fig. S9E). Nearly twofold increase in the 6MSA titer (from 4.7 mg/L to 8.1 mg/L) (Fig. 4C) was achieved without additional metabolic engineering approaches beyond RppA-based library screening. To show that further metabolic engineering can increase the production titer, the acetyl-CoA carboxylase gene was overexpressed in the 6MSA-producing strain. The 6MSA concentration could be further increased to 440.3 ± 30.2 mg/L (mean ± SD) (Fig. 4 C and D) by fed-batch culture of the BAP1 harboring pTac-Pg6MSAS, pWAS–anti-pabA and pBBR1-accBCD1 strain developed by applying this simple metabolic engineering strategy (Discussion and SI Appendix, Text S4).
Fig. 4.
Application of the screened knockdown gene targets for the increased production of polyketides. (A) The biosynthetic pathways of 6MSA and aloesone. Blue X (along with gene written in bold letters surrounded by yellow box) indicates that the corresponding gene was knocked down. Red bold arrow indicates that the corresponding gene was overexpressed for 6MSA production; blue bold arrow indicates that the corresponding gene was overexpressed for aloesone production. Gray dotted lines indicate omitted pathways. The upper box describes the heterologous 6MSA biosynthetic pathway. The lower box describes the heterologous aloesone biosynthetic pathway. Each gene or gene cluster encodes the following: accBCD1, acetyl-CoA carboxylase from C. glutamicum; pabA, para-aminobenzoate synthetase; zwf, glucose 6-phosphate 1-dehydrogenase. (B) Results of combinatorial 6MSA production in test tubes. The 6 E. coli strains selected among the initial 16 strains for their higher 6MSA production (SI Appendix, Fig. S9D) were introduced with each of the 14 synthetic sRNAs for enhanced malonyl-CoA accumulation and thus increased 6MSA production. The gene names written horizontally represent the 14 knockdown gene targets. CT, control (without sRNA). The displayed average values were calculated from two biological replicates. (C) Enhanced production of 6MSA by flask cultivation. The +/− signs for BL21/BAP1 indicate host strain selection. The +/− signs for pabA KD/accBCD1 mean whether pabA knockdown or accBCD1 overexpression was implemented in the production host. Error bars, mean ± SD (n = 3). **P < 0.01, ***P < 0.001, determined by two-tailed Student’s t test. (D) Fed-batch fermentation of the best 6MSA producer (E. coli BAP1 harboring pTac-Pg6MSAS, pWAS−anti-pabA and pBBR1-accBCD1). The red arrow denotes isopropyl β-d-1-thiogalactopyranoside (IPTG) induction time point. The red line denotes 6MSA production and the blue line denotes cell growth represented as OD600. The displayed average values were calculated from two biological replicates. Error bars, mean ± SD (n = 2). (E) Enhanced production of aloesone by flask cultivation. The +/− signs for pabA KD/zwf mean whether pabA knockdown or zwf overexpression was implemented in the production host. Error bars, mean ± SD (n = 3). *P < 0.05, ***P < 0.001, determined by two-tailed Student’s t test. (F) Results of combinatorial aloesone production in test tubes. Two E. coli DE3 strains were introduced with 14 synthetic sRNAs for enhanced malonyl-CoA accumulation and thus increased aloesone production. The gene names written horizontally represent the knockdown gene targets. CT, control (without sRNA). The displayed average values were calculated from two biological replicates. Abbreviations: 1,3BPG, 1,3-bisphosphoglycerate; 2PG, 2-phosphoglycerate; 3PG, 3-phosphoglycerate; 6MSAS, 6MSA synthase; AcCoA, acetyl-CoA; ACP, acyl carrier protein; ALS, aloesone synthase; AT, acyltransferase; CHOR, chorismate; DAHP, 3-deoxy-d-arabinoheptulosonate 7-phosphate; DH, dehydratase; DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; E4P, d-erythrose 4-phosphate; F1,6BP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; G3P, glyceraldehyde 3-phosphate; G6P, glucose 6-phosphate; Glc, glucose; Gly, glycerol; Gly-3P, glycerol 3-phosphate; KR, ketoreductase; KS, ketosynthase; MalCoA, malonyl-CoA; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PYR, pyruvate.
Aloesin, a polyketide produced by a type III PKS from Rheum palmatum or A. arborescens, is widely used in cosmetic industry as skin-whitening agent by inhibiting tyrosinase activity and melanogenesis (40). Aloesin is also reported to possess antiinflammatory and free-radical scavenging activity (41). However, the amount of aloesin extracted from Aloe species is notably low. When we extracted metabolites from commercially available dried aloe powder (Aloe vera), 0.000264% (wt/wt) of aloesin was obtained. Because the metabolic pathway leading to aloesin from its direct aglycone precursor aloesone is currently unknown, we decided to focus on producing aloesone. Aloesone is produced by condensation of one molecule of acetyl-CoA and six molecules of malonyl-CoA (Fig. 4A). Two enzyme candidates, R. palmatum ALS (aloesone synthase; RpALS) (42) and A. arborescens PKS3 (aloesone synthase; AaPKS3) (32), are reported to be responsible for these reactions. Initially, RpALS and AaPKS3 were each cloned into plasmid pTac15K, but neither aloesone production nor enzyme expression was observed from these constructs. Thus, RpALS and AaPKS3 were each cloned under the strong T7 promoter in pCDFDuet-1 plasmid, resulting in plasmids pCDF-RpALS and pCDF-AaPKS3. SDS/PAGE analysis showed that the heterologous enzymes were successfully expressed (SI Appendix, Fig. S10A). Aloesone was successfully produced from both enzyme candidates, and the authenticity of aloesone was confirmed by LC-MS and MS/MS analysis (SI Appendix, Fig. S10B). From 20 g/L of glucose, 20.5 mg/L and 4.7 mg/L of aloesone were produced by the strains expressing RpALS and AaPKS3, respectively (Fig. 4E and SI Appendix, Fig. S10C). When 20 g/L of glycerol was used, slightly less (19.7 mg/L and 3.1 mg/L, respectively) aloesone titers were obtained; this was different from the 6MSA case showing better production using glycerol (SI Appendix, Text S8). To test the effects of the 14 synthetic sRNAs on aloesone production in different E. coli DE3 strains, each sRNA was introduced into both E. coli BL21(DE3) harboring pCDF-RpALS and E. coli W3110(DE3) harboring pCDF-RpALS. Test tube scale aloesone production from the resultant 28 strains are shown in Fig. 4F (Dataset S2). Among these strains, E. coli BL21(DE3) harboring pCDF-RpALS and pWAS–anti-pabA produced the highest titer (18.5 mg/L) of aloesone. Shake flask cultivation of this strain produced 27.1 mg/L of aloesone, which was 32.2% higher than that obtained with the control strain without sRNA (Fig. 4E and SI Appendix, Fig. S10D). It is notable that the same knockdown gene target (pabA) introduced for the best 6MSA producer yielded the highest aloesone titer as well. The aloesone concentration could be increased to 30.9 ± 1.5 mg/L (mean ± SD) (Fig. 4E) by further simple metabolic engineering [E. coli BL21(DE3) harboring pCDF-RpALS, pWAS–anti-pabA and pBBR1-zwf] (SI Appendix, Text S4). This represents a demonstration of microbial production of aloesone.
Enhanced Production of Phenylpropanoids by Knocking Down the Screened Gene Targets.
Phenylpropanoids are a large group of natural compounds that originated from aromatic amino acids (9, 11). They are categorized into several subgroups, including stilbenoids and flavonoids (11). Resveratrol is one of the most popularly studied stilbenoids, which displays diverse biological activities, including antioxidant, antiaging, and anticancer properties (43). Resveratrol is produced by condensation of one molecule of p-coumaroyl-CoA and three molecules of malonyl-CoA (SI Appendix, Fig. S11). Many studies on the heterologous production of this compound have been reported, both in E. coli (44) and S. cerevisiae (45); 304.5 mg/L of resveratrol was produced in E. coli. To achieve resveratrol production in E. coli, we first constructed the downstream resveratrol biosynthetic pathway starting from p-coumaric acid, composed of mutant Arabidopsis thaliana 4-coumarate:CoA ligase 1 (At4CL1m) and Vitis vinifera stilbene synthase (VvSTS) (SI Appendix, Text S5 and Figs. S11 and S12 A–C). Because the conversion of p-coumaric acid (2 mM; 328.1 mg/L) to resveratrol (21.2 mg/L) in E. coli BL21(DE3) harboring pTacCDF-VvSTS-At4CL1m (which expresses the VvSTS and At4CL1m genes separately from the tac promoter) was low, it was speculated that intracellular malonyl-CoA pool could be a potential bottleneck for resveratrol production. To construct an E. coli strain capable of producing resveratrol from simple carbon sources, a p-coumaric acid producer BTY5 harboring pTY13-HisTAL was constructed (SI Appendix, Text S6) based on the previously reported l-tyrosine–overproducing E. coli strain BTY5.13 (46). This strain produced 0.35 g/L of p-coumaric acid from 20 g/L glycerol (SI Appendix, Fig. S12D). Plasmid pTacCDF-VvSTS-At4CL1 harboring the downstream resveratrol biosynthetic pathway was transformed into the p-coumaric acid producer, resulting in production of 12.4 mg/L of resveratrol from 20 g/L of glycerol (SI Appendix, Fig. S12C, strain 5, and SI Appendix, Fig. S13). When glucose was used as a carbon source, only 0.2 mg/L of resveratrol was produced by the same strain. After the successful construction of a resveratrol producer, the previously screened 14 synthetic sRNAs were transformed into this strain. Among the resultant strains, the best strain produced 51.8 ± 3.7 mg/L (mean ± SD) of resveratrol when pabA was knocked down, which was 4.2-fold increase in titer compared with that of the control strain without sRNA (Fig. 5A). It is notable that pabA knockdown also yielded the best 6MSA producer as well as the best aloesone producer. Next, we chose six knockdown gene targets showing more than 2.5-fold increase in resveratrol titer to test the effects of combinatorial double knockdown on resveratrol production. However, testing the 15 double knockdown combinations did not further enhance resveratrol production, as the highest titer obtained was 50.0 mg/L when yfiD and purB were simultaneously knocked down (SI Appendix, Fig. S12E).
Fig. 5.
Application of the screened knockdown gene targets for the increased production of phenylpropanoids. (A) Effects of introducing each of the 14 synthetic sRNAs to the initial resveratrol producer (BTY5 harboring pTY13-HisTAL and pTacCDF-VvSTS-At4CL1m) on resveratrol production. The gene names written along the x axis represent the knockdown gene targets. CT, control (without sRNA), is noted as the gray bar. Six knockdown gene targets displayed as deeper red bars are selected for further experiments according to the set threshold (2.5-fold increase in titer compared with the control strain without sRNA). **P < 0.01, ***P < 0.001, determined by two-tailed Student’s t test. (B) The naringenin biosynthetic pathway from glycerol. A red X indicates that the corresponding gene was knocked out. A blue X (along with gene written in bold letters surrounded by yellow box) indicates that the corresponding gene was knocked down. Gray dotted lines indicate omitted pathways. Thick black arrows along with genes written in bold letters denote overexpressed metabolic fluxes. Red dotted lines (along with − signs in circles) indicate transcriptional repression. Black dotted lines (along with + signs in circles) indicate transcriptional activation. The upper box describes the introduced plasmids. The genetic construct shown at the lower left is on the chromosome. bla, β-lactamase gene; CDF, replication origin; ColE1, replication origin; kanR, kanamycin-resistance gene; p15A, replication origin; PBAD, arabinose-inducible promoter; PR, PR promoter; Ptac, tac promoter; Ptrc, trc promoter; rrnB, rrnBT1T2 terminator; spcR, spectinomycin-resistance gene; T1/TE, terminator. Each gene encodes the following: aroA, 3-phosphoshikimate 1-carboxyvinyltransferase; aroC, chorismate synthase; aroG, 3-deoxy-7-phosphoheptulonate synthase; aroL, shikimate kinase; aspC, aspartate aminotransferase; fabA, β-hydroxydecanoyl thioester dehydrase; fadR, negative regulator for fad regulon and positive regulator of fabA; fbr, feedback resistant; tyrA, chorismate mutase/prephenate dehydrogenase; tyrB, aromatic-amino acid transaminase; tyrP, tyrosine-specific transport protein; tyrR, transcriptional regulator of aroF, aroG and tyrA; xapA, xanthosine phosphorylase; xapB, xanthosine transporter; xapR, transcriptional activator of xapAB. (C) Effects of introducing each of the 14 synthetic sRNAs to the initial naringenin producer (BTY5 harboring pTY13-HisTAL and pTrcCDF-At4CL1m-AtCHI-PhCHS) on naringenin production. The gene names written along the x axis represent the knockdown gene targets. CT, control (without sRNA). Three knockdown gene targets displayed as deeper red bars are selected for further experiments according to the set threshold (15% increase in titer compared with the control strain without sRNA). *P < 0.05, determined by two-tailed Student’s t test. (D) Combinatorial double knockdown of the three gene targets selected from C for naringenin production. Control (without sRNA) is noted as the gray bar. The genes corresponding to the filled squares under each bar denote knocked down genes. All error bars, mean ± SD (n = 3). **P < 0.01, determined by two-tailed Student’s t test. Abbreviations: 1,3BPG, 1,3-bisphosphoglycerate; 2PG, 2-phosphoglycerate; 3PG, 3-phosphoglycerate; AcCoA, acetyl-CoA; CHOR, chorismate; COU, p-coumaric acid; CouCoA, p-coumaroyl-CoA; DAHP, 3-deoxy-d-arabinoheptulosonate 7-phosphate; DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; E4P, d-erythrose 4-phosphate; FOR, formate; G3P, glyceraldehyde 3-phosphate; Gly, glycerol; Gly-3P, glycerol 3-phosphate; HPP, 4-hydroxyphenylpyruvate; MalCoA, malonyl-CoA; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PPHN, prephenate; PYR, pyruvate; SHIK, shikimate; TYR, l-tyrosine.
Naringenin is a representative flavonoid that is also an important precursor to many complex flavonoids possessing various medicinal activities. Naringenin itself is also reported to possess many pharmacological activities, such as anticancer, antioxidant, and antibacterial properties (47). Many studies on the heterologous production of this compound have been reported, both in E. coli (48) and in S. cerevisiae (49); 100.64 mg/L of naringenin was produced in E. coli. Naringenin is produced by condensation of one molecule of p-coumaroyl-CoA and three molecules of malonyl-CoA (Fig. 5B). Because we developed the p-coumaric acid producer as described above, the downstream naringenin biosynthetic pathway from p-coumaric acid, comprising At4CL1m, chalcone synthase from Petunia x hybrida (PhCHS) and chalcone isomerase from A. thaliana (AtCHI) was constructed (SI Appendix, Text S7 and Fig. S14). Plasmid pTrcCDF-At4CL1m-AtCHI-PhCHS (which expresses the At4CL1m-AtCHI-PhCHS artificial operon under the trc promoter) was introduced into the p-coumaric acid producer (BTY5 harboring pTY13-HisTAL). This engineered strain produced 37.2 and 64.5 mg/L of naringenin from glucose and glycerol, respectively. It is notable that using glycerol as a carbon source resulted in improved titers for 6MSA, resveratrol, and naringenin (SI Appendix, Text S8 and Fig. S15).
Having constructed the naringenin producer (BTY5 harboring pTY13-HisTAL and pTrcCDF-At4CL1m-AtCHI-PhCHS), the 14 synthetic sRNAs screened earlier were introduced to test their effects on naringenin production. Among the tested strains, the best naringenin producer was the strain with fadR knockdown. This strain could produce 92.3 mg/L of naringenin, which corresponds to 43% increase in titer compared with that of the control strain without sRNA (Fig. 5C). The fadR gene encodes a transcriptional regulator that activates fabA expression and down-regulates fad regulon. Therefore, fadR knockdown results in reduced fabA expression and increased fad regulon expression. Because fabA encodes β-hydroxyacyl-ACP dehydratase/isomerase, an enzyme responsible for fatty acid biosynthesis, reduced fabA expression results in reduced fatty acid production and thus increased malonyl-CoA accumulation. In addition, increased fad regulon expression results in activation of the β-oxidation pathway, thus contributing to the increased intracellular acetyl-CoA concentration (Fig. 5B). This allows increased metabolic flux toward malonyl-CoA. Including fadR, 3 of 14 knockdown gene targets (fadR, fabH, and fabF) screened in the previous section are involved in the fatty acid metabolism (SI Appendix, Table S3). We then selected three knockdown gene targets showing more than 15% increase in naringenin titer to test the effects of combinatorial knockdown on naringenin production (Fig. 5C). The strain with both fadR and xapR knocked down showed the highest naringenin titer, 103.8 ± 2.7 mg/L (mean ± SD) (Fig. 5D). This corresponds to 61% increase in naringenin titer compared with that of the control strain without sRNA. The xapR gene encodes a transcriptional activator of xapAB, which are responsible for nucleoside metabolism and transport (50), but the exact mechanism of how its knockdown contributed to the increased malonyl-CoA pool is currently not understood. It is notable that 3 of 14 screened knockdown gene targets (xapR, cytR, and purB) are involved in nucleoside metabolism (SI Appendix, Table S3).
The RppA Biosensor Is also Functional in P. putida and C. glutamicum.
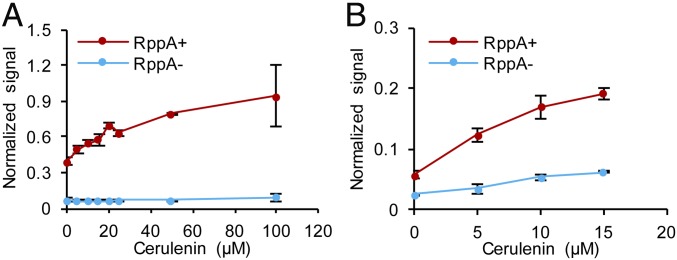
The RppA biosensor was also tested in two other industrially important bacteria, P. putida and C. glutamicum; they also represent Gram-negative and Gram-positive bacteria, respectively. Different vector constructs for RppA expression in P. putida were compared, among which pBBR1-rppA showed the best performance in flaviolin production (44.7 mg/L) (SI Appendix, Fig. S16A). Then, cerulenin titration for the constructed sensor strain confirmed that the strain could respond to a broad range of intracellular malonyl-CoA levels (Fig. 6A and SI Appendix, Fig. S16B). Flaviolin was also successfully produced in C. glutamicum by introducing the plasmid pCES-His-rppA (3.9 mg/L) (SI Appendix, Fig. S16C). Plasmid pCES-His-rppA harboring the gene encoding N-terminal poly-histidine(His)-tagged RppA was selected because flaviolin was not produced without the N-terminal poly-His-tag (SI Appendix, Materials and Methods). Cerulenin titration experiments were performed for the constructed sensor strain, confirming that the RppA biosensor is functional in C. glutamicum as well (Fig. 6B and SI Appendix, Fig. S16D). In conclusion, the RppA biosensor seems to be generally useful in both Gram-negative and Gram-positive bacteria, suggesting its wide applicability.
Fig. 6.
The RppA biosensor is also functional in P. putida and C. glutamicum. Normalized signals generated from the RppA biosensors in (A) P. putida and in (B) C. glutamicum showed dose-dependent responses to intracellular malonyl-CoA abundance. Intracellular malonyl-CoA abundance was titrated using different concentrations of cerulenin added to the medium. Signals normalized with cell growth are plotted either with (RppA+) or without (RppA−) RppA expression. Error bars, mean ± SD (n = 3).
Discussion
In this study, we developed a malonyl-CoA biosensor using the repurposed type III PKS RppA. RppA can convert five molecules of malonyl-CoA into red-colored flaviolin, which was used as an indicator of intracellular malonyl-CoA abundance. Also, based on cerulenin titration studies, the RppA biosensor was found to have a broad dynamic response range with respect to the intracellular malonyl-CoA level. In addition to RppA, we show that other type III PKSs can also be used as malonyl-CoA biosensors. By using the E. coli genome-scale synthetic sRNA library constructed, it was possible to screen 14 knockdown gene targets that generally increased intracellular malonyl-CoA abundance. These knockdown gene targets were applied for the enhanced production of representative proof-of-concept natural products derived from malonyl-CoA. Four different engineered E. coli strains could be rapidly developed for the improved production of two polyketide products, 6MSA and aloesone, and two phenylpropanoid products, resveratrol and naringenin, by applying a RppA biosensor system coupled with synthetic sRNA library screening. This report on the production of aloesone by an engineered E. coli strain is unique. It should be emphasized that these strains were developed easily in a short period of time (∼3 d) by taking a simple approach of colorimetric screening. Furthermore, the works described in this paper do not involve any systems metabolic engineering (1–3) approaches beyond pathway construction and RppA-based library screening. Thus, the titers, yields, and productivities reported in this paper can be further enhanced by metabolic engineering. For example, after the successful high-throughput screening of the highest 6MSA producer, a simple metabolic engineering strategy coupled with fed-batch fermentation allowed a significant increase in the 6MSA titer (from 8.0 mg/L to 440.3 mg/L) (SI Appendix, Text S4). Furthermore, it was demonstrated that the RppA biosensor is also functional in P. putida and C. glutamicum. Taken together, these results demonstrate general applicability of the RppA biosensor in rapidly developing strains efficiently producing malonyl-CoA–derived products. While we were revising our manuscript, we found a recent paper describing the use of flaviolin as an indicator of intracellular malonyl-CoA abundance (51). The authors of that paper used this system to screen gene knockdown targets for increased production of 3-hydroxypropionic acid, which is produced from malonyl-CoA by one-step conversion, which further proves the usefulness of RppA-based biosensor for developing strains overproducing malonyl-CoA–derived products.
Although there have been many studies on rationally increasing intracellular malonyl-CoA concentration (52, 53), prediction of effective gene targets to be manipulated is often difficult due to the complexity of biological systems. In this regard, high-throughput screening of malonyl-CoA overproducers using the E. coli genome-scale synthetic sRNA library enabled identification of beneficial targets that were difficult to be rationally predicted. For example, down-regulation of pabA had a strong and widely observed impact on increased production of malonyl-CoA–derived natural products 6MSA, aloesone, and resveratrol. The pabA gene encodes aminodeoxychorismate synthase, which is responsible for the production of p-aminobenzoate, which is used together with purine bases to form folate. Because folate biosynthesis requires two moles of ATP (54), knocking down pabA might increase availability of ATP required for conversion of acetyl-CoA to malonyl-CoA. However, further study is needed to understand the exact reason. This demonstrates the usefulness of sRNA-based screening that identifies gene knockdown targets like pabA, which is not possible to be rationally selected. In addition, it is notable that three transcriptional regulators (xapR, cytR, and fadR) (SI Appendix, Table S3) were selected among the 14 final knockdown gene targets, which are difficult to predict due to insufficient studies on their complete regulatory networks. Furthermore, identification and knockdown of essential genes were possible by employing the synthetic sRNA system. Because most metabolic engineering studies still involve gene knockouts rather than gene knockdowns, identification of the pabA gene would have not been possible if synthetic sRNA knockdown system was not employed.
The most popular malonyl-CoA sensor reported to date involves the transcription factor FapR, which binds to the operator site fapO located between the promoter and the gene encoding a fluorescence reporter protein, thereby blocking the reporter gene expression. Upon coupling with malonyl-CoA (16), FapR is detached from the operator, allowing the intracellular malonyl-CoA level to be monitored through the output fluorescence signal. On the other hand, the RppA system directly converts malonyl-CoA to flaviolin, allowing direct signal generation. This simple and robust sensor system enables unbiased analysis of the intracellular malonyl-CoA level with a wide dynamic response range. Moreover, because the RppA malonyl-CoA biosensor does not involve fluorescence, it can be easily applied to microorganisms displaying autofluorescence, such as Pseudomonas species (18). Due to its simple mechanism and easiness of constructing the system, the RppA biosensor is expected to be widely applicable to other microorganisms beyond those demonstrated in this study.
The red-colored flaviolin is easily detectable by the naked eyes, which facilitates rapid colorimetric prescreening even without colorimetric equipment if one does not have it. As was demonstrated by previous studies (55, 56), colorimetric screening employing colored metabolites (e.g., lycopene and β-carotene) as indicators of the abundance of precursor metabolites was shown to be effective for developing high-level production strains. Although the colorimetric screening is slower than fluorescence screening (57), it allows easy and rapid prescreening through the naked eyes (SI Appendix, Fig. S5B), which can further increase the throughput. As demonstrated in this study, a robotic high-throughput screening system obviously allows efficient screening of a large library. In high-throughput screening of libraries, false-positives and false-negatives can affect the efficiency and accuracy of screening. In the cases of colorimetric screening, however, false-positives are of particular concern because false-negatives are not usually monitored during the screening. Such false-positive issues have also been observed for colorimetric screening of carotenoids (55, 56). We observed that a notable number (161 of 231 strains) of initial high-throughput screened strains did not give high enough signals when cultured again in test tubes. Although they are not exactly false-positives because we applied the least stringent threshold condition during high-throughput screening, such false-positives can be reduced by setting the threshold of high-throughput screening more stringently. Nonetheless, we do not recommend it as selecting true-positives after high-throughput screening is simple and efficient.
In conclusion, the RppA biosensor allows development of microbial strains capable of enhanced production of malonyl-CoA–derived products. Either target-specific metabolic engineering strategies or large-scale library approaches can be applied during the screening process. It is expected that the RppA biosensor will allow rapid development of strains capable of efficiently producing malonyl-CoA–derived products, which will significantly contribute to the pharmaceutical, chemical, cosmetics, and food industries.
Materials and Methods
All of the materials and methods conducted in this study are detailed in SI Appendix, Materials and Methods: materials, plasmid construction, E. coli genome-scale synthetic sRNA library construction, strains, media and culture conditions, flaviolin production from P. putida and C. glutamicum, RppA malonyl-CoA biosensor characterization, high-throughput screening of malonyl-CoA overproducers, test tube scale cultivation, malonyl-CoA quantification, analytical procedures, genome engineering, in silico analysis, SDS/PAGE analysis, and statistical analysis.
Supplementary Material
Acknowledgments
We thank Seon Young Park, Jae Sung Cho, In Jin Cho, Yoo Jin Choi, Hye Mi Kim, Seung Woo Chun, Kyeong Rok Choi, and Prof. Hyun Uk Kim for valuable advice; Minho Noh, Jung Ae Lim, Jae Eun Lee, So Hee Park, and Yu Hyun Lee for their help in constructing the Escherichia coli genome-scale synthetic sRNA library; and Kyeong Rok Choi for the construction of the plasmid pBBR1TaC. This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and by the Intelligent Synthetic Biology Center through the Global Frontier Project (Grant 2011-0031963) of the Ministry of Science and ICT (MSIT) through the National Research Foundation of Korea. Synthetic sRNA library construction was supported by Commercialization Promotion Agency for R&D Outcomes (Grant COMPA-2015K000365) of MSIT. D.Y. and S.Y.L. were also supported by Novo Nordisk Foundation Grant NNF16OC0021746.
Footnotes
Conflict of interest statement: S.Y.L., D.Y., and S.M.Y. declare that the sRNA technology described here is patent filed including, but not limited to, KR 10-1575587, US 9388417, EP 13735942.8, CN 201380012767.X, KR 10-1690780, KR 10-1750855, US 15317939, CN 201480081132.X for potential commercialization. Additionally, S.Y.L. and D.Y. have conflict of interest as the RppA biosensor technology is of commercial interest and is patent filed including, but not limited to KR 10-2018-0066323.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MH473344, MH473345, MH488902–MH488953, and MH651713–MH651726).
See QnAs on page 9816.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808567115/-/DCSupplemental.
References
- 1.Lee SY, Kim HU. Systems strategies for developing industrial microbial strains. Nat Biotechnol. 2015;33:1061–1072. doi: 10.1038/nbt.3365. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen J, Keasling JD. Engineering cellular metabolism. Cell. 2016;164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Choi KR, Shin JH, Cho JS, Yang D, Lee SY. 2016. Systems metabolic engineering of Escherichia coli. Ecosal Plus, 7, 10.1128/ecosalplus.ESP-0010-2015.
- 4.Rogers JK, Church GM. Genetically encoded sensors enable real-time observation of metabolite production. Proc Natl Acad Sci USA. 2016;113:2388–2393. doi: 10.1073/pnas.1600375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, Si T, Wang M, Zhao H. Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening. ACS Synth Biol. 2015;4:1308–1315. doi: 10.1021/acssynbio.5b00069. [DOI] [PubMed] [Google Scholar]
- 6.Lin JL, Wagner JM, Alper HS. Enabling tools for high-throughput detection of metabolites: Metabolic engineering and directed evolution applications. Biotechnol Adv. 2017;35:950–970. doi: 10.1016/j.biotechadv.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci USA. 2014;111:11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLoache WC, et al. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat Chem Biol. 2015;11:465–471. doi: 10.1038/nchembio.1816. [DOI] [PubMed] [Google Scholar]
- 9.Park SY, Yang D, Ha SH, Lee SY. Metabolic engineering of microorganisms for the production of natural compounds. Adv Biosyst. 2018;2:1700190. [Google Scholar]
- 10.Weber T, et al. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015;33:15–26. doi: 10.1016/j.tibtech.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Pandey RP, Parajuli P, Koffas MAG, Sohng JK. Microbial production of natural and non-natural flavonoids: Pathway engineering, directed evolution and systems/synthetic biology. Biotechnol Adv. 2016;34:634–662. doi: 10.1016/j.biotechadv.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Pfleger BF, Kim SW. Reassessing Escherichia coli as a cell factory for biofuel production. Curr Opin Biotechnol. 2017;45:92–103. doi: 10.1016/j.copbio.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Gao L, et al. Simultaneous quantification of malonyl-CoA and several other short-chain acyl-CoAs in animal tissues by ion-pairing reversed-phase HPLC/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;853:303–313. doi: 10.1016/j.jchromb.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 14.Zha W, Rubin-Pitel SB, Shao Z, Zhao H. Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering. Metab Eng. 2009;11:192–198. doi: 10.1016/j.ymben.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 15.De Spiegeleer BM, Sintobin K, Desmet J. High performance liquid chromatography stability study of malonyl-coenzyme A, using statistical experimental designs. Biomed Chromatogr. 1989;3:213–216. doi: 10.1002/bmc.1130030508. [DOI] [PubMed] [Google Scholar]
- 16.Johnson AO, et al. Design and application of genetically-encoded malonyl-CoA biosensors for metabolic engineering of microbial cell factories. Metab Eng. 2017;44:253–264. doi: 10.1016/j.ymben.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Chen W, Jin R, Jin JM, Tang SY. Biosensor-aided high-throughput screening of hyper-producing cells for malonyl-CoA-derived products. Microb Cell Fact. 2017;16:187. doi: 10.1186/s12934-017-0794-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeschcke A, Thies S. Pseudomonas putida—A versatile host for the production of natural products. Appl Microbiol Biotechnol. 2015;99:6197–6214. doi: 10.1007/s00253-015-6745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funa N, et al. A new pathway for polyketide synthesis in microorganisms. Nature. 1999;400:897–899. doi: 10.1038/23748. [DOI] [PubMed] [Google Scholar]
- 20.Funa N, Ohnishi Y, Ebizuka Y, Horinouchi S. Properties and substrate specificity of RppA, a chalcone synthase-related polyketide synthase in Streptomyces griseus. J Biol Chem. 2002;277:4628–4635. doi: 10.1074/jbc.M110357200. [DOI] [PubMed] [Google Scholar]
- 21.Austin MB, et al. Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J Biol Chem. 2004;279:45162–45174. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- 22.Funa N, Funabashi M, Yoshimura E, Horinouchi S. A novel quinone-forming monooxygenase family involved in modification of aromatic polyketides. J Biol Chem. 2005;280:14514–14523. doi: 10.1074/jbc.M500190200. [DOI] [PubMed] [Google Scholar]
- 23.Cortés J, et al. Identification and cloning of a type III polyketide synthase required for diffusible pigment biosynthesis in Saccharopolyspora erythraea. Mol Microbiol. 2002;44:1213–1224. doi: 10.1046/j.1365-2958.2002.02975.x. [DOI] [PubMed] [Google Scholar]
- 24.Ghimire GP, Oh TJ, Liou K, Sohng JK. Identification of a cryptic type III polyketide synthase (1,3,6,8-tetrahydroxynaphthalene synthase) from Streptomyces peucetius ATCC 27952. Mol Cells. 2008;26:362–367. [PubMed] [Google Scholar]
- 25.Winter JM, et al. Molecular basis for chloronium-mediated meroterpene cyclization: Cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. J Biol Chem. 2007;282:16362–16368. doi: 10.1074/jbc.M611046200. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Grüschow S, Dordick JS, Sherman DH. Molecular analysis of the role of tyrosine 224 in the active site of Streptomyces coelicolor RppA, a bacterial type III polyketide synthase. J Biol Chem. 2007;282:12765–12772. doi: 10.1074/jbc.M700393200. [DOI] [PubMed] [Google Scholar]
- 27.Seo SW, et al. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab Eng. 2013;15:67–74. doi: 10.1016/j.ymben.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Romero-Martinez R, Wheeler M, Guerrero-Plata A, Rico G, Torres-Guerrero H. Biosynthesis and functions of melanin in Sporothrix schenckii. Infect Immun. 2000;68:3696–3703. doi: 10.1128/iai.68.6.3696-3703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Omura S. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol Rev. 1976;40:681–697. doi: 10.1128/br.40.3.681-697.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe I, et al. A plant type III polyketide synthase that produces pentaketide chromone. J Am Chem Soc. 2005;127:1362–1363. doi: 10.1021/ja0431206. [DOI] [PubMed] [Google Scholar]
- 31.Abe I, Oguro S, Utsumi Y, Sano Y, Noguchi H. Engineered biosynthesis of plant polyketides: Chain length control in an octaketide-producing plant type III polyketide synthase. J Am Chem Soc. 2005;127:12709–12716. doi: 10.1021/ja053945v. [DOI] [PubMed] [Google Scholar]
- 32.Mizuuchi Y, et al. Novel type III polyketide synthases from Aloe arborescens. FEBS J. 2009;276:2391–2401. doi: 10.1111/j.1742-4658.2009.06971.x. [DOI] [PubMed] [Google Scholar]
- 33.Na D, et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol. 2013;31:170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 34.Noh M, Yoo SM, Kim WJ, Lee SY. Gene expression knockdown by modulating synthetic small RNA expression in Escherichia coli. Cell Syst. 2017;5:418–426.e4. doi: 10.1016/j.cels.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Karp PD, et al. The EcoCyc database. Ecosal Plus. 2014;6 doi: 10.1128/ecosalplus.ESP-0009-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park JM, et al. Flux variability scanning based on enforced objective flux for identifying gene amplification targets. BMC Syst Biol. 2012;6:106. doi: 10.1186/1752-0509-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dimroth P, Ringelmann E, Lynen F. 6-Methylsalicylic acid synthetase from Penicillium patulum. Some catalytic properties of the enzyme and its relation to fatty acid synthetase. Eur J Biochem. 1976;68:591–596. doi: 10.1111/j.1432-1033.1976.tb10847.x. [DOI] [PubMed] [Google Scholar]
- 38.Kealey JT, Liu L, Santi DV, Betlach MC, Barr PJ. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc Natl Acad Sci USA. 1998;95:505–509. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wattanachaisaereekul S, Lantz AE, Nielsen ML, Nielsen J. Production of the polyketide 6-MSA in yeast engineered for increased malonyl-CoA supply. Metab Eng. 2008;10:246–254. doi: 10.1016/j.ymben.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Jones K, Hughes J, Hong M, Jia Q, Orndorff S. Modulation of melanogenesis by aloesin: A competitive inhibitor of tyrosinase. Pigment Cell Res. 2002;15:335–340. doi: 10.1034/j.1600-0749.2002.02014.x. [DOI] [PubMed] [Google Scholar]
- 41.Yagi A, et al. Antioxidant, free radical scavenging and anti-inflammatory effects of aloesin derivatives in Aloe vera. Planta Med. 2002;68:957–960. doi: 10.1055/s-2002-35666. [DOI] [PubMed] [Google Scholar]
- 42.Abe I, Utsumi Y, Oguro S, Noguchi H. The first plant type III polyketide synthase that catalyzes formation of aromatic heptaketide. FEBS Lett. 2004;562:171–176. doi: 10.1016/S0014-5793(04)00230-3. [DOI] [PubMed] [Google Scholar]
- 43.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: The in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 44.Wu J, Zhou P, Zhang X, Dong M. Efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli. J Ind Microbiol Biotechnol. 2017;44:1083–1095. doi: 10.1007/s10295-017-1937-9. [DOI] [PubMed] [Google Scholar]
- 45.Li M, Schneider K, Kristensen M, Borodina I, Nielsen J. Engineering yeast for high-level production of stilbenoid antioxidants. Sci Rep. 2016;6:36827. doi: 10.1038/srep36827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim B, Binkley R, Kim HU, Lee SY. Metabolic engineering of Escherichia coli for the enhanced production of l-tyrosine. Biotechnol Bioeng. July 18, 2018 doi: 10.1002/bit.26797. [DOI] [PubMed] [Google Scholar]
- 47.Kanno S, et al. Inhibitory effects of naringenin on tumor growth in human cancer cell lines and sarcoma S-180-implanted mice. Biol Pharm Bull. 2005;28:527–530. doi: 10.1248/bpb.28.527. [DOI] [PubMed] [Google Scholar]
- 48.Wu J, Zhou T, Du G, Zhou J, Chen J. Modular optimization of heterologous pathways for de novo synthesis of (2S)-naringenin in Escherichia coli. PLoS One. 2014;9:e101492. doi: 10.1371/journal.pone.0101492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koopman F, et al. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb Cell Fact. 2012;11:155. doi: 10.1186/1475-2859-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seeger C, Poulsen C, Dandanell G. Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli. J Bacteriol. 1995;177:5506–5516. doi: 10.1128/jb.177.19.5506-5516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarasava K, Liu R, Garst A, Gill RT. Combinatorial pathway engineering using type I-E CRISPR interference. Biotechnol Bioeng. 2018;115:1878–1883. doi: 10.1002/bit.26589. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Lin Y, Li L, Linhardt RJ, Yan Y. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products. Metab Eng. 2015;29:217–226. doi: 10.1016/j.ymben.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 53.Bhan N, Xu P, Khalidi O, Koffas MAG. Redirecting carbon flux into malonyl-CoA to improve resveratrol titers: Proof of concept for genetic interventions predicted by OptForce computational framework. Chem Eng Sci. 2013;103:109–114. [Google Scholar]
- 54.Sybesma W, et al. Increased production of folate by metabolic engineering of Lactococcus lactis. Appl Environ Microbiol. 2003;69:3069–3076. doi: 10.1128/AEM.69.6.3069-3076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leonard E, et al. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci USA. 2010;107:13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Özaydın B, Burd H, Lee TS, Keasling JD. Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production. Metab Eng. 2013;15:174–183. doi: 10.1016/j.ymben.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 57.Dietrich JA, McKee AE, Keasling JD. High-throughput metabolic engineering: Advances in small-molecule screening and selection. Annu Rev Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.